Abstract
Secretin is a linear 27-residue peptide hormone that stimulates pancreatic and biliary ductular bicarbonate and water secretion by acting at its family B G protein-coupled receptor. While, like other family members, the carboxyl-terminal region of secretin is most important for high affinity binding and its amino-terminal region is most important for receptor selectivity and receptor activation, determinants for these activities are distributed throughout the entire length of this peptide. In this work, we have systematically investigated changing each residue within secretin to alanine, and evaluating the impact on receptor binding and biological activity. The residues most critical for receptor binding were His1, Asp3, Gly4, Phe6, Thr7, Ser8, Leu10, Asp15, Leu19, and Leu23. The residues most critical for biological activity included His1, Gly4, Thr7, Ser8, Glu9, Leu10, Leu19, Leu22, and Leu23, with Asp3, Phe6, Ser11, Leu13, Asp15, Leu26 and Val27 also contributing. While the importance of residues in positions analogous to His1, Asp3, Phe6, Thr7, and Leu23 is conserved for several closely-related members of this family, Leu19 is uniquely important for secretin. We, therefore, have further studied this residue by molecular modeling and molecular dynamics simulations. Indeed, the molecular dynamics simulations showed that mutation of Leu19 to alanine was destabilizing, with this effect greater than that observed for the analogous position in the other close family members. This could reflect reduced contact with the receptor or an increase in the solvent-accessible surface area of the hydrophobic residues in the carboxyl terminus of secretin as bound to its receptor.
Secretin, a 27-residue peptide hormone produced by the S cells in the small intestinal mucosa, is secreted in response to luminal acid and stimulates biliary and pancreatic ductular bicarbonate and water secretion, thereby regulating the pH of duodenal contents (1). The secretin receptor is a prototypic member of the B family of G protein-coupled receptors (GPCRs)1 that includes many important drug targets, such as receptors for vasoactive intestinal polypeptide (VIP), pituitary adenylate cyclase-activating peptide (PACAP), gluacagon-like peptide 1 (GLP1), parathyroid hormone, and calcitonin (2). All of these hormone-receptor systems share common structural and functional themes. Therefore, understanding the molecular basis of interaction between secretin and its receptor may facilitate the rational design and refinement of drugs acting at the orthosteric sites within these receptors.
One of the characteristics of members of this family of receptors is the structurally complex amino terminus that represents the predominant natural ligand-binding domain. NMR and crystal structures of the amino-terminal domain of several members of this receptor family have recently been solved, demonstrating that the carboxyl-terminal region of their ligands dock within a binding cleft in the structure (3–11). Mutagenesis, peptide structure-activity, and photoaffinity labeling studies are consistent with this, and suggest that the amino terminus of the peptide ligands bind to the core portions of these receptors (12–14). Thus, a tethering mechanism involving two domains of binding, with the carboxyl-terminal region of the ligand binding to the receptor amino terminus, and the amino-terminal region of the ligand possibly interacting with the receptor body, has been proposed for activation of this family of receptors (12–14). Although high resolution structures of the receptor amino-terminal domain suggest similarity in structural motifs and ligand binding modes among several members, there are inconsistencies in the absolute sites of ligand binding and in the positioning of the ligands in these structures (15), suggesting some variation in binding mechanisms across this family. Further, divergent molecular mechanisms have been proposed for docking the flexible amino-terminal regions of ligands to these receptors, and even the relative orientation between the receptor amino terminus and its core is unknown.
Natural ligands for family B GPCRs are all moderately long peptides having diffuse pharmacophoric domains that contribute to the complexity of their interactions with their receptors (16). While the amino-terminal portions of these ligands are known to be critical for biological activity and the carboxyl-terminal portions are critical for binding, residues scattered throughout the entire sequence of secretin are required for high affinity binding and full biological activity (1, 16). Alanine-scanning mutagenesis has been performed for several family B GPCR peptide ligands (17–21), but not yet for secretin.
In this work, we have systematically studied the role of each of the 27 residues of secretin in ligand binding and activation of the secretin receptor by alanine scanning. Multiple residues throughout the length of this hormone were found to be important for both binding and biological activity. While there were some consistent themes for all of these ligand-receptor systems, there was one unique functionally-important residue within secretin that was not present in the other members of this family that have been similarly studied. Molecular modeling was also utilized to examine and extend these data, with the basis for the importance of secretin residue Leu19 experimentally tested and validated.
MATERIALS AND METHODS
Materials
Amino acids for peptide synthesis were purchased from Advanced ChemTech (Louisville, KY). Ham’s F-12 medium and soybean trypsin inhibitor were from Invitrogen (Carlsbad, CA). Fetal clone II culture medium supplement was from Hyclone laboratories (Logan, UT). 3-Isobutyl-1-methylxanthine and bacitracin were from Sigma-Aldrich. Bovine serum albumin was from Serologicals Corp. (Norcross, GA). The solid-phase oxidant, N-chlorobenzenesulfonamide (Iodo-beads), was purchased from Pierce Chemical Co (Rockford, IL). All other reagents were of analytical grade.
Peptide Synthesis and Radioiodination
Rat secretin and the secretin-like radioligand, [125I-Tyr10]rat secretin-27, were synthesized as we previously described (22). Natural rat secretin has 27 residues, including an endogenous alanine in position 17. Therefore, 26 secretin analogues representing single alanine substitutions for each of the remaining positions were synthesized for the current work. All peptides were synthesized by manual solid-phase techniques and purified by reversed-phase HPLC using procedures described previously (23). Expected molecular masses were verified by matrix-assisted laser desorption/ionization-time of flight mass spectrometry.
The [Tyr10]rat secretin-27 analogue was radioiodinated oxidatively using 1 mCi Na125I and exposure for 15 s to the solid-phase oxidant, Iodo-beads, as we described previously (23). The radioiodinated peptide was purified by reversed-phase HPLC to yield specific radioactivity of approximately 2,000 Ci/mmol (23).
Receptor Preparations
A Chinese hamster ovary (CHO) cell line stably expressing the wild type rat secretin receptor (CHO-SecR) that was previously established (22) was used as a source of receptor for the current study. Cells were cultured at 37 °C in an environment containing 5% CO2 on tissue culture plasticware in Ham's F-12 medium supplemented with 5% Fetal Clone II, and were passaged approximately twice a week.
Receptor Binding Assay
Whole cell binding in 24-well tissue culture plates was performed to determine the binding affinity of each of the secretin analogues using a radioligand competition-binding assay. For this, approximately 50,000 CHO-SecR cells per well were grown for approximately 72 h prior to the assay. After being washed twice with Krebs-Ringers/HEPES (KRH) medium (25 mM HEPES, pH 7.4, 104 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM KH2PO4, 1.2 mM MgSO4) containing 0.01% soybean trypsin inhibitor and 0.2% bovine serum albumin, cells were incubated with a constant amount of radioligand, [125I-Tyr10]rat secretin-27 (3–5 pM, approximately 20,000 cpm) and increasing concentrations of secretin or each of the alanine-replacement secretin analogues (ranging from 0 to 1 µM) for 60 min at room temperature. Cells were then washed twice with ice-cold KRH medium to separate free from cell-bound radioligand before being lysed with 0.5 M NaOH. Membrane-bound radioactivity was quantified with a γ-spectrometer. Nonspecific binding was determined in the presence of 1 µM secretin and represented less than 15% of total binding.
cAMP Assay
The biological activity of each of the alanine-replacement analogues of secretin was determined by examining its ability to stimulate cAMP responses in CHO-SecR cells. After cells (~8,000 per well) were grown in 96-well plates for two days, they were washed twice with phosphate-buffered saline and stimulated with increasing concentrations of secretin or each of the alanine-replacement analogues (0 to 1 µM) in KRH medium containing 0.01% soybean trypsin inhibitor, 0.2% bovine serum albumin, 0.1% bacitracin, and 1 mM 3-isobutyl-1-methylxanthine for 30 min at 37°C. After incubation, the reaction solution was aspirated and cells were lyzed with 6% ice-cold perchloric acid for 15 min with vigorous shaking. The cell lysates were adjusted to pH 6 with 30% NaHCO3 and assayed for cAMP levels in a 384-well white Optiplate using a LANCE kit from PerkinElmer (Boston, MA).
Molecular Modeling
A homology model of secretin docked with the amino-terminal domain of the secretin receptor was built based on the crystal structure of the complex of glucagon-like peptide 1 and its receptor amino-terminal domain (PDB: 3IOL) (11). The Internal Coordinate Mechanics (ICM) program (version 3.6, Molsoft LLC) was used for the homology modeling, wherein the disulfide bonds and the conserved residues within the sequences were aligned and tethered to the GLP1 peptide-receptor structure. The secretin peptide-receptor structure was annealed to the tethers and the conformational space of the complex was sampled using Biased-Probability Monte Carlo (24) simulations for 5×107 function calls.
For the computational alanine scanning of the secretin model, ICM was used to predict the change in free energy due to the replacement of each residue within the peptide to alanine. Biased-Probability Monte Carlo (24) simulations at T = 700 K were used to determine the side chain torsion angles of each mutated residue (and its corresponding residue in natural secretin) and all residues with side chain heavy atoms within 4.0 Å of any side chain heavy atoms in the mutated residue. The backbone was fixed throughout the simulation and each simulation had 2×105 function calls with up to 2000 steps of conjugate gradient minimization for each accepted Monte Carlo move. An energy function with electrostatic, van der Waals, hydrogen bonding and torsional terms from ECEPP/3 force field (25–27), as well as solvation term with parameters from Wesson and Eisenberg (28) and side chain entropy term (24) were used. The lowest energy conformation from each simulation was used to determine the change in binding free energy (ΔΔG) using a protocol described previously (29). A critical residue was identified as one having a ΔΔG > 1.0 kcal/mol.
Molecular dynamics (MD) simulations were utilized to explore the structural basis for one particularly important residue within secretin (Leu19) in which the functional impact of alanine replacement was distinct from analogous studies of other family members. The GROMACS software package (version 4.5.3) (30) with OPLS all-atom parameters (31) was used to perform all simulations. The starting coordinates of the secretin peptide-receptor and the L19A secretin analogue-receptor models were obtained from the homology model described above. In addition, a homology model of the VIP peptide-VPAC2 receptor and one with the analogous alanine-replacement VIP analogue were built using the same homology modeling procedure and template structure. The starting structure for the GLP1 peptide-receptor model was obtained from the crystal structure of GLP1 bound to the amino-terminal domain of its receptor (11). Each complex was solvated in pre-equilibrated water using the SPC/E water model (32). In both natural and modified systems, 4 Na+ and 5 Cl− were added to the secretin peptide-secretin receptor complex, 6 Na+ and 4 Cl− were added to the VIP peptide-VPAC2 receptor complex, and 9 Na+ and 2 Cl− were added to the GLP1 peptide-GLP1 receptor complex to make the respective systems neutral.
All systems were equilibrated under a canonical (NVT) ensemble for 500 ps using the Berendsen thermostat (33) with a relaxation time of 0.1 ps for temperature control. The systems were further equilibrated in an isothermal-isobaric (NPT) ensemble for another 500 ps using the Parrinello-Rahman barostat (34) with a relaxation time of 2.0 ps. The reference temperature and pressure for all systems were 300 K and 0.1 MPa, respectively. Position restraints (with spring constant of 1000 kJ/mol˙ nm2) were applied to all heavy atoms of the protein complex during the equilibration. Following equilibration, 12 ns production MD using 2-fs time steps and the linear constraint solver method (35) to constrain all bond lengths was conducted per system using an NPT ensemble. The Lennard-Jones interactions were cut-off at 1.0 nm and the neighbor list was updated every 10 fs. Long-range electrostatic interactions were treated using the particle mesh Ewald method (36) with fourth-order spline interpolation and 0.16 nm grid spacing. Coordinates were saved every 1 ps for analysis using the built-in analysis tools in GROMACS.
Statistical Analysis
All assays were performed in duplicate in a minimum of three independent experiments and are expressed as the means ± S.E.M. Both competition-binding and cAMP concentration-response curves were analyzed and plotted using the non-linear regression analysis program in the Prism software suite v3.0 (GraphPad Software, San Diego, CA). Binding kinetics were determined by analysis with the LIGAND program of Munson and Rodbard (37). Two-tailed P value tests were performed to determine the significance of differences using InStat3 (GraphPad Software, San Diego, CA).
RESULTS
In order to understand the role of each residue of secretin in its binding and biological activity, twenty-six secretin analogues were prepared, systematically replacing each residue with an alanine (except for position 17 which has an endogenous alanine). Peptides were synthesized by solid-phase techniques and purified to homogeneity (>99%) by reversed-phase HPLC, and their identities were verified by mass spectrometry.
Receptor Binding of Alanine-Replacement Analogues of Secretin
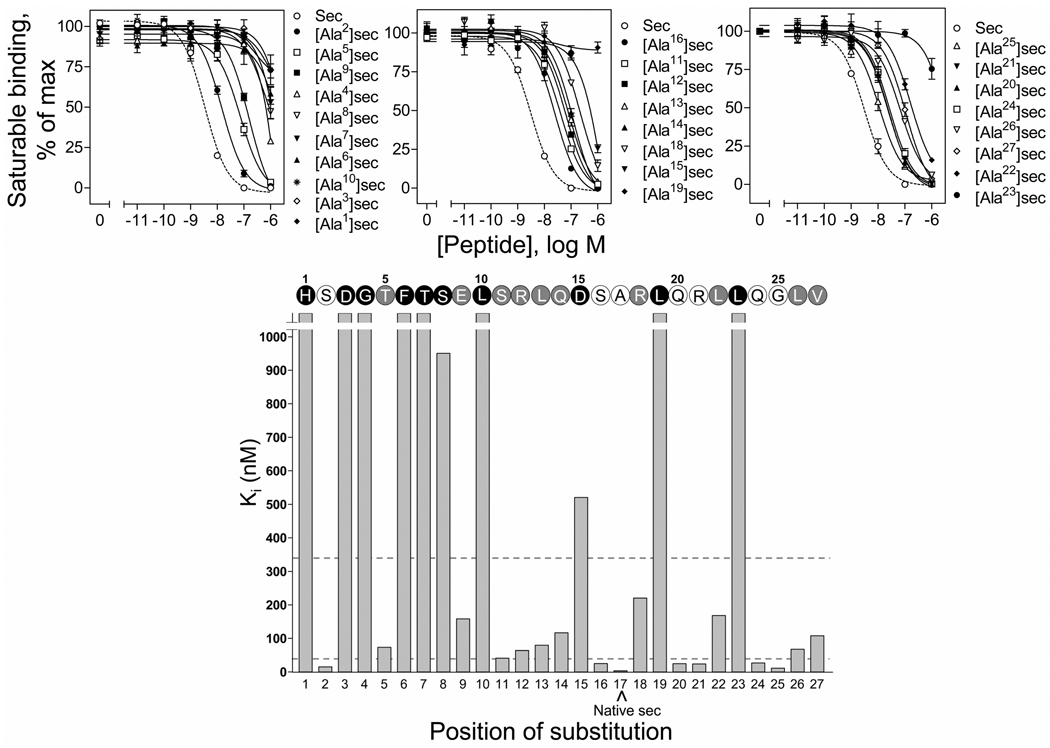
The ability of each of the secretin analogues to bind to the secretin receptor was determined by competition-binding to the secretin receptor-bearing CHO-SecR cells. These cells had 114,000 ± 10,000 secretin binding sites per cell (Bmax). The competition-binding curves are shown in Figure 1 and the calculated Ki values are provided in Table 1. As shown, multiple residues throughout the entire length of secretin were critical for its binding to this receptor. The most pronounced decreases in binding affinity (>100-fold) were observed for alanine substitution of residues His1, Asp3, Gly4, Phe6, Thr7, Ser8, Leu10, Asp15, Leu19, and Leu23. While alanine substitution of residues Thr5, Glu9, Ser11, Arg12, Leu13, Gln14, Arg18, Leu22, Leu26 and Val27 also resulted in an important decrease in binding affinity (>10-fold, but <100-fold), substitution of residues Ser2, Ser16, Gln20, Arg21, Gln24, and Gly25 resulted in minimal decrease (<10-fold).
FIGURE 1.
Binding activities of the alanine-replacement secretin analogues. Top, binding curves of increasing concentrations of secretin and alanine-replacement secretin analogues to compete for binding of radioligand [125I-Tyr10]rat secretin-27 to secretin receptor-bearing CHO-SecR cells. Values illustrated represent saturable binding as percentages of maximal binding observed in the absence of the competing peptide and are expressed as the means ± S.E.M. of duplicate values from a minimum of three independent experiments. Data are presented in three groups, i.e. amino-terminal (top left panel, positions 1 to 10), mid-region (top middle panel, positions 11 to 19) and carboxyl-terminal (top right panel, positions 21 to 27) based on the positions of incorporating alanine in secretin, with the affinities in each group being illustrated in the order of high to low. Bottom, role of each residue in secretin binding to its receptor. Shown are the Ki values of each of the alanine-replacement secretin analogues and the secretin sequence illustrating the role of each residue in binding to the secretin receptor. Open circles represent residues whose replacement by alanine resulted in less than 10-fold in binding affinity comparing to native secretin. Grey and black circles represent residues whose replacement with alanine resulted in more than 10-fold but less than 100-fold, and more than 100-fold increase in binding affinity (dashed lines), respectively, comparing to native secretin. Sec, secretin.
Table 1.
Binding and biological activities of secretin and alanine-replacement analogues. Shown are the Ki values of binding of secretin or each of its analogues to CHO-SecR cells and EC50 values of the intracellular cAMP responses stimulated by these ligands. All values represent the means ± S.E.M. of data from three independent experiments performed in duplicate. Shown also are relative affinity (KiAla/KiSec), relative potency (EC50Ala/EC50Sec) and ratio of both for each alanine-replacement analogue compared to data for natural secretin (Ki = 3.4 ± 0.4 nM and EC50 = 0.05 ± 0.01 nM).
| Peptides | Binding | cAMP response | Relative affinity/ Relative potency |
||||
|---|---|---|---|---|---|---|---|
| Amino acid |
Position | Substitution | Ki (nM) |
Relative affinity |
EC50 (nM) |
Relative potency |
|
| His | 1 | Ala | >1,000 | > 300 | 55.2 ± 8.7 | 1104 | |
| Ser | 2 | Ala | 14.9 ± 2.4 | 4.4 | 0.3 ± 0.1 | 6 | 0.7 |
| Asp | 3 | Ala | >1,000 | > 300 | 3.9 ± 0.6 | 48 | |
| Gly | 4 | Ala | >1,000 | > 300 | 5.1 ± 0.8 | 102 | |
| Thr | 5 | Ala | 73.4 ± 7.3 | 21.6 | 0.2 ± 0.05 | 4 | 5.4 |
| Phe | 6 | Ala | >1,000 | > 300 | 4.6 ± 0.5 | 92 | |
| Thr | 7 | Ala | >1,000 | > 300 | 9.0 ± 1.5 | 180 | |
| Ser | 8 | Ala | 950.5 ± 84.3 | 279.6 | 6.1 ± 1.4 | 122 | 2.3 |
| Glu | 9 | Ala | 158.4 ± 12.2 | 46.6 | 8.5 ± 1.8 | 170 | 0.3 |
| Leu | 10 | Ala | >1,000 | > 300 | 6.1 ± 1.3 | 122 | |
| Ser | 11 | Ala | 40.8 ± 5.6 | 12 | 1.0 ± 0.2 | 20 | 0.6 |
| Arg | 12 | Ala | 63.8 ± 7.2 | 18.8 | 0.3 ± 0.09 | 6 | 3.1 |
| Leu | 13 | Ala | 79.4 ± 8.9 | 23.4 | 0.8 ± 0.2 | 16 | 1.5 |
| Gln | 14 | Ala | 116.5 ± 13.3 | 34.3 | 0.2 ± 0.07 | 4 | 8.6 |
| Asp | 15 | Ala | 520.5 ± 46.4 | 153.1 | 2.6 ± 0.6 | 52 | 2.9 |
| Ser | 16 | Ala | 24.9 ± 4.5 | 7.3 | 0.1 ± 0.04 | 2 | 3.7 |
| Ala | 17a | - | 3.4 ± 0.4 | 1.0 | 0.05 ± 0.01 | 1 | 1 |
| Arg | 18 | Ala | 220.2 ± 26.6 | 64.8 | 0.4 ± 0.1 | 8 | 8.1 |
| Leu | 19 | Ala | >1,000 | > 300 | 34.5 ± 5.5 | 690 | |
| Gln | 20 | Ala | 24.6 ± 3.2 | 7.2 | 0.2 ± 0.04 | 4 | 1.8 |
| Arg | 21 | Ala | 23.6 ± 3.5 | 6.9 | 0.2 ± 0.03 | 4 | 1.8 |
| Leu | 22 | Ala | 168.3 ± 20.0 | 49.5 | 11.7 ± 2.1 | 234 | 0.2 |
| Leu | 23 | Ala | >1,000 | > 300 | 33.2 ± 6.2 | 664 | |
| Gln | 24 | Ala | 26.8 ± 4.4 | 7.9 | 0.2 ± 0.05 | 4 | 2.0 |
| Gly | 25 | Ala | 10.9 ± 1.4 | 3.2 | 0.2 ± 0.06 | 4 | 0.8 |
| Leu | 26 | Ala | 67.6 ± 8.4 | 19.9 | 1.3 ± 0.2 | 26 | 0.8 |
| Val | 27 | Ala | 107.8 ± 15.4 | 31.7 | 0.5 ± 0.2 | 10 | 3.2 |
The residue at position 17 of natural secretin is an alanine.
Biological Activity of Alanine-Replacement Analogues of Secretin
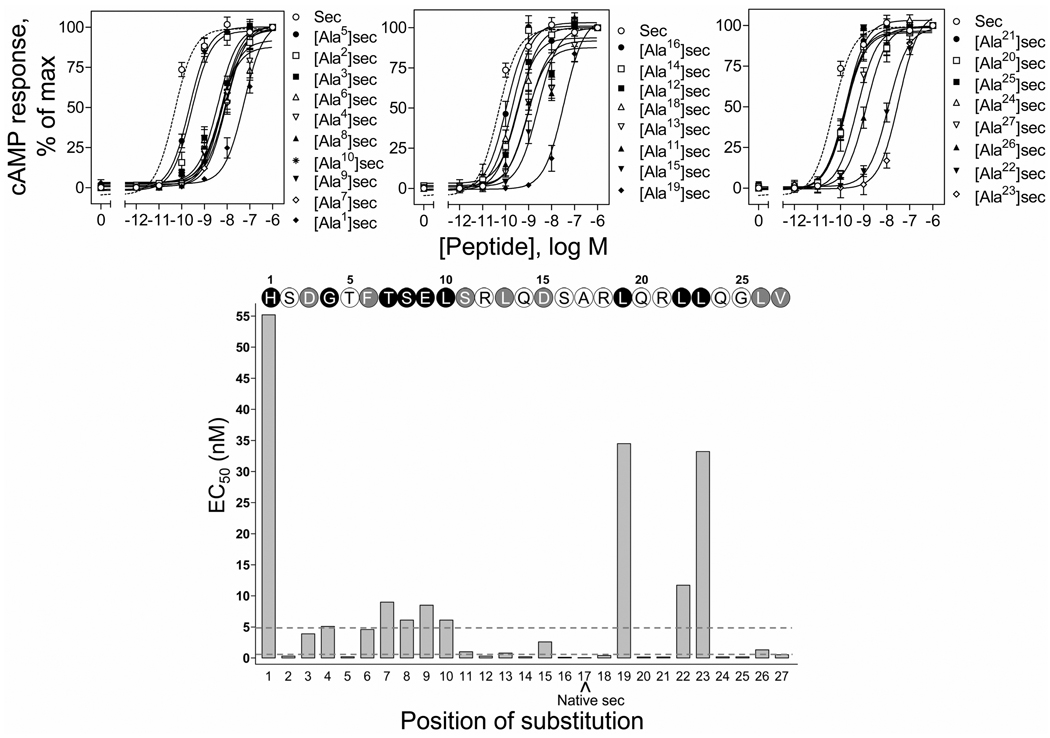
The biological activity of each of the secretin analogues was determined by its ability to stimulate intracellular cAMP responses in the secretin receptor-bearing CHO-SecR cells. The cAMP concentration-response curves are shown in Figure 2 and the calculated EC50 values are provided in Table 1. Although some of the alanine-replacement secretin analogues had very low binding affinity, they all represented full agonists, with maximal cAMP stimulation in CHO-SecR cells similar to that stimulated by natural secretin. Figure 2 and Table 1 show that multiple residues spread throughout the entire length of secretin were critical for its biological activity at this receptor. The most pronounced decreases in stimulating cAMP responses in CHO-SecR cells (>100-fold) were observed for alanine substitution of residues His1, Gly4, Thr7, Ser8, Glu9, Leu10, Leu19, Leu22, and Leu23. While alanine substitution of residues Asp3, Phe6, Ser11, Leu13, Asp15, Leu26 and Val27 also resulted in an important decrease in biological activity (>10-fold, but <100-fold), substitution of residues Ser2, Thr5, Arg12, Gln14, Ser16, Arg18, Gln20, Arg21, Arg22, Gln24, and Gly25 only resulted in a slight decrease (<10-fold).
FIGURE 2.
Biological activities of the alanine-replacement secretin analogues. Top, curves of intracellular cAMP responses in secretin receptor-bearing CHO-SecR cells stimulated by increasing concentrations of the alanine-replacement secretin analogues. Data points represent the means ± S.E.M. of three independent experiments performed in duplicate, normalized relative to the maximal response to secretin. Basal and maximal cAMP levels by secretin were 4.2 ± 1.1 and 197 ± 44 pmol/million cells, respectively. Data are presented in three groups, i.e. amino-terminal (top left panel, positions 1 to 10), mid-region (top middle panel, positions 11 to 19) and carboxyl-terminal (top right panel, positions 21 to 27) based on the positions of incorporating alanine in secretin, with the potencies in each group being illustrated in the order of high to low. Bottom, role of each secretin residue in its biological activity. Shown are the EC50 values of each of the alanine-replacement analogues of secretin and the secretin sequence illustrating the role of each residue in their biological activity. Open circles represent residues whose replacement by alanine resulted in less than 10-fold in biological activity comparing to native secretin. Grey and black circles represent residues whose replacement with alanine resulted in more than 10-fold but less than 100-fold, and more than 100-fold increase in biological activity (dashed lines), respectively, comparing to native secretin. Sec, secretin.
Relationship between Receptor Binding and Biological Activity
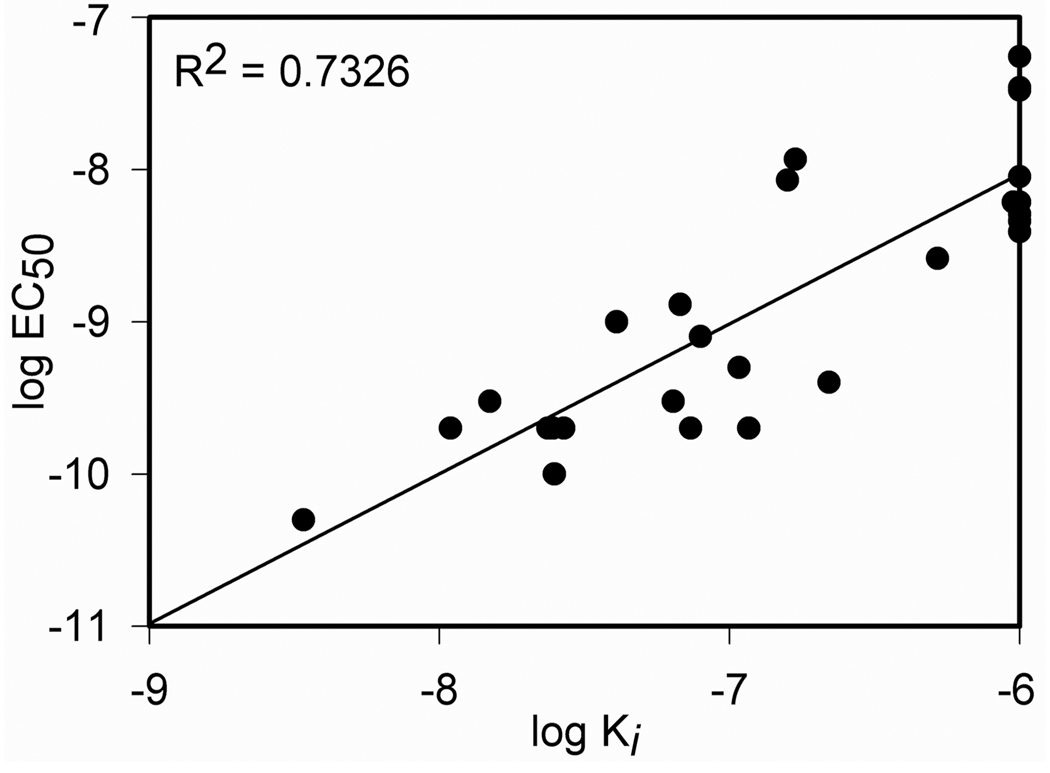
With these functional studies of ligand binding and ligand-stimulated biological activity supporting the contributions of residues spread throughout the entire length of secretin, it was important to determine whether these two functions were correlated with each other or distinct. Figure 3 illustrates the strong correlation between ligand binding affinity and biological activity for these secretin analogues (R2 = 0.7326). Thus, the negative impact of specific ligand modifications was likely the result of disrupted binding, rather than any exclusive effect on receptor activation.
FIGURE 3.
Relationship between binding affinity and biological activity of alanine-replacement analogues of secretin. Shown is the plot of the logarithmic transformations of Ki and EC50 values for these constructs, as well as their correlation coefficient.
Computational Analysis of Secretin Binding to the Receptor Amino Terminus
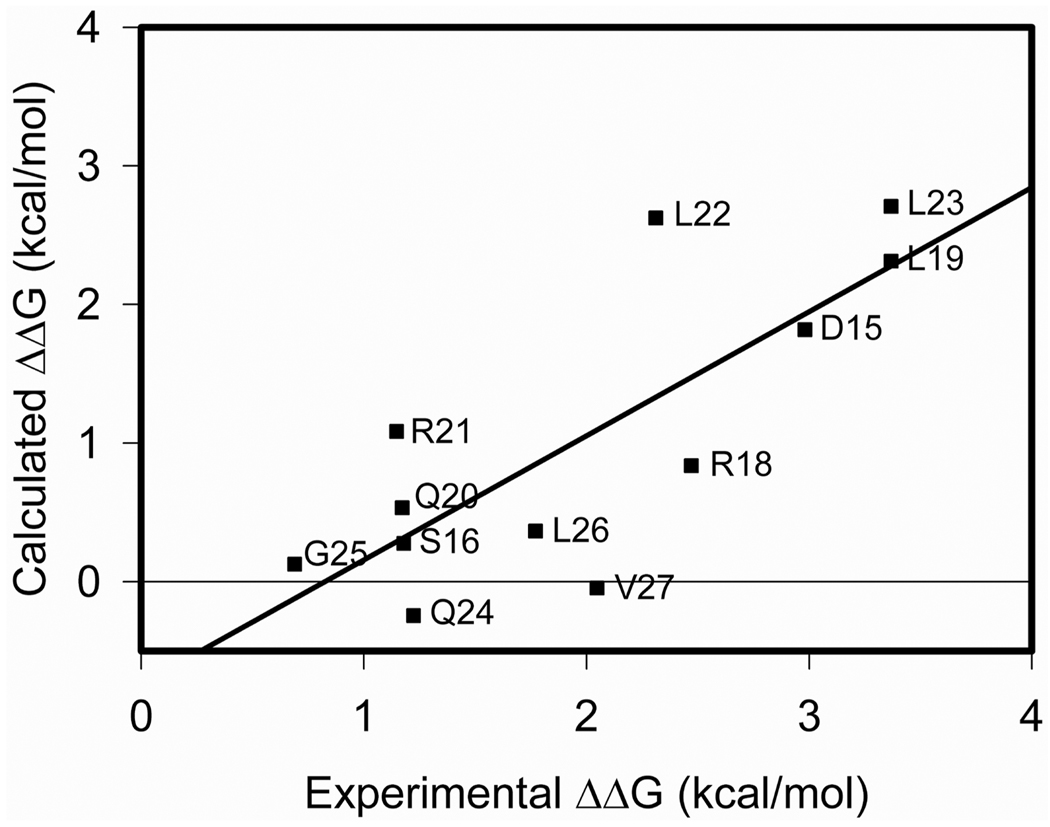
Computational modeling was also used to analyze the alanine replacements within secretin as docked to its receptor amino terminus. This could only be applied to peptide residues 15 to 27, the region of the peptide present within the peptide-binding cleft of this structure. The fixed backbone protocol in ICM was utilized to predict the change in binding free energy (ΔΔG) for residues 15 to 27 of secretin (Figure 4). It is noteworthy that the key residues in positions 19, 22, and 23 that were identified experimentally had the greatest impact of alanine replacement in this computational analysis as well.
FIGURE 4.
Calculated ΔΔG (in kcal/mol) for the alanine mutation of residue 15 to 27 of secretin compared to the experimental ΔΔG. The computational ΔΔG is given for the fixed backbone simulation while the experimental ΔΔG was obtained from ΔΔG = RT ln (KiAla/KiSec), where R is the gas constant and T is the temperature.
Molecular Dynamics Simulation of Secretin and the L19A Secretin Analogue
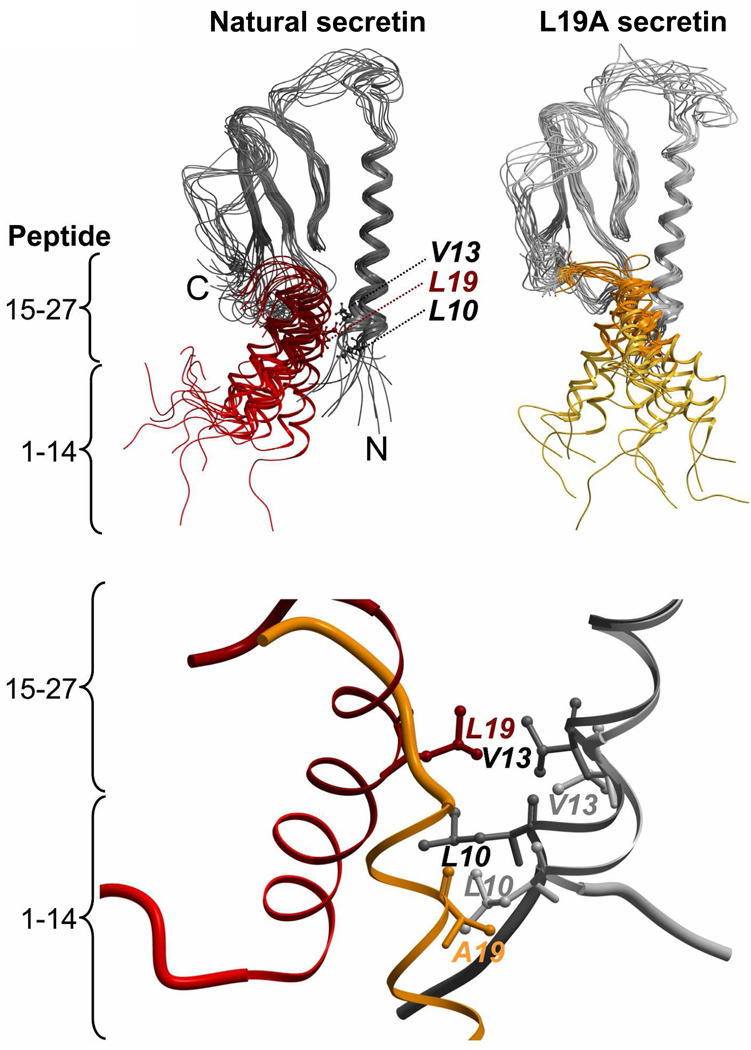
To further study the effects of replacement of Leu19 with alanine in secretin, a 12-ns MD simulation was performed for natural secretin and the L19A secretin analogue in complex with the secretin receptor. A snapshot of every 1 ns time point in the simulation is shown in Figure 5 (top panel). When the receptor backbone structures for all snapshots were superimposed, it was observed that the α-helix in natural secretin was oriented approximately parallel to the amino-terminal α-helix of the receptor. On the other hand, the L19A secretin analogue shifted away from the parallel orientation, demonstrating greater variation in the position of the peptide compared to natural secretin. In the MD simulations of peptide-amino-terminal receptor complexes of other family members, including VIP binding to the VPAC2 receptor, GLP1 binding to the GLP1 receptor and the binding of the V19A analogue of VIP to the VPAC2 receptor, the peptide ligands were oriented parallel to the α-helix of each receptor, similar to situation in which natural secretin was bound to the secretin receptor (data not shown).
FIGURE 5.
Structural snapshots of the complex of secretin with the amino-terminal domain of its receptor during the 12 ns simulation. Top, superimposed structure for every 1-ns time point in the molecular dynamics simulation for the natural secretin peptide (red, left) and the L19A variant of secretin (gold, right) bound to the receptor amino terminus (shades of black and gray). The complexes started with both peptides in the same conformation. The receptors are colored black (with natural secretin docked, left) and gray (with the L19A variant docked, right), with the regions of the peptides representing amino-terminal residues 1–14 (lighter colors) and carboxyl-terminal residues 15–27 (darker colors) identified. Bottom, a structural snapshot taken at the identical time point (10 ns) in the simulations showing the approximation of residue 19 of the peptides with Leu10 and Val13 of the secretin receptor.
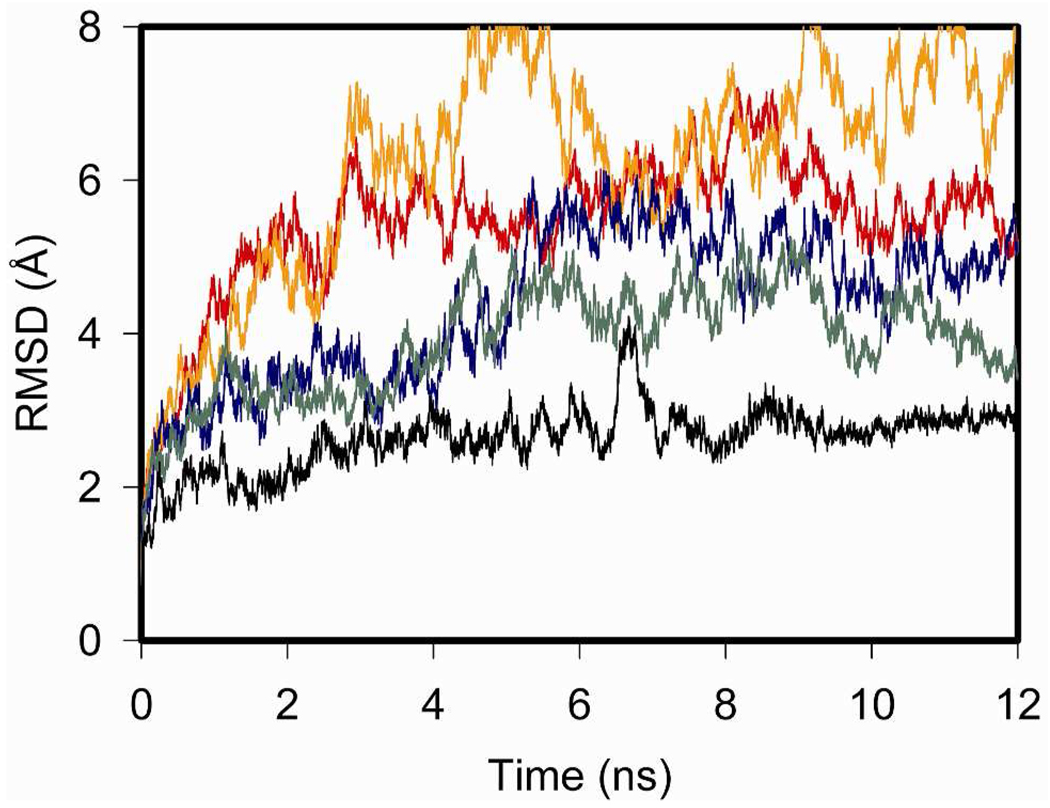
The root-mean-square deviations (RMSD) of the backbone residues for the complex relative to the starting structure for all five MD simulations were also determined (Figure 6). Not surprisingly, the GLP1 peptide-receptor complex, where the starting structure was taken from the crystal structure, exhibited the least deviation throughout the 12-ns simulation. Interestingly, the RMSD data reflected the instability of the complex of the L19A analogue of secretin with its receptor, particularly during the last 8 ns of the simulation. The erratic RMSD of this simulation reflected the greater mobility of the peptide analogue when complexed with its receptor, which is consistent with the snapshots of the predicted structure shown in Figure 5.
FIGURE 6.
RMSD of the GPCR complex simulation. Shown are the root-mean-square deviations of the backbone of the peptide-receptor complex for Sec-SecR (red), L19A/Sec-SecR (orange), VIP-VPAC2 (blue), V19A/VIP-VPAC2 (green), and GLP1-GLP1R (black).
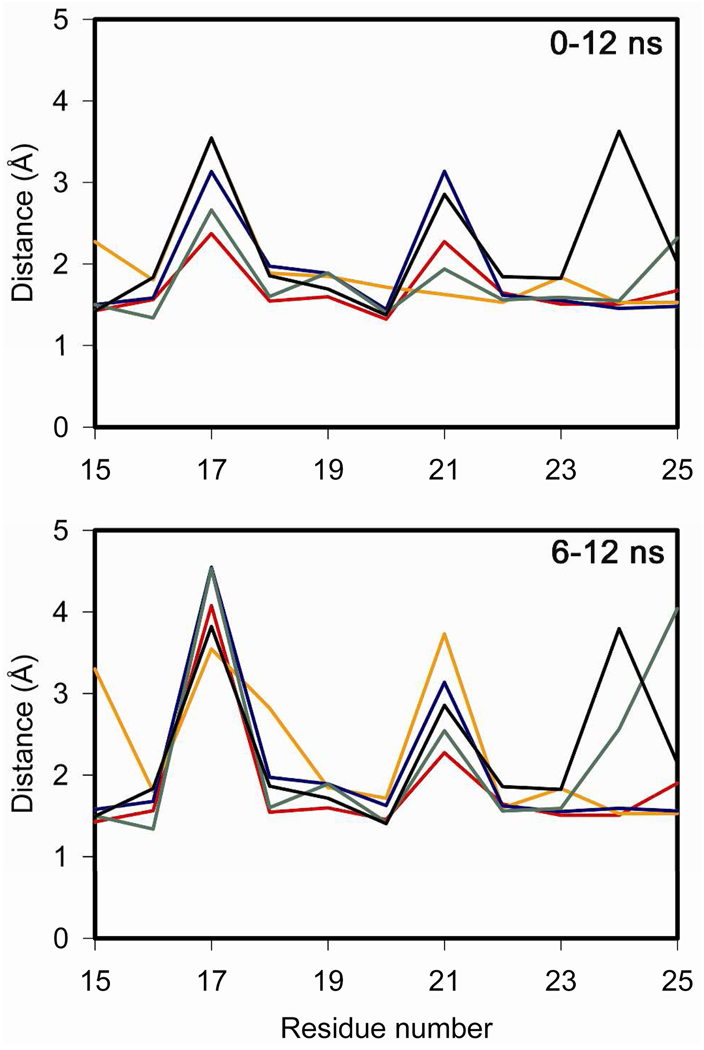
The top panel of Figure 7 shows the minimum distance between any atom of a given peptide residue and any atom in the receptor where this could be meaningfully measured, based on homology models with the existing crystal structures. Particular attention was focused on residues 19 and 23, which are among the hydrophobic residues within the peptide that are predicted to be facing the binding cleft within the amino-terminal domain of the receptor. Of these residues, the distances established by the L19A secretin analogue appeared to be greater than those for docked natural secretin, while natural VIP and the V19A VIP analogue had similar minimum distances in these positions. Since the starting structure (at t = 0 ns) could well influence the result for the minimum distance analysis, the same analysis was performed for the last 6 ns of each simulation, where natural secretin again exhibited closer association with the receptor for residues 19 and 23 than the L19A analogue (Figure 7, bottom panel). However, this analysis tool can not establish the duration of any possible interaction, so we also determined the minimum distance as a function of time between any atom of secretin residue 19 and any atom of receptor residue Val13, representing the closest point of contact for peptide residue 19 in the 12-ns simulation. Interestingly, the minimum distance between these residues was significantly longer for the L19A secretin analogue than for the natural peptide (Table 2).
FIGURE 7.
Minimum distances established by the peptide residues to the receptor. Shown are the minimum distances between any atom of a specific residue number in the peptide and any atom in the receptor for the entire 12-ns simulation (top) and for the last 6 ns of the simulation (bottom). The different complexes are Sec-SecR (red), L19A/Sec-SecR (orange), VIP-VPAC2 (blue), V19A/VIP-VPAC2 (green), and GLP1/GLP1R (black). The residue numbering for both panels is based on the secretin peptide.
Table 2.
The average minimum distances (± standard deviation, in Ångstroms) as a function of time between any atom in residue 19 of the peptide and any atom of Val13 in the secretin receptor or Phe27 in the VPAC2 receptor.
| Full MD simulation | Last 6 ns of the simulation | |
|---|---|---|
| Sec | 2.43 ± 0.28 | 2.40 ± 0.24 |
| L19A Sec | 6.70 ± 1.13 | 6.51 ± 0.61 |
| VIP | 2.50 ± 0.24 | 2.47 ± 0.22 |
| V19A VIP | 2.71 ± 0.51 | 2.60 ± 0.34 |
Based on sequence alignments among the most closely-related receptors, Val13 in the secretin receptor is equivalent to Phe27 in the VPAC2 receptor. The average minimum distances between residue 19 in the VIP peptide and Phe27 in VPAC2 were similar for natural VIP and the V19A VIP analogue (Table 2). The average minimum distances (± standard deviation, in Ångstroms) between residue 19 in the peptide and Leu10 in the secretin receptor for the entire 12-ns simulation and the last 6 ns of the simulation, respectively, were 3.09 ± 0.99 and 2.84 ± 0.75 for natural secretin and 2.66 ± 0.67 and 2.47 ± 0.32 for the L19A secretin analogue. A snapshot of the simulation reflected the close approximation between residue 19 in the L19A analogue and Leu10 of the receptor (Figure 5, bottom panel).
Furthermore, the L19A analogue of secretin also affected the solvent-accessible surface area (SASA) of the hydrophobic residues of the peptide in the carboxyl terminus. In comparing the natural peptide and the L19A analogue of secretin, the SASA was greater for the analogue than for the natural peptide throughout the simulation, except during the first nanosecond of the simulation when the analogue had not yet diverged from the starting structure. The average SASA values (± standard deviation, in Ångstroms2) for the natural peptide and the L19A analogue of secretin were 760 ± 33 and 842 ± 32, respectively, during the entire 12-ns simulation. During the last 6 ns of the simulation, the average SASA values were 756 ± 32 and 855 ± 27 for the natural peptide and the L19A analogue of secretin, respectively. The higher observed solvent exposure of hydrophobic residues in the L19A analogue of secretin compared with natural secretin is expected to be energetically unfavorable, and thus contributes to its reduced binding affinity to the receptor. In contrast, the SASA values of both natural VIP and its analogue were similar, particularly in the last 6 ns of the simulation where the average SASA values were 1036 ± 41 and 1014 ± 32 for the natural VIP and its analogue, respectively.
DISCUSSION
Understanding the structural basis for interaction between a ligand and its receptor may facilitate the rational design and refinement of receptor-active drugs. However, our understanding of the molecular mechanisms of ligand binding and activation of family B GPCRs is limited, due to the current absence of high resolution structures of intact receptors in this family. Multiple experimental approaches including structure-activity relationships for ligands, receptor mutagenesis, photoaffinity labeling, and NMR and crystallographic studies of soluble extracellular domains have been applied to these receptors to yield general themes that seem to be common across this family (3–11). These studies have established the folded, disulfide-bonded structure of the amino-terminal domains of these receptors that includes a peptide-binding cleft for the carboxyl terminus of the natural ligands (15). They also demonstrated the importance of the amino terminus of the ligands for biological activity, mediated by interaction with the core helical bundle domains of these receptors. However, high resolution insights are not yet available (15).
Much is known about the prototypic family B GPCR, the secretin receptor. This was the first member of this receptor family to be cloned and it serves important roles in stimulating biliary and pancreatic ductular bicarbonate and water secretion (1). Each of the general themes for the family is also applicable to this receptor (38–40). Additionally, we have identified eleven pairs of spatial approximations between residues along this peptide ligand and its receptor, as normally docked (41). These provide low resolution spatial approximation constraints that can be further refined by systematic mutagenesis, such as we have achieved in the current study.
In the current project, we have explored the functional impact of replacing each of the non-alanine residues within secretin with an alanine residue. A similar approach has been applied to the VPAC1 and VPAC2 receptors, and the glucagon-like peptide 1 receptor (17–21). These studies have generated consistent themes, as well as some receptor-specific observations.
Multiple secretin residues spread throughout the peptide ligand are important for binding and biological activity. This is consistent with previous studies exploring structure-activity relationships using various fragments and analogues of this hormone (40, 42–44). In these studies, binding is possible using a carboxyl-terminal fragment of secretin with truncation of up to six amino acids, but complementary addition of the amino-terminal residues to the carboxyl-terminal segment of this peptide is critical to establish high affinity binding and full agonist activity (40, 42–44). This theme is consistent for many members of this family (45–47).
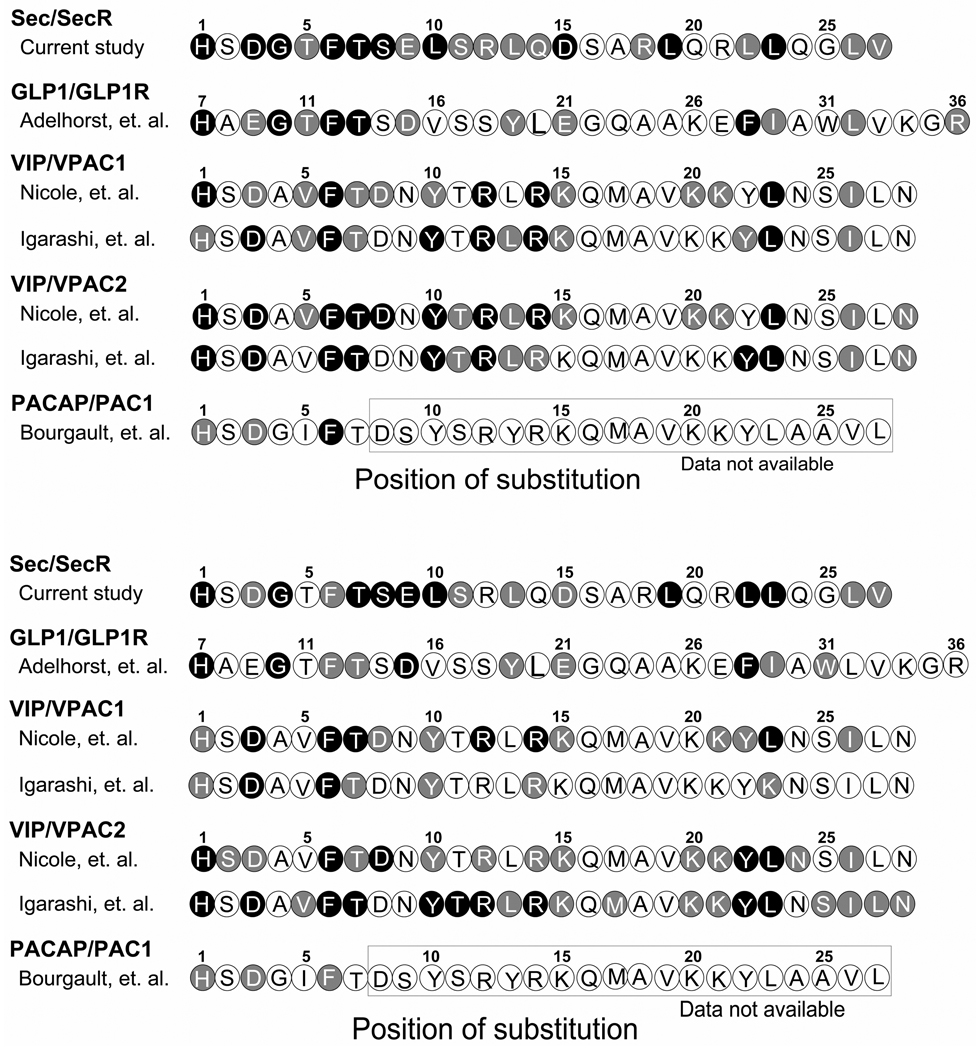
As expected, many of the residues currently found to be functionally important are within the amino-terminal region of this hormone, including His1, Asp3, Gly4, Phe6, Thr7, Ser8, and Leu10. His1 is conserved in several closely-related members of this family, and the current study confirms its importance for both secretin binding and biological activity, consistent with the previous studies (13, 48). This position has been previously demonstrated by photoaffinity labeling studies to be spatially approximated to the receptor residue Phe338 at the top of transmembrane segment six of the secretin receptor (13). The functional importance of this residue is also present in the ligands most closely related to secretin, including VIP, GLP1 and PACAP (Figure 8). Asp3 is conserved in most members and is critical for binding of secretin, VIP, GLP1, and PACAP, but this position is not critical for the biological activity of GLP1 (Figure 8). This residue has been demonstrated to be implicated in interaction with receptor residue Tyr128 in the first transmembrane domain and Lys173 in the first extracellular loop domain of the secretin receptor in mutagenesis studies (49, 50). Gly4 is functionally important for binding and biological activity only in secretin and GLP1, with the analogous residues present in VIP and PACAP shown to not be important for function at their receptors (Figure 8). The importance of Phe6 and Thr7 for secretin binding and biological activity is also conserved throughout the family (Figure 8), although Phe6 in glucose-dependent insulinotropic polypeptide appears to be less critical (51). The secretin Phe6 residue has been demonstrated to be spatially close to receptor residue Val4 in the distal amino-terminal end of the secretin receptor in photoaffinity labeling studies (52). Ser8 is important for secretin binding and biological activity, but not for GLP1 (Figure 8). The role of position 8 for VIP binding has been inconsistent, with one study (21) showing it is critical for both binding and biological activity, while another study failed to confirm this (19, 20). The importance of residue Leu10 in binding and biological activity is present in secretin and VIP, but not in GLP1 (Figure 8). Although alanine substitution of Leu10 in secretin significantly impairs its binding and biological activity, replacement with a tyrosine is well tolerated and this replacement has been extensively utilized to generate a secretin radioligand (22).
FIGURE 8.
Comparison of the roles of each secretin residue in binding and biological activity with that of other family B GPCR ligands. Top, role in binding affinity. Bottom, role in biological activity. Open circles, little importance (Ki or EC50 values are less than 10-fold of that of natural ligand). Grey circles, important (Ki or EC50 values are more than 10-fold but less than 100-fold of that of natural ligand). Black circles, critical (Ki or EC50 values are more than 100-fold of that of natural ligand). Sec, secretin; SecR, secretin receptor; GLP1R, GLP1 receptor.
Of note, the amino-terminal region of natural peptide ligands for family B GPCRs has been proposed as contributing a helix N-capping motif to several members of this family (53). The residues within secretin proposed as contributing to such a motif are Phe6, Thr7, and Leu10, all of which have substantial impact on both binding and biological activity of this peptide. It is not clear whether these residues contribute their effects directly by influencing binding or biological activity, or indirectly by stabilizing the helical structure of the middle and carboxyl terminus of the peptide.
The mid-region of secretin and family B GPCRs has also been shown to be important for function. Asp15 is in a position where it is an acidic residue in secretin and a basic residue in VIP and PACAP. This residue is the focus of a series of cross-chimeric studies (54). It is indeed critical for binding and biological activity for secretin, GLP1, and VIP (Figure 8). Most interesting is the non-conserved Leu19 since it appears to be critical for secretin binding and biological activity, but not to be important for other family members in which it has been studied (Figure 8). The importance of this residue is further supported by our inability to incorporate a photolabile benzoyl phenylalanine in this position for photoaffinity labeling studies, due to very low affinity binding (data not shown).
The importance of the carboxyl-terminal residue Leu23 in binding and biological activity is also conserved throughout the family (Figure 8). This residue has been demonstrated in photoaffinity labeling studies to be spatially close to receptor residue Arg21 within the amino terminus of the secretin receptor (55).
It was noteworthy that the alanine-replacement analysis of secretin differed from GLP1, VIP, and PACAP in having negative impact of modification of Leu19. For this reason, we applied molecular dynamics simulations to determine the effects of Leu19 on the stability of the structure. Results of our molecular dynamics simulations suggest that the distance between the peptide and the receptor and the solvent exposure of hydrophobic residues might contribute to the instability of the L19A mutant in the complex. Since the important residues in secretin are hydrophobic residues that are facing the binding cleft of the receptor, the van der Waals interactions, which are strongest at short separation distances, are important. In the simulation, while the L19A analogue of secretin makes contact with Leu10 in the secretin receptor, it does not establish close contacts with the next residue in the α-helix of the receptor that is facing the peptide, namely Val13, which on average was the closest point of contact in the 12-ns simulation for natural secretin, natural VIP, and the V19A VIP analogue. The short length of the alanine side chain might allow only close van der Waals interactions with Leu10 in the receptor, rather than the hydrophobic environment of both Leu10 and Val13 in the secretin receptor. The simulations seem to indicate that Leu19, which is a relatively large hydrophobic side chain, is maintaining closer contacts with the appropriate residues in the receptor and as a result is more stable than the secretin analogue. In addition, the change from Leu to Ala increased the solvent-accessible surface area of the hydrophobic residues in the carboxyl terminus of the peptide. Since hydrophobic residues prefer the hydrophobic environment (lower SASA), this increase might further contribute to the instability of the peptide analogue in the complex.
ACKNOWLEDGEMENTS
The authors thank M.L. Augustine for her technical assistance.
Footnotes
This work was supported by a grant from the National Institutes of Health (DK46577).
Abbreviations: CHO-SecR, Chinese hamster ovary cell line stably expressing the wild type rat secretin receptor; GLP1, gluacagon-like peptide 1; GPCR, G protein-coupled receptors; ICM, internal coordinate mechanics; MD, molecular dynamics; PACAP, pituitary adenylate cyclase-activating peptide; RMSD, root-mean-square deviation; SASA, solvent-accessible surface area; VIP, vasoactive intestinal polypeptide.
REFERENCES
- 1.Dong M, Miller LJ. Molecular pharmacology of the secretin receptor. Receptors Channels. 2002;8:189–200. [PubMed] [Google Scholar]
- 2.Mayo KE, Miller LJ, Bataille D, Dalle S, Goke B, Thorens B, Drucker DJ. International Union of Pharmacology. XXXV. The glucagon receptor family. Pharmacol. Rev. 2003;55:167–194. doi: 10.1124/pr.55.1.6. [DOI] [PubMed] [Google Scholar]
- 3.Grace CR, Perrin MH, DiGruccio MR, Miller CL, Rivier JE, Vale WW, Riek R. NMR structure and peptide hormone binding site of the first extracellular domain of a type B1 G protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12836–12841. doi: 10.1073/pnas.0404702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grace CR, Perrin MH, Gulyas J, Digruccio MR, Cantle JP, Rivier JE, Vale WW, Riek R. Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand. Proc. Natl. Acad. Sci. U.S.A. 2007;104:4858–4863. doi: 10.1073/pnas.0700682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parthier C, Kleinschmidt M, Neumann P, Rudolph R, Manhart S, Schlenzig D, Fanghanel J, Rahfeld JU, Demuth HU, Stubbs MT. Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13942–13947. doi: 10.1073/pnas.0706404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pioszak AA, Parker NR, Suino-Powell K, Xu HE. Molecular recognition of corticotropin-releasing factor by its G-protein-coupled receptor CRFR1. J. Biol. Chem. 2008;283:32900–32912. doi: 10.1074/jbc.M805749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pioszak AA, Xu HE. Molecular recognition of parathyroid hormone by its G protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5034–5039. doi: 10.1073/pnas.0801027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Runge S, Thogersen H, Madsen K, Lau J, Rudolph R. Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain. J. Biol. Chem. 2008;283:11340–11347. doi: 10.1074/jbc.M708740200. [DOI] [PubMed] [Google Scholar]
- 9.Sun C, Song D, Davis-Taber RA, Barrett LW, Scott VE, Richardson PL, Pereda-Lopez A, Uchic ME, Solomon LR, Lake MR, Walter KA, Hajduk PJ, Olejniczak ET. Solution structure and mutational analysis of pituitary adenylate cyclase-activating polypeptide binding to the extracellular domain of PAC1-RS. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7875–7880. doi: 10.1073/pnas.0611397104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ter Haar E, Koth CM, Abdul-Manan N, Swenson L, Coll JT, Lippke JA, Lepre CA, Garcia-Guzman M, Moore JM. Crystal structure of the ectodomain complex of the CGRP receptor, a class-B GPCR, reveals the site of drug antagonism. Structure. 2010;18:1083–1093. doi: 10.1016/j.str.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Underwood CR, Garibay P, Knudsen LB, Hastrup S, Peters GH, Rudolph R, Reedtz-Runge S. Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J. Biol. Chem. 2010;285:723–730. doi: 10.1074/jbc.M109.033829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bisello A, Adams AE, Mierke DF, Pellegrini M, Rosenblatt M, Suva LJ, Chorev M. Parathyroid hormone-receptor interactions identified directly by photocross-linking and molecular modeling studies. J. Biol. Chem. 1998;273:22498–22505. doi: 10.1074/jbc.273.35.22498. [DOI] [PubMed] [Google Scholar]
- 13.Dong M, Li Z, Pinon DI, Lybrand TP, Miller LJ. Spatial approximation between the amino terminus of a peptide agonist and the top of the sixth transmembrane segment of the secretin receptor. J. Biol. Chem. 2004;279:2894–2903. doi: 10.1074/jbc.M310407200. [DOI] [PubMed] [Google Scholar]
- 14.Dong M, Pinon DI, Cox RF, Miller LJ. Molecular approximation between a residue in the amino-terminal region of calcitonin and the third extracellular loop of the class B G protein-coupled calcitonin receptor. J. Biol. Chem. 2004;279:31177–31182. doi: 10.1074/jbc.M404113200. [DOI] [PubMed] [Google Scholar]
- 15.Parthier C, Reedtz-Runge S, Rudolph R, Stubbs MT. Passing the baton in class B GPCRs: peptide hormone activation via helix induction? Trends Biochem. Sci. 2009;34:303–310. doi: 10.1016/j.tibs.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Ulrich CD, 2nd, Holtmann M, Miller LJ. Secretin and vasoactive intestinal peptide receptors: members of a unique family of G protein-coupled receptors. Gastroenterology. 1998;114:382–397. doi: 10.1016/s0016-5085(98)70491-3. [DOI] [PubMed] [Google Scholar]
- 17.Adelhorst K, Hedegaard BB, Knudsen LB, Kirk O. Structure-activity studies of glucagon-like peptide-1. J. Biol. Chem. 1994;269:6275–6278. [PubMed] [Google Scholar]
- 18.Bourgault S, Vaudry D, Segalas-Milazzo I, Guilhaudis L, Couvineau A, Laburthe M, Vaudry H, Fournier A. Molecular and conformational determinants of pituitary adenylate cyclase-activating polypeptide (PACAP) for activation of the PAC1 receptor. J. Med. Chem. 2009;52:3308–3316. doi: 10.1021/jm900291j. [DOI] [PubMed] [Google Scholar]
- 19.Igarashi H, Ito T, Hou W, Mantey SA, Pradhan TK, Ulrich CD, 2nd, Hocart SJ, Coy DH, Jensen RT. Elucidation of vasoactive intestinal peptide pharmacophore for VPAC(1) receptors in human, rat, and guinea pig. J. Pharmacol. Exp. Ther. 2002;301:37–50. doi: 10.1124/jpet.301.1.37. [DOI] [PubMed] [Google Scholar]
- 20.Igarashi H, Ito T, Pradhan TK, Mantey SA, Hou W, Coy DH, Jensen RT. Elucidation of the vasoactive intestinal peptide pharmacophore for VPAC(2) receptors in human and rat and comparison to the pharmacophore for VPAC(1) receptors. J. Pharmacol. Exp. Ther. 2002;303:445–460. doi: 10.1124/jpet.102.038075. [DOI] [PubMed] [Google Scholar]
- 21.Nicole P, Lins L, Rouyer-Fessard C, Drouot C, Fulcrand P, Thomas A, Couvineau A, Martinez J, Brasseur R, Laburthe M. Identification of key residues for interaction of vasoactive intestinal peptide with human VPAC1 and VPAC2 receptors and development of a highly selective VPAC1 receptor agonist. Alanine scanning and molecular modeling of the peptide. J. Biol. Chem. 2000;275:24003–24012. doi: 10.1074/jbc.M002325200. [DOI] [PubMed] [Google Scholar]
- 22.Ulrich CD, 2nd, Pinon DI, Hadac EM, Holicky EL, Chang-Miller A, Gates LK, Miller LJ. Intrinsic photoaffinity labeling of native and recombinant rat pancreatic secretin receptors. Gastroenterology. 1993;105:1534–1543. doi: 10.1016/0016-5085(93)90162-6. [DOI] [PubMed] [Google Scholar]
- 23.Powers SP, Pinon DI, Miller LJ. Use of N,O-bis-Fmoc-D-Tyr-ONSu for introduction of an oxidative iodination site into cholecystokinin family peptides. Int. J. Pept. Protein Res. 1988;31:429–434. doi: 10.1111/j.1399-3011.1988.tb00899.x. [DOI] [PubMed] [Google Scholar]
- 24.Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 25.Momany F, McGuire R, Burgess A, HA S. Energy parameters in polypeptides. VII. Geometric parameters, partial atomic charges, nonbonded interactions, hydrogen bond interactions, and intrinsic torsional potentials for the naturally occurring amino acids. J. Phys. Chem. 1975;79:2361–2381. [Google Scholar]
- 26.Nemethy G, Gibson KD, Palmer KA, Yoon CN, Paterlini G, Zagari A, Rumsey S, Scheraga HA. Energy Parameters in Polypeptides. 10. Improved Geometrical Parameters and Nonbonded Interactions for Use in the Ecepp/3 Algorithm, with Application to Proline-Containing Peptides. J. Phys. Chem. 1992;96:6472–6484. [Google Scholar]
- 27.Nemethy G, Pottle M, Scheraga H. Energy parameters in polypeptides. 9. Updating of gemetrical parameters, nonbonded interactions and hydrogen bond interactions for the naturally occurring amino acids. J. Phys. Chem. 1983;87:1883–1887. [Google Scholar]
- 28.Wesson L, Eisenberg D. Atomic solvation parameters applied to molecular dynamics of proteins in solution. Protein Sci. 1992;1:227–235. doi: 10.1002/pro.5560010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bordner AJ, Abagyan R. Ab initio prediction of peptide-MHC binding geometry for diverse class I MHC allotypes. Proteins. 2006;63:512–526. doi: 10.1002/prot.20831. [DOI] [PubMed] [Google Scholar]
- 30.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 31.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J. Phys. Chem. B. 2001;105:6474–6487. [Google Scholar]
- 32.Berendsen HJC, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269–6271. [Google Scholar]
- 33.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 34.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 2009;52:7182–7190. [Google Scholar]
- 35.Hess B, Bekker H, Berendsen HJC, Fraaije J. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 36.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 37.Munson PJ, Rodbard D. Ligand: a versatile computerized approach for characterization of ligand-binding systems. Anal. Biochem. 1980;107:220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- 38.Holtmann MH, Ganguli S, Hadac EM, Dolu V, Miller LJ. Multiple extracellular loop domains contribute critical determinants for agonist binding and activation of the secretin receptor. J. Biol. Chem. 1996;271:14944–14949. doi: 10.1074/jbc.271.25.14944. [DOI] [PubMed] [Google Scholar]
- 39.Holtmann MH, Hadac EM, Miller LJ. Critical contributions of amino-terminal extracellular domains in agonist binding and activation of secretin and vasoactive intestinal polypeptide receptors. Studies of chimeric receptors. J. Biol. Chem. 1995;270:14394–14398. doi: 10.1074/jbc.270.24.14394. [DOI] [PubMed] [Google Scholar]
- 40.Vilardaga JP, Ciccarelli E, Dubeaux C, De Neef P, Bollen A, Robberecht P. Properties and regulation of the coupling to adenylate cyclase of secretin receptors stably transfected in Chinese hamster ovary cells. Mol. Pharmacol. 1994;45:1022–1028. [PubMed] [Google Scholar]
- 41.Dong M, Lam PC, Pinon DI, Orry A, Sexton PM, Abagyan R, Miller LJ. Secretin occupies a single protomer of the homodimeric secretin receptor complex: insights from photoaffinity labeling studies using dual sites of covalent attachment. J. Biol. Chem. 2010;285:9919–9931. doi: 10.1074/jbc.M109.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Izzo RS, Praissman M. Effect of N-terminal iodination on the biological, immunological and receptor binding properties of secretion. A role for the alpha-amino group of histidine in stabilizing hormone-receptor interactions. Int. J. Pept. Protein Res. 1984;23:292–299. [PubMed] [Google Scholar]
- 43.Konig W, Bickel M, Karch K, Teetz V, Uhmann R. Analogues and fragments of secretin. Peptides. 1984;5:189–193. doi: 10.1016/0196-9781(84)90205-5. [DOI] [PubMed] [Google Scholar]
- 44.Robberecht P, De Neef P, Waelbroeck M, Camus JC, Scemama JL, Fourmy D, Pradayrol L, Vaysse N, Christophe J. Secretin receptors in human pancreatic membranes. Pancreas. 1988;3:529–535. doi: 10.1097/00006676-198810000-00004. [DOI] [PubMed] [Google Scholar]
- 45.Chorev M. Parathyroid hormone 1 receptor: insights into structure and function. Receptors Channels. 2002;8:219–242. [PubMed] [Google Scholar]
- 46.Laburthe M, Couvineau A, Marie JC. VPAC receptors for VIP and PACAP. Receptors Channels. 2002;8:137–153. [PubMed] [Google Scholar]
- 47.Purdue BW, Tilakaratne N, Sexton PM. Molecular pharmacology of the calcitonin receptor. Receptors Channels. 2002;8:243–255. [PubMed] [Google Scholar]
- 48.Hefford MA, Kaplan H. Chemical properties of the histidine residue of secretin: evidence for a specific intramolecular interaction. Biochim. Biophys. Acta. 1989;998:267–270. doi: 10.1016/0167-4838(89)90283-5. [DOI] [PubMed] [Google Scholar]
- 49.Di Paolo E, Petry H, Moguilevsky N, Bollen A, De Neef P, Waelbroeck M, Robberecht P. Mutations of aromatic residues in the first transmembrane helix impair signalling by the secretin receptor. Receptors Channels. 1999;6:309–315. [PubMed] [Google Scholar]
- 50.Vilardaga JP, di Paolo E, de Neef P, Waelbroeck M, Bollen A, Robberecht P. Lysine 173 residue within the first exoloop of rat secretin receptor is involved in carboxylate moiety recognition of Asp 3 in secretin. Biochem. Biophys. Res. Commun. 1996;218:842–846. doi: 10.1006/bbrc.1996.0150. [DOI] [PubMed] [Google Scholar]
- 51.Alana I, Parker JC, Gault VA, Flatt PR, O'Harte FP, Malthouse JP, Hewage CM. NMR and alanine scan studies of glucose-dependent insulinotropic polypeptide in water. J. Biol. Chem. 2006;281:16370–16376. doi: 10.1074/jbc.M510414200. [DOI] [PubMed] [Google Scholar]
- 52.Dong M, Wang Y, Hadac EM, Pinon DI, Holicky E, Miller LJ. Identification of an interaction between residue 6 of the natural peptide ligand and a distinct residue within the amino-terminal tail of the secretin receptor. J. Biol. Chem. 1999;274:19161–19167. doi: 10.1074/jbc.274.27.19161. [DOI] [PubMed] [Google Scholar]
- 53.Neumann JM, Couvineau A, Murail S, Lacapere JJ, Jamin N, Laburthe M. Class-B GPCR activation: is ligand helix-capping the key? Trends Biochem. Sci. 2008;33:314–319. doi: 10.1016/j.tibs.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 54.Park CG, Ganguli SC, Pinon DI, Hadac EM, Miller LJ. Cross-chimeric analysis of selectivity of secretin and VPAC(1) receptor activation. J. Pharmacol. Exp. Ther. 2000;295:682–688. [PubMed] [Google Scholar]
- 55.Dong M, Lam PC, Gao F, Hosohata K, Pinon DI, Sexton PM, Abagyan R, Miller LJ. Molecular approximations between residues 21 and 23 of secretin and its receptor: Development of a model for peptide docking with the amino terminus of the secretin receptor. Mol. Pharmacol. 2007;72:280–290. doi: 10.1124/mol.107.035402. [DOI] [PubMed] [Google Scholar]