Abstract
Since the vast majority of infections occur at mucosal surfaces, accurate characterization of mucosal immune cells is critically important for understanding transmission and control of infectious diseases. Standard flow cytometric analysis of cells obtained from mucosal tissues can provide valuable information on the phenotype of mucosal leukocytes and their relative abundance, but does not provide absolute cell counts of mucosal cell populations. We developed a bead-based flow cytometry assay to determine the absolute numbers of multiple mononuclear cell types in colorectal biopsies of rhesus macaques. Using 10-color flow cytometry panels and pan-fluorescent beads, cells were enumerated in biopsy specimens by adding a constant ratio of beads per mg of tissue and then calculating cell numbers/mg of tissue based on cell-to-bead ratios determined at the time of sample acquisition. Testing in duplicate specimens showed the assay to be highly reproducible (Spearman R = 0.9476, P < 0.0001). Using this assay, we report enumeration of total CD45+ leukocytes, CD4+ and CD8+ T cells, B cells, NK cells, CD14+ monocytes, and myeloid and plasmacytoid dendritic cells in colorectal biopsies, with cell numbers in normal rhesus macaques varying from medians of 4486 cells/mg (T cells) to 3 cells/mg (plasmacytoid dendritic cells). This assay represents a significant advancement in rapid, accurate quantification of mononuclear cell populations in mucosal tissues and could be applied to provide absolute counts of a variety of different cell populations in diverse tissues.
Keywords: fluorescent beads, lymphocyte enumeration, flow cytometry, mucosal lymphocytes
1. Introduction
Because most infections occur at mucosal surfaces, quantification of mucosal immune cells is critical to understand transmission and control of infectious diseases. Two of the most common approaches used to study mucosal specimens are flow cytometry and histological techniques. Although flow cytometry is a powerful tool to phenotypically characterize isolated mucosal immune cells, it is typically employed to determine the relative abundance of specific cell types but not absolute numbers of cells. While relative cellular quantification is adequate in many circumstances, such as addressing the frequency of expression of a phenotypic activation marker, these data can be misleading, because frequencies of the cell population of interest may be affected by influx or efflux of other cell populations. Histological techniques such as immunohistochemistry or immunofluorescence microscopy are the preferred techniques for determining the anatomic localization of cell populations in situ, but these approaches are relatively time-consuming and generally only approximate cell numbers in two dimensions such as mm2 or by the area occupied by a given immunohistochemical marker (Li et al. 2005; Estes et al., 2008).
Given the limitations of existing approaches for the absolute quantification of cells in mucosal tissues, the need for a simplified, rapid assay which can accurately enumerate mucosal cells using minimal amounts of tissue is evident. We have developed a fluorescent bead-based method for quantifying absolute counts of lymphocytes in colorectal biopsies of rhesus macaques that could have significant applicability to both basic mucosal research and clinical diagnostics.
2. Material and Methods
2.1 Animals and biopsy collection
A total of 12 rhesus macaques (Macaca mulatta) of Indian genetic background were used for sampling in this study. All animals were free of SIV and were housed at the New England Primate Research Center and maintained in accordance with the guidelines of the Committee on Animals of the Harvard Medical School and the Guide for the Care and Use of Laboratory Animals. Macaques were anesthetized with ketamine HCl (10–20 mg/kg, intramuscularly) and then ten-to-twelve rectal punch biopsies were collected using 1.9 mm fenestrated endoscopic biopsy forceps (Olympus America, Center Valley, PA). Biopsy pieces were weighed in bulk (Mettler balances MS104S or PB602, Mettler-Toledo, Columbus, OH) and had a mean mass of 22.9 mg (range 20–50 mg, 30 samples).
2.2 Biopsy processing and cell isolation
Mononuclear cells were isolated from rectal biopsy samples using both enzymatic and mechanical disruption following standard protocols for our laboratory as described previously (Reeves et al., 2010). Briefly, after weighing, biopsies were first incubated for 30 min in 5 mM EDTA, washed, and then incubated in media with 15 U/ml collagenase (type II, Sigma) for 1 h on an orbital shaker. Samples were then mechanically disrupted with an 18-gauge feeding needle. To avoid cell loss, no density-based separation was performed.
2.2 Bead-based quantification assay
Absolute quantification of cell subsets in mucosal samples was performed using a fluorescent bead-based assay modified from a peripheral blood assay optimized in our laboratory (Reeves et al., 2009a; Reeves et al., 2010). After mononuclear cell isolation, all cells were resuspended in 2% FBS in PBS and then each cellular suspension was spiked with 10 μm SPHERO AccuCount Rainbow Fluorescent Particles (Spherotech, Lake Forest, IL) at a concentration of 1,000 beads/mg of absolute tissue weight (measured before processing). Polychromatic flow cytometry (PFC) staining was then performed on each sample using fluorchrome-conjugated monoclonal antibodies (Table 1). All samples also included an Aqua Live/Dead stain (Invitrogen) to discriminate live cells and all acquisitions were made on an LSR II (BD Biosciences) then analyzed using FlowJo 9.0 software (Tree Star Inc., Ashland, OR).
Table 1.
Antibodies used in flow cytometric bead assay.
| Antibody | Clone | Fluorochrome | Manufacturer | Panel |
|---|---|---|---|---|
| anti-CD3 | SP34.2 | APC-Cy7 | BD Biosciences (La Jolla, CA) | I, II |
| anti-CD4 | L200 | PE | BD Biosciences | I |
| anti-CD8α | RPA-T8 | Alexa700 | BD Biosciences | I |
| anti-CD11c | S-HCL-3 | APC | BD Biosciences | II |
| anti-CD14 | TUK4 | Alexa700 | Invitrogen (Carlsbad, CA) | II |
| anti-CD20 | L27 | PerCp-Cy5.5 | BD Biosciences | II |
| anti-CD45 | D058-1283 | FITC | BD Biosciences | I |
| anti-CD45 | D058-1283 | Pacific Blue | BD Biosciences | II |
| anti-CD68 | Y1/82A | PE | BD Biosciences | II |
| anti-CD123 | 7G3 | PE-Cy7 | BD Biosciences | II |
| anti-HLA-DR | Immu-357 | PE-Texas Red | Beckman-Coulter (Fullerton, CA) | I, II |
| anti-NKG2A | Z199 | Pacific Bluea | Beckman-Coulter | I |
In-house custom conjugate
3. Results and Discussion
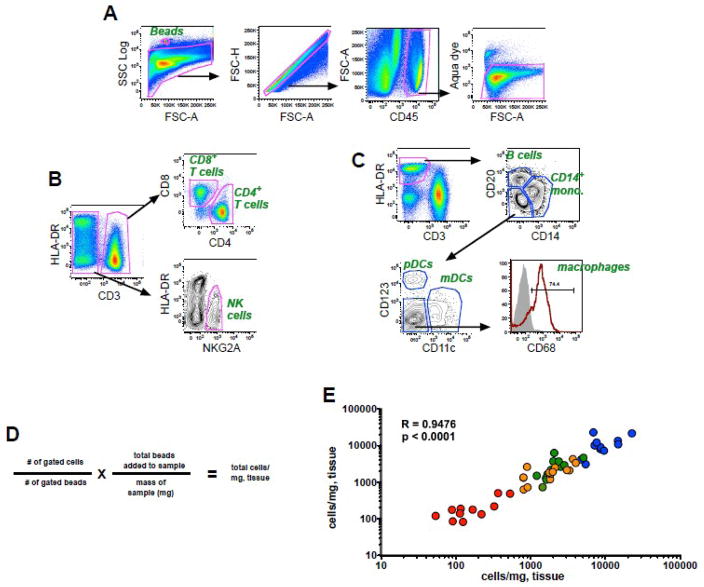
As described in the Methods, 10-color flow cytometry panels optimized for use in rhesus macaques were used in combination with pan-fluorescent beads to calculate absolute cell numbers of mononuclear cells in colorectal biopsies based on cell-to-bead ratios. As shown in Fig. 1A, fluorescent beads were readily identified by their high SSC properties, and live leukocytes were identified in a CD45+ inclusion gate excluding contaminating epithelial cells and debris that are common in mucosal preparations. While all assays contained markers to distinguish live CD45+ cells, one of two different panels was used to identify T cells and NK cells (Fig. 1B) (Panel I), or B cells, monocytes and dendritic cells (Fig. 1C) (Panel II). CD4+ and CD8+ T cells were identified among the CD3+ cell population, and NK cells were identified based on expression of NKG2A and lack of CD3, a strategy previously used for identification of these subpopulations in rhesus macaques (Reeves et al., 2010). B cells and monocytes were identified as CD20+HLA-DR+ and CD14+ HLA-DR+, respectively, and plasmacytoid (pDCs) and myeloid dendritic cells (mDCs) were enumerated among HLA-DR+CD3−CD14−CD20− cells and positively identified by mutually exclusive expression of CD123 and CD11c, as described (Brown et al., 2007; Reeves et al., 2009b). The absolute number of cells/mg of tissue was calculated based on the number of gated cells and the number of bead events using the formula shown in Fig. 1D. In a subgroup of experiments, independent biopsy preparations were processed in parallel and these data were highly correlated, demonstrating a high level of reproducibility (Fig. 1E).
Figure 1. Bead-based cellular quantification in rectal biopsies.
Representative gating strategies showing fluorescence profiles of (a) beads and live cells and subsequent gating for (b) T cells and NK cells or (c) monocytes, B cells, and dendritic cells. (d) Formula for calculation of cell numbers/milligram of tissue. (e) Correlation of absolute cell counts obtained from duplicate colorectal biopsies from 12 animals. Color coding: blue, CD45+ leukocytes; orange, CD8+ T cells; green, CD4+ T cells; red, NK cells. The correlation between the replicate tests was determined using the Spearman rank correlation coefficient.
In colorectal biopsies we found a median number of total CD45+ leukocytes of 8731 cells/mg of colorectal tissue (Fig. 2), and not surprisingly, the total numbers of CD45+ leukocytes strongly correlated with sample mass (R = 0.5452, P < 0.0018, Spearman correlation). While no previous assays have enumerated mucosal lymphocytes in this manner, Sopper et al. (2003), estimated total lamina propria lymphocytes to be between 1500–3700 lymphocytes per milligram of gut tissue, and because our analyses include total mucosal lymphocytes, both lamina proprial and intraepithelial, our estimates appear well in range. In regards to specific subpopulations of mucosal mononuclear cells, we found that approximately half of gut mononuclear cells were CD3+ T cells (median, 4486 cells/mg). Furthermore, CD4+ and CD8+ T cells were found at a roughly 1:1 ratio (Fig. 2), similar to ratios previously reported in rhesus macaque gut mucosa by flow cytometry analyses (Sopper et al., 2003), but in contrast to the 2:1 or 3:1 ratios normally found in peripheral blood of both macaques and humans. Total NKG2A+ NK cells were also found in colorectal biopsies at a median of 152 cells/mg, consistent with our previous observations of a relative abundance of ~5% of total CD45+ colorectal mononuclear cells (Reeves et al., 2010).
Figure 2. Enumeration of leukocyte subsets in the rectal mucosa.
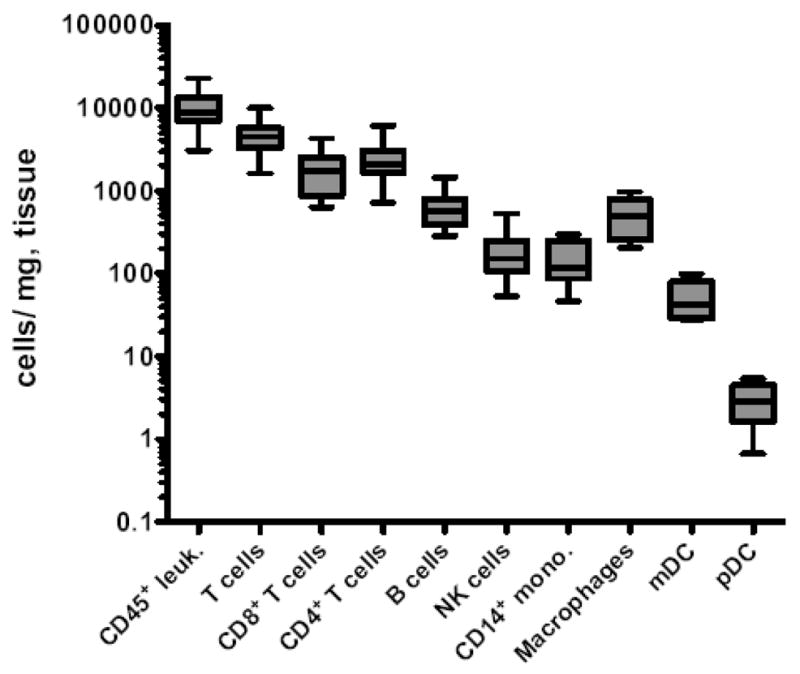
Horizontal lines indicate means and whiskers represent minimum to maximum of 6 to 12 animals per group. Macrophages were defined as CD68+CD3−CD11c−CD14−CD20−CD123−HLA-DR+. Leuk., leukocytes; mono., monocytes.
Using Panel II (Fig. 1C), we enumerated HLA-DR+ B cells, monocytes, and dendritic cells in colorectal biopsies (Fig. 2). B cells were relatively abundant in the rectal mucosa (median, 574 cells/mg). CD14+ monocytes were detectable with a median value of 115 cells/mg, while mDCs and pDCs were less frequent, with median values of 43 cells/mg and 3 cells/mg, respectively. Interestingly, CD3−CD14−CD20−HLA-DR+CD123−CD11c− cells were relatively frequent in colorectal specimens with a median number of 489 cells/mg of tissue. This population that has been observed in macaques previously (Reeves et al, 2009b; Brown et al., 2008), and is most likely dominated by macrophages, based on intracellular staining for CD68 (Fig. 1C). It is, however, possible that this population could contain other DC or monocyte subsets not phenotypically identifiable in our assay.
The development of this relatively simple, bead-based assay for mucosal mononuclear cell enumeration represents a significant step forward in analysis of small mucosal specimens. While beads have been previously used to calculate absolute counts for leukocyte populations in whole blood (Barnett et al., 1999; Montes et al., 2006) and track cell yields from biopsies (Anton et al., 2008), to our knowledge beads have never been used to enumerate mucosal cells based on tissue mass. It is important to note that cell frequencies calculated using this approach may be affected by factors such as cell loss during tissue processing. Nonetheless, the calculation of bead-normalized absolute counts based on tissue weights represents a significant advance over the current standard practice of reporting the relative frequency of cells in mucosal biopsies. This technique is suitable for application to the processing of relatively large numbers of specimens and should prove especially useful in settings such as HIV or SIV infection that result in substantial perturbations of lymphocyte populations in mucosal sites. Finally, this technique could have broad implications not only in basic mucosal immunology studies in animal models, but also in clinical research or human diagnostic studies.
Acknowledgments
The authors thank Elaine Roberts for dedicated animal care and Yi Yu for quality technical assistance. This work was supported through NIH grants AI071306, AI090735, RR00168, and a CHAVI/HVTN Early Career Investigator award, grant number U19 AI 067854-04, to R.K.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anton PA, Ibarrondo FJ, Boscardin WJ, Zhou Y, Schwartz EJ, Ng HL, Hausner MA, Shih R, Elliott J, Hultin PM, Hultin LE, Price C, Fuerst M, Adler A, Wong JT, Yang OO, Jamieson BD. Differential immunogenicity of vaccinia and HIV-1 components of a human recombinant vaccine in mucosal and blood compartments. Vaccine. 2008;26:4617. doi: 10.1016/j.vaccine.2008.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett D, Granger V, Whitby L, Storie I, Reilly JT. Absolute CD4+ T-lymphocyte and CD34+ stem counts by single-platform flow cytometry: the way forward. Br J Haematol. 1999;106:1059. doi: 10.1046/j.1365-2141.1999.01632.x. [DOI] [PubMed] [Google Scholar]
- Brown KN, Trichel A, Barratt-Boyes SM. Parallel loss of myeloid and plasmacytoid dendritic cells from blood and lymphoid tissue in simian AIDS. J Immunol. 2007;178:6958. doi: 10.4049/jimmunol.178.11.6958. [DOI] [PubMed] [Google Scholar]
- Estes J, Baker JV, Brenchley JM, Khoruts A, Barthold JL, Bantle A, Reilly CS, Beilman GJ, George ME, Douek DC, Haase AT, Schacker TW. Collagen deposition limits immune reconstitution in the gut. J Infect Dis. 2008;198:456. doi: 10.1086/590112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Duan L, Estes JD, Zhong-Min M, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, Haase AT. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- Montes M, Jaensson EA, Orozco AF, Lewis DE, Cory DB. A general method for bead-enhanced quantitation by flow cytometry. J Immunol Methods. 2006;317:45. doi: 10.1016/j.jim.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves RK, Gillis J, Wong FE, Johnson RP. Vaccination with SIVmac239 nef activates CD4+ T cells in the absence of CD4 T-cell loss. J Med Primatol. 2009;38(Suppl 1):8. doi: 10.1111/j.1600-0684.2009.00370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves RK, Wei Q, Stallworth J, Fultz PN. Systemic dendritic cell mobilization associated with administration of FLT3 ligand to SIV- and SHIV-infected macaques. AIDS Res Hum Retroviruses. 2009;25:1313. doi: 10.1089/aid.2009.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves RK, Gillis J, Wong FE, Yu Y, Connole M, Johnson RP. CD16− natural killer cells: enrichment in mucosal and secondary lymphoid tissues and altered function during chronic SIV infection. Blood. 2010;115:4439. doi: 10.1182/blood-2010-01-265595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopper S, Nierwetberg Y, Halbach H, Sauer U, Scheller C, Stahl-Hennig C, Matz-Rensing K, Schafer F, Schneider T, ter Meulen V, Muller JG. Impact of simian immunodeficiency virus (SIV) infection on lymphocyte numbers and T-cell turnover in different organs of rhesus monkeys. Blood. 2003;101:1213. doi: 10.1182/blood-2002-06-1644. [DOI] [PubMed] [Google Scholar]