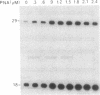
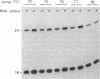
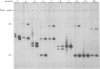
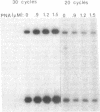
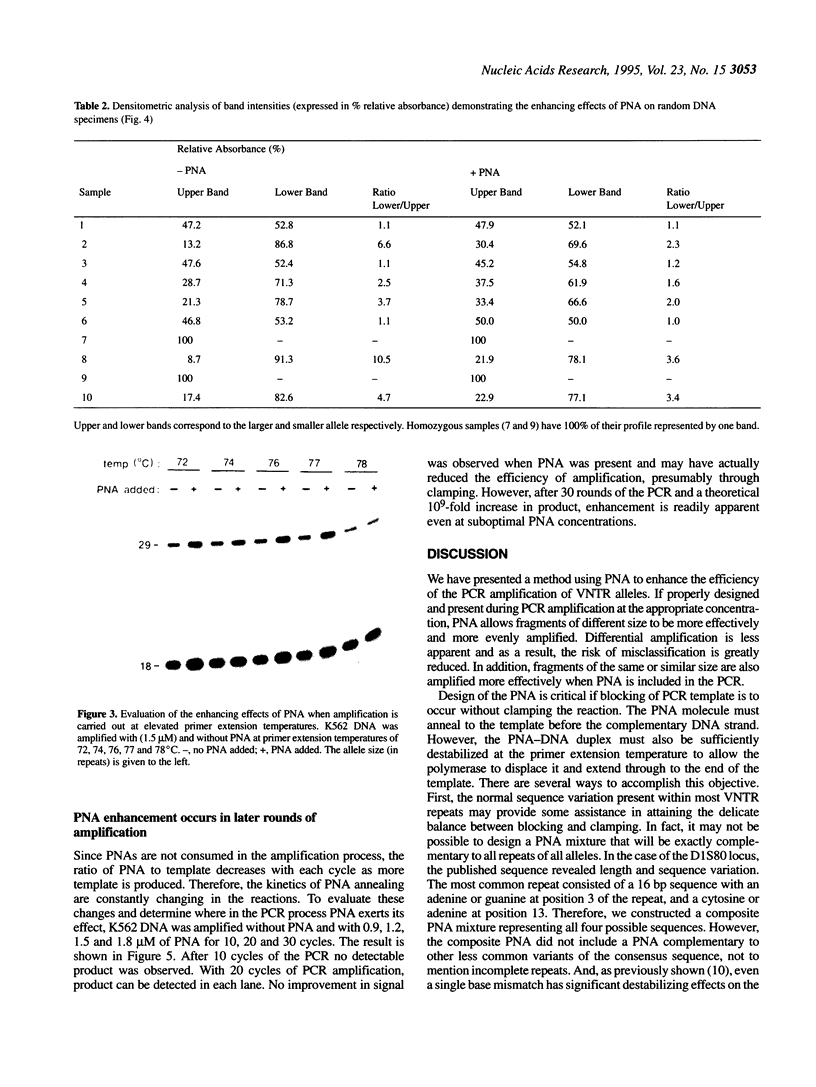
Abstract
Use of the polymerase chain reaction (PCR) to amplify variable numbers of tandem repeat (VNTR) loci has become widely used in genetic typing. Unfortunately, preferential amplification of small allelic products relative to large allelic products may result in incorrect or ambiguous typing in a heterozygous sample. The mechanism for preferential amplification has not been elucidated. Recently, PNA oligomers (peptide nucleic acids) have been used to detect single base mutations through PCR clamping. PNA is a DNA mimic that exhibits several unique hybridization characteristics. In this report we present a new application of PNA which exploits its unique properties to provide enhanced amplification. Rather than clamping the PCR, PNA is used to block the template making it unavailable for interstrand and intrastrand interactions while allowing polymerase to displace the PNA molecules and extend the primer to completion. Preferential amplification is reduced and overall efficiency is enhanced.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnes W. M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budowle B., Chakraborty R., Giusti A. M., Eisenberg A. J., Allen R. C. Analysis of the VNTR locus D1S80 by the PCR followed by high-resolution PAGE. Am J Hum Genet. 1991 Jan;48(1):137–144. [PMC free article] [PubMed] [Google Scholar]
- Cheng S., Fockler C., Barnes W. M., Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracopoli N. C., Meisler M. H. Mapping the human amylase gene cluster on the proximal short arm of chromosome 1 using a highly informative (CA)n repeat. Genomics. 1990 May;7(1):97–102. doi: 10.1016/0888-7543(90)90523-w. [DOI] [PubMed] [Google Scholar]
- Egholm M., Buchardt O., Christensen L., Behrens C., Freier S. M., Driver D. A., Berg R. H., Kim S. K., Norden B., Nielsen P. E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993 Oct 7;365(6446):566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- Hu G. DNA polymerase-catalyzed addition of nontemplated extra nucleotides to the 3' end of a DNA fragment. DNA Cell Biol. 1993 Oct;12(8):763–770. doi: 10.1089/dna.1993.12.763. [DOI] [PubMed] [Google Scholar]
- Kasai K., Nakamura Y., White R. Amplification of a variable number of tandem repeats (VNTR) locus (pMCT118) by the polymerase chain reaction (PCR) and its application to forensic science. J Forensic Sci. 1990 Sep;35(5):1196–1200. [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987 May;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Litt M., Luty J. A. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet. 1989 Mar;44(3):397–401. [PMC free article] [PubMed] [Google Scholar]
- Marton A., Delbecchi L., Bourgaux P. DNA nicking favors PCR recombination. Nucleic Acids Res. 1991 May 11;19(9):2423–2426. doi: 10.1093/nar/19.9.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhans A., Vartanian J. P., Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990 Apr 11;18(7):1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odelberg S. J., White R. A method for accurate amplification of polymorphic CA-repeat sequences. PCR Methods Appl. 1993 Aug;3(1):7–12. doi: 10.1101/gr.3.1.7. [DOI] [PubMed] [Google Scholar]
- Orum H., Nielsen P. E., Egholm M., Berg R. H., Buchardt O., Stanley C. Single base pair mutation analysis by PNA directed PCR clamping. Nucleic Acids Res. 1993 Nov 25;21(23):5332–5336. doi: 10.1093/nar/21.23.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena S. D., Chakraborty R. Paternity testing in the DNA era. Trends Genet. 1994 Jun;10(6):204–209. doi: 10.1016/0168-9525(94)90257-7. [DOI] [PubMed] [Google Scholar]
- Petersen M. B., Economou E. P., Slaugenhaupt S. A., Chakravarti A., Antonarakis S. E. Linkage analysis of the human HMG14 gene on chromosome 21 using a GT dinucleotide repeat as polymorphic marker. Genomics. 1990 May;7(1):136–138. doi: 10.1016/0888-7543(90)90531-x. [DOI] [PubMed] [Google Scholar]
- Sajantila A., Budowle B., Ström M., Johnsson V., Lukka M., Peltonen L., Ehnholm C. PCR amplification of alleles at the DIS80 locus: comparison of a Finnish and a North American Caucasian population sample, and forensic casework evaluation. Am J Hum Genet. 1992 Apr;50(4):816–825. [PMC free article] [PubMed] [Google Scholar]
- Tanaka J., Kasai M., Imamura M., Higa T., Kobayashi S., Hashino S., Sakurada K., Miyazaki T. Evaluation of mixed chimerism by two-step polymerase chain reaction amplification of hypervariable region MCT118 after allogeneic bone marrow transplantation. Ann Hematol. 1994 Apr;68(4):189–193. doi: 10.1007/BF01834365. [DOI] [PubMed] [Google Scholar]
- Tautz D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989 Aug 25;17(16):6463–6471. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P. S., Erlich H. A., Higuchi R. Preferential PCR amplification of alleles: mechanisms and solutions. PCR Methods Appl. 1992 May;1(4):241–250. doi: 10.1101/gr.1.4.241. [DOI] [PubMed] [Google Scholar]
- Wittung P., Nielsen P. E., Buchardt O., Egholm M., Nordén B. DNA-like double helix formed by peptide nucleic acid. Nature. 1994 Apr 7;368(6471):561–563. doi: 10.1038/368561a0. [DOI] [PubMed] [Google Scholar]