Abstract
A hydrogen bond network has been identified that adjusts protein-substrate contacts in cytochrome P450cam (CYP101A1). Replacing the native substrate camphor with adamantanone or norcamphor causes perturbations in NMR-detected NH correlations assigned to the network, which includes portions of aβ sheet and an adjacent helix that is remote from the active site. A mutation in this helix reduces enzyme efficiency and perturbs the extent of substrate-induced spin state changes at the haem iron that accompany substrate binding. In turn, the magnitude of the spin state changes induced by alternate substrate binding parallel the NMR-detected perturbations observed near the haem in the enzyme active site.
Keywords: enzyme, haem, nuclear magnetic resonance, site-directed mutagenesis
One of the distinguishing characteristics of enzymes is their ability to selectively bind a particular substrate. The origin of this selectivity remains a puzzle. Typically, a group of related enzymes (isozymes) with a common function exhibit a well-conserved fold, with little variation in tertiary structure. Furthermore, catalytically important residues in the active site are usually strongly conserved and mutation of these residues often renders the enzyme inactive. But if active site residues and overall tertiary structure are conserved, how is substrate selectivity and product specificity enforced? The answers must lie in the complex interactions between protein structure and dynamics that are only now being investigated.
The issue of substrate recognition and selectivity is of particular interest for the superfamily of cytochrome P450 monooxygenases. These enzymes are found in all kingdoms of life, and oxidize a bewildering variety of substrates. While many P450s are non-specific (particularly those involved in xenobiotic metabolism in vertebrates), other members of the superfamily catalyze highly regio- and stereospecific reactions. Examples include P450 EryF and EryK, which catalyze specific macrolide ring oxidations in the biosynthetic pathway of erythromycin,1, 2 sterol biosynthetic enzymes such as aromatase3 and bacterial catabolic enzymes, including the well-known examples of cytochrome P450cam4 and P450terp.5 Despite the multiplicity of P450 enzyme-substrate combinations, the P450 fold is highly conserved, with identical folding topology for all of the enzymes examined to date.6 Structurally characterized P450s also show remarkable consistency in secondary structure as well, with major structural features having recognizable analogs in most members of the superfamily. Furthermore, the available P450 structures with substrates or substrate analogs bound in the enzyme active sites exhibit substrate-enzyme interactions that typically involve the same active site features, with some exceptions for large or unusual substrates and co-factors.6
Cytochrome P450cam (CYP101A1) is ideal for investigating issues of substrate recognition and selectivity. Originally isolated from Pseudomonas putida strains that can use (1R)-(+) camphor 1 as sole carbon and energy source, CYP101A1 catalyzes the 5-exo-hydroxylation of 1 as the first step leading into normal aerobic catabolic pathways. This enzyme has been extensively characterized, and considerable data are available from structural, mutational and chemical studies regarding CYP101A1-substrate interactions.7–11 We have reported extensive sequential 1H, 15N and 13C resonance assignments for oxidized and reduced forms of CYP101A1,12, 13 and have shown that NMR spectroscopy can provide residue-specific information regarding structural and dynamic perturbations in this enzyme.12 We demonstrated that the solution conformation adopted by CYP101A1 in the absence of the effector protein putidaredoxin differs significantly from crystallographic structures.13–15 We now show that NMR methods can provide insight into enzyme-substrate interactions by identifying structural features that are remote from the active site but nevertheless are involved in substrate recognition and binding. Specifically, we have compared NMR fingerprint 1H,15N TROSY HSQC spectra of reduced and CO-bound CYP101A1 saturated with camphor 1, adamantanone 2 and norcamphor 3 in order to identify regions of the CYP101A1 structure that respond to changes in substrate structure, and have used site-directed mutagenesis and comparison with published results of other groups to confirm that changes in these regions indeed perturb the interactions between substrate and enzyme.
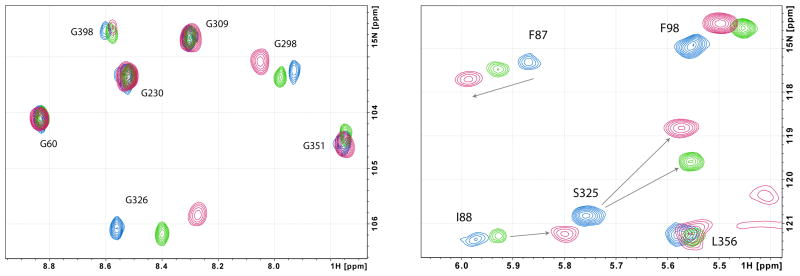
Analysis of 1H,15N-TROSY-HSQC spectra of CYP-1-CO, CYP-2-CO and CYP-3-CO identified two regions of the CYP101A1 backbone as being most sensitive to the identity of bound substrate (Figure 1). The first includes residues Gly 248 to Val 254 in the I helix, which forms one wall of the enzyme active site. These residues span the I helix “kink” thought to accommodate O2 bound to the haem iron. Of this group, Val 254 is the most strongly perturbed, in that the Val 254 NH correlation cannot be identified in the spectra of either the admantanone-bound or norcamphor-bound forms. The conserved Thr 252 shows the largest traceable perturbations in the I helix (see Table I). Thr 252 is in close proximity to bound substrate in the CYP101A1 active site, and the side chain hydroxyl group of this residue is critical in stabilizing the I helix “kink”.
Figure 1.
Superimposed 1H, 15N TROSY-HSQC spectra of CYP-S-CO with camphor 1 (blue), admanatanone 2 (green) and norcamphor 3 (red). At left the region containing glycine NH correlations is shown, and at right, the upfield region containing the Ser 325 correlation.
Table I.
Combined chemical shift perturbations to NH (amide) correlations of selected residues in CYP-S-CO relative to the shifts of CYP-1-CO (camphor-bound) as a function of substrate. Perturbations were calculated from individual 1H and 15N shifts using the equation (Ref 18).
| Residue | |Δδ| (1H, 15N) ppm adamantanone (CYP-2-CO) |
|Δδ| (1H, 15N) ppm norcamphor (CYP-3-CO) |
|---|---|---|
| Val 247 (I helix) | 0.14 | 0.22 |
| Gly 248 (I helix) | 0.07 | 0.14 |
| Gly 249 (I helix) | 0.16 | 0.31 |
| Thr 252 (I helix) | 0.32 | 0.42 |
| Val 254 (I helix) | Large change | Large change |
| Val 295 (β3) | Small or none | Small or none |
| Ala 296 (β3) | 0.06 | 0.13 |
| Gly 298 (β3) | 0.03 | 0.13 |
| Met 323 | overlapped | overlapped |
| Leu 324 | None or small | None or small |
| Ser 325 (K’ helix) | 0.33 | 0.45 |
| Gly 326 (K’ helix) | 0.16 | 0.29 |
| Cys 357 (haem ligand) | 0.03 | 0.11 |
The second region includes two residues in the β3 sheet adjacent to the active site (Ala 296 and Gly 298) and the short proximal K’ helix, residues Leu 324 to Asp 328. With the exception of the I helix residues noted above, Ser 325 and Gly 326 in the K’ helix show the most profound sensitivity of any residue in the protein to substrate replacement (Table I). The K’ helix is not in the active site, but follows the second strand of the β3 sheet and forms the rough apex of a pyramidal arrangement of the A, B and K helices. The turn that ends the K’ helix (Glu 329-Asn 332) initiates the β-meander, a region of irregular structure that precedes the loop containing the haem axial ligand, Cys 357. It is worth noting that in the β-meander and the loop only the NH resonance of the haem iron axial ligand Cys 357 thiolate shows any significant effect upon changing substrate, and then only for the smallest substrate (norcamphor). The magnitudes of chemical shift changes for the most perturbed residues are shown in Table I, while the structural distribution of the affected regions are shown in Figure 2.
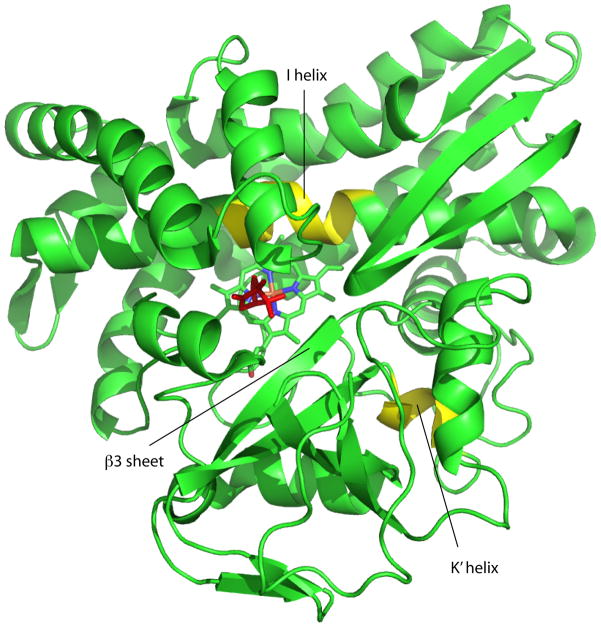
Figure 2.
Structure of CYP101A1 (PDB entry 2CPP) with regions discussed in text highlighted and labeled. The I helix “kink” and K’ helix are shown in yellow. Susbstrate (camphor) is shown in red, and the haem in cyan. Figure generated using PyMOL.20
The finding that the NH correlation in CYP-S-CO most sensitive to changing substrates (Ser 325) is well removed from the active site (the amide nitrogen of Ser 325 is 14.1 A from the nearest carbon atom of bound camphor in 2CPP) was somewhat surprising. However, portions of the K’ helix are strongly conserved in related P450 sequences. While Ser 325 is most commonly a His residue in related sequences, the sequence GLD (Gly 326-Leu 327-Asp 328 in CYP101A1) is uniformly conserved in the sequences detected in BLAST homology searches with CYP101A1 (Fig. 3). We made the replacement mutation G326A in order to decrease the flexibility of the polypeptide at that point, and an insertion mutant (G327’) in order to determine whether the length of the helix played a role in the sensitivity of the K’ helix to substrate. The insertion mutant failed to express, but the G326A mutant was successfully isolated and purified. Differences were noted in the ratio of high spin to low spin UV absorbance in the presence of the three substrates for the WT and G326A mutants. With camphor bound, WT CYP101A1 gave 85% HS, and G326A gave 90% HS. With adamantanone, we observed 73% HS for the WT, versus 78% for G326A. Norcamphor showed the greatest difference, with 23% HS for WT versus 12% HS for G326A. Spin state fractions were calculated from relative heights of peaks in second-derivative UV-visible spectra with maxima at 417 nm (low-spin) and 391 nm (high-spin), with estimated uncertainties of +/−2%. The G326A mutation did not result in a significant shift in the absorbance maximum of either spin state. While the observed spin-state differences are not large, they suggest that the flexibility of the K’ helix plays a role in determining the spin state equilibrium.
Figure 3.
BLAST21 alignment of related P450 sequences from GenBank, identified by species. P. putida is the CYP101A1 sequence. Conserved substrate contact Val (295 in CYP101) is highlighted in green, the Arg anchor discussed in the text (299 in CYP101) is highlighted in cyan, and the conserved GLD sequence in the K’ helix (Gly 326-Leu 327-Asp 328) is highlighted in gold. Glu 331 in the CYP101A1 sequence (highlighted in red) was mutated to Lys in mutants that oxidize naphthalene (see Ref. 17).
The mutation also affects the rate of NADH consumption and product production in CYP101. The G326A mutant showed ~60% of the initial rate of NADH consumption relative to WT in reconstituted reaction mixtures, regardless of the substrate present. If the reactions were allowed to proceed to completion (using a fixed amount of NADH), the WT enzyme was more efficient in the production of hydroxycamphor as calculated for the relative areas of product and substrate peaks in GC traces (SOH/S = 0.27 ± 0.04 for WT versus 0.18 ± 0.01 for G326A). The product ratios were difficult to establish for norcamphor due to the complexity of the GC trace, but for adamantanone, similar conversion efficiencies were observed for both WT and mutant (SOH/S = 0.025 ± 0.001 for WT versus 0.023 ± 0.005 for G326A).
In all of the P450 enzymes for which structures have been determined, residues in the β3 strand provide at least one active site contact with bound substrate.6 In CYP101, the side chain of Val 295 provides primary contacts for the camphor gem-methyl groups, and mutation of this residue has been shown to alter the ratios of hydroxylation products of norcamphor.8 The NH of Val 295 forms a hydrogen bond with the carbonyl of Met 323 as part of the regular anti-parallel hydrogen bonding of the β3 sheet. However, the amide NH correlation of Val 295 or any other β3 residue involved in regular anti-parallel hydrogen bonding show little or no substrate-induced perturbation in our experiments. This suggests to us that the β3 sheet responds to substrate binding as a unit. This conclusion is supported by the observations of relatively small but detectable perturbations at Ala 296 and Gly 298, also in the β3 sheet. The NH groups of these residues do not take part in anti-parallel sheet hydrogen bonding, but instead are exposed to the active site cavity.
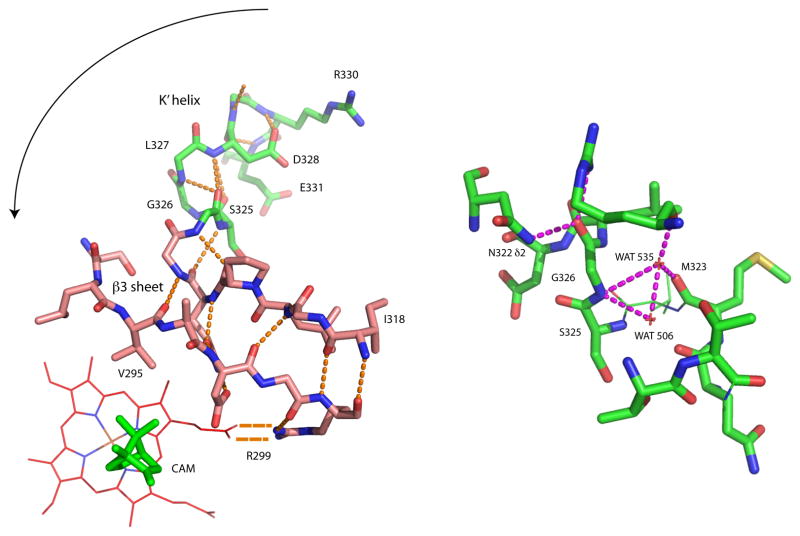
The β3 sheet is anchored in the active site at one end by salt bridge between Arg 299 and a haem propionate. This interaction is strongly conserved in related P450 sequences, and provides a flexible pivot tethering one end of the β3 sheet to the haem and active site, while the other end (including Val 295) is unconstrained relative to the active site cavity. Instead, hydrogen bonding interactions via the carbonyl oxygen of Met 323 link this end of the β3 sheet into the network of hydrogen bonds between peptide and water that define the N-terminus of the K’ helix (see Table II and Fig. 4). In particular, the NH of Gly 326 shares polar interactions to a single water molecule (WAT 535 in 2CPP) with the carbonyl of Met 323. The change in chemical shifts for Ser 325 and Gly 326 indicate an increase in nuclear shielding upon replacing camphor with either norcamphor or adamantanone. Such changes suggest a decrease in the linearity and/or increase in length of polar interactions involving the amide NH groups of Ser 325 and Gly 326.
Table II.
Distances for selected hydrogen bonds in the β3 sheet and K’ helix measured from crystallographic structures of CYP101A1 with bound camphor (RCSB PDB entry 2CPP)19, adamantanone (entry 5CPP)16 and norcamphor (entry 7CPP)16. Water numbering is from 2CPP.
| H-donor | H-acceptor | Distance (A) camphor | Distance (A) adamananone | Distance (A) norcamphor |
|---|---|---|---|---|
| M323 N | V295 O | 3.0 | 3.0 | 3.0 |
| WAT 535 | M323 O | 2.6 | 2.6 | 3.0 |
| G326 N | WAT 535 | 3.2 | 3.2 | 3.2 |
| G326 N | WAT 506 | 3.0 | 3.1 | 3.1 |
| S325 Oγ | T348 Oγ1 | 3.0 | 2.8 | 2.9 |
| R290 Nη1 | G326 O | 2.9 | 2.8 | 2.8 |
| N332 Nδ | G326 O | 2.9 | 2.9 | 2.8 |
| N332 N | D328 O | 2.9 | 2.8 | 2.8 |
| L327 N | L324 O | 3.2 | 3.0 | 3.1 |
| D328 N | L324 O | 3.2 | 3.3 | 3.2 |
Figure 4.
Left, hydrogen bonding patterns in the β3 sheet and K’ helix, shown in relation to the haem and camphor in the CYP101A1 active site (from PDB entry 2CPP). Direction of movement of β3 sheet into active site upon conversion to of K’ to 310 conformation is indicated by arrow. Right, details of the hydrogen bonding involving Gly 326, WAT 535, WAT 506 and Met 323. See text for details. Figure generated using PyMOL.20
Crystallographic structures of CYP101A1 with both adamantanone and norcamphor bound are available.16 With the caveat that we believe these structures (along with that of the camphor-bound CYP101A1) represent a more compact “closed” form of the enzyme than we are observing by solution NMR methods,13 it is still instructive to examine the β3 and K’ helix regions of these structures in some detail. The structures of CYP101A1 with bound camphor (2CPP), adamantanone (5CPP) and norcamphor (7CPP) do not show the NH of Ser 325 involved in any obvious hydrogen bond, while the NH of Gly 326 interacts with two bound water molecules, one of which is shared with the carbonyl of Met 323. Met 323 is at the end of theβ3 strand, antiparallel with Val 295. Comparison of hydrogen bond distances in this region from the three structures shows relatively few differences (Table II). The hydrogen bonding between the two strands of the β3 sheet are unperturbed, supporting the assumption that the β3 sheet moves as a unit in response to changes in the active site. The most intriguing difference involves hydrogen bonding between the carbonyl oxygen of Leu 324 and the amides of Leu 327 and Asp 328. In the presence of camphor (2CPP), the distances are equal (3.2 A), but in the presence of adamantanone (5CPP), the L324 O—N| Leu 327 decreases to 3.0 A, while the corresponding distance to the N of Asp 328 increases to 3.3 A. A similar though smaller change is noted in the norcamphor-bound form. This suggests an increased preference for a 310 helix conformation in the presence of adamantanone for this region (i, i+3 hydrogen bonding) relative to α-helical conformation (i, i+4 bonding) for the Leu 324-Asp 328 turn of the helix in the presence of non-native substrates.
Keeping in mind that the crystallographic structures may not clearly delineate changes in this region that might exist in the open conformations of the enzyme, the differences seem to suggest that the K’ helix can alter its hydrogen bonding patterns both internally and to adjacent water molecules so as to form either a turn of α-helix or 310 helix, depending upon the steric interactions between the substrate and contacts on the β3 sheet. In the case of adamantanone and norcamphor, neither of which present the steric interactions with the side chain of Val 295 that are observed between that residue and camphor, the 310 configuration would be preferred, since this lengthens the K’ helix relative to the α-helix conformation, and assuming one end of the K’ helix remains fixed, would tend to move the β3 sheet into the active site. In short, we propose that the K’ helix can act as a spring, adjusting the position of the unconstrained end of the β3 sheet in the active site depending upon the steric bulk of the substrate. The sense of this proposed motion is indicated in Figure 4. The results from the G326A mutation demonstrate that changes in the K’ helix have an effect on the efficiency of the enzyme. Even more intriguing are the results reported by the Arnold group. These researchers used directed evolution to identify mutants of CYP101A1 that show increased ability to hydroxylate naphthalene using hydrogen peroxide as the oxidant.17 Two CYP101A1 variants were isolated showing the desired activity, both of which contained the E331K mutation. While those workers could not explain the role of the E331K mutation in promoting the observed activity, we can now speculate as to the origin of the effect. Glu 331 lies in the type 1β-turn that terminates the K’ helix. The Glu 331 carboxylate forms a salt bridge with Arg 72 in the B helix in 2CPP, and likely plays a role in anchoring the K’ helix relative to the B helix. By negating (or replacing) that interaction, it is possible that the β3 sheet-K’ helix complex can move with greater ease into the active site, permitting a planar substrate (naphthalene) to be bound in place of the sterically bulkier camphor.
It is also worth considering the perturbations observed in the I helix “kink” (Val 247-Val 254) upon changing substrate. These residues surround the axial haem ligand binding site, so perturbations here reflect a changed environment for the axial ligand as a function of substrate. Given that in all cases where it could be measured, the chemical shift perturbations for residues 247–254 due to norcamphor are larger than those due to adamantanone, and the haem spin state equilibrium due to binding of these substrates follows a similar trend, it may be that the spin state equilibrium in cytochromes P450 is more complicated than a simple presence or absence of an axial water molecule, as has been commonly assumed. At least under the conditions that we observe the NMR signals described here, the presence of a carbonmonoxy axial ligand is unquestioned due to the lack of paramagnetism that would accompany the loss of the ligand, generating the S=2 spin state for ferrous iron.
Supplementary Material
Acknowledgments
This work was supported by a grant from the US National Institutes of Health (R01-GM44191, TCP).
References cited
- 1.Cupp-Vickery JR, Poulos TL. Steroids. 1997;62:112–116. doi: 10.1016/s0039-128x(96)00168-7. [DOI] [PubMed] [Google Scholar]
- 2.Savino C, Montemiglio LC, Sciara G, Miele AE, Kendrew SG, Jemth P, Gianni S, Vallone B. J Biol Chem. 2009;22:22. doi: 10.1074/jbc.M109.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh D, Griswold J, Erman M, Pangborn W. Nature. 2009;457:219–U119. doi: 10.1038/nature07614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sligar SG. Biochemistry. 1976;15:5399–5406. doi: 10.1021/bi00669a029. [DOI] [PubMed] [Google Scholar]
- 5.Hasemann CA, Ravichandran KG, Peterson JA, Deisenhofer J. J Mol Biol. 1994;236:1169–1185. doi: 10.1016/0022-2836(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 6.Pochapsky TC, Kazanis S, Dang M. Antioxidants and Redox Signaling. 2010;13:1273–1296. doi: 10.1089/ars.2010.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell SG, Chen X, Xu F, Rao Z, Wong L. Biochemical Society Transactions. 2003;31:558–562. doi: 10.1042/bst0310558. [DOI] [PubMed] [Google Scholar]
- 8.Atkins WM, Sligar SG. J Am Chem Soc. 1989 Mar 29;111:2715–2717. [Google Scholar]
- 9.Vidakovic M, Sligar SG, Li HY, Poulos TL. Biochemistry. 1998;37:9211–9219. doi: 10.1021/bi980189f. [DOI] [PubMed] [Google Scholar]
- 10.Lee H, de Montellano PRO, McDermott AE. Biochemistry. 1999;38:10808–10813. doi: 10.1021/bi990463l. [DOI] [PubMed] [Google Scholar]
- 11.DeVoss JJ, Sibbesen O, Zhang ZP, deMontellano PRO. J Am Chem Soc. 1997;119:5489–5498. [Google Scholar]
- 12.Pochapsky SS, Pochapsky TC, Wei JW. Biochemistry. 2003;42:5649–5656. doi: 10.1021/bi034263s. [DOI] [PubMed] [Google Scholar]
- 13.Asciutto EK, Madura JD, Pochapsky SS, OuYang B, Pochapsky TC. Journal of Molecular Biology. 2009;388:801–814. doi: 10.1016/j.jmb.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei JY, Pochapsky TC, Pochapsky SS. J Am Chem Soc. 2005;127:6974–6976. doi: 10.1021/ja051195j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.OuYang B, Pochapsky SS, Dang M, Pochapsky TC. Structure. 2008;16:916–923. doi: 10.1016/j.str.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raag R, Poulos TL. Biochemistry. 1989;28:917–922. doi: 10.1021/bi00428a077. [DOI] [PubMed] [Google Scholar]
- 17.Joo H, Lin ZL, Arnold FH. Nature. 1999;399:670–673. [Google Scholar]
- 18.Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE. Biochemistry. 2009;48:2115–2124. doi: 10.1021/bi802055v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poulos TL, Finzel BC, Howard AJ. J Mol Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 20.DeLano WL. The PyMOL Molecular Visualization Program. 2008. [Google Scholar]
- 21.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.