Abstract
Rates of antibiotic resistance in Pseudomonas aeruginosa are increasing worldwide. The multidrug-resistant (MDR) phenotype in P. aeruginosa could be mediated by several mechanisms including multidrug efflux systems, enzyme production, outer membrane protein (porin) loss and target mutations. Currently, no international consensus on the definition of multidrug resistance exists, making direct comparison of the literature difficult. Inappropriate empirical therapy has been associated with increased mortality in P. aeruginosa infections; delays in starting appropriate therapy may contribute to increased length of hospital stay and persistence of infection. In addition, worse clinical outcomes may be associated with MDR infections owing to limited effective antimicrobial options. This article aims to summarize the contemporary literature on patient outcomes following infections caused by drug-resistant P. aeruginosa. The impact of antimicrobial therapy on patient outcomes, mortality and morbidity; and the economic impact of MDR P. aeruginosa infections will be examined.
Keywords: antipseudomonal agents, infection, morbidity, mortality, outcomes, resistance, virulence
Pseudomonas aeruginosa is an important pathogen frequently implicated in healthcare-associated infections (HAIs), particularly in critically ill or immunocompromised patients [1,2]. It is a versatile pathogen with the ability to cause diverse infection types. Data from the National Nosocomial Infections Surveillance system from 1986–2003 reported P. aeruginosa as the second most common cause of pneumonia (18.1%), the third most common cause of urinary tract infection (16.3%) and the eighth most frequently isolated pathogen from the bloodstream (3.4%) [3]. While the overall proportion of infections caused by P. aeruginosa has remained stable during 1986–2003, the proportion of resistant isolates had alarming increases in 2003 compared with 1998 through 2002 [4]. Rates of resistance to imipenem, quinolones and third-generation cephalosporins increased by 15, 9 and 20%, respectively. Similarly, a national surveillance study of intensive care unit (ICU) patients from 1993 to 2002, reported a significant increase in multidrug-resistant (MDR; defined as resistance to at least three of four agents: imipenem, ceftazidime, ciprofloxacin and tobramycin) P. aeruginosa isolates [5].
The true prevalence of MDR P. aeruginosa is not well established, presumably for several reasons: first, there is considerable disagreement within the medical community as to the definition of multidrug resistance. Multidrug resistance is a heterogeneous phenotype, which could result from different (a combination of) resistance mechanism(s). A review of studies reporting on MDR and ‘pan-drug resistant’ P. aeruginosa infections revealed considerably different definitions used in the literature, ranging from resistance to a single antibiotic agent/class to resistance to all tested antibiotics [6]. In the majority of the published studies, multidrug resistance was defined as resistance to at least three drugs from a variety of antibiotic classes, mainly aminoglycosides, antipseudomonal penicillins, cephalosporins, carbapenems and fluoroquinolones. Although there have been attempts to establish a precise definition for multidrug resistance, there is currently no international consensus. Second, there is no international surveillance system specifically designed to track MDR organisms. The SENTRY antimicrobial surveillance program is designed to track antimicrobial resistance trends nationally and internationally. However, annual variations in geographic regions and participating centers limit the ability to track the true prevalence of MDR P. aeruginosa [7]. Data from our own institution revealed the prevalence rate of multidrug resistance (defined as resistance to all agents in at least three out of four classes: fluoroquinolones, aminoglycosides, carbapenems, antipseudomonal penicillins/cephalosporins) in P. aeruginosa bloodstream isolates to be approximately 10–17% from 2005 to 2007 [8]. Furthermore, diverse resistance mechanisms were found in these MDR isolates.
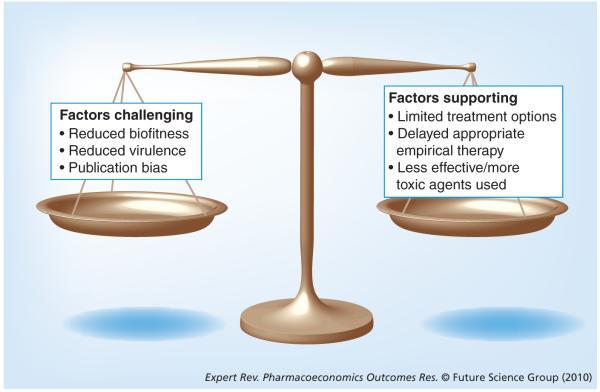
Broad spectrum antimicrobial resistance in MDR isolates significantly limits effective therapeutic options. Commonly, the agents of last resort for MDR organisms include the aminoglycosides and polymyxins. Recent articles have highlighted that these agents may or may not be as effective as first-line agents, but may also be associated with more significant adverse effects (i.e., nephrotoxicity, ototoxicity and neurotoxicity) [9-15]. This contributes (at least partially) to our difficulty in assessing whether MDR pathogens are truly associated with worse clinical outcomes (Figure 1). Available clinical data suggest that MDR P. aeruginosa infections may be associated with poorer outcomes; however, these investigations are often confounded by varied definitions of multidrug resistance and publication bias.
Figure 1.
Factors challenging and supporting the argument that multidrug-resistant pathogens are associated with worse clinical outcomes.
Resistance mechanisms & their effect on bacterial fitness
Multidrug resistance in P. aeruginosa results from the bacterium’s notable inherent antibiotic resistance, in addition to its ability to acquire and harbor diverse resistance determinants (Table 1) [8,16,17]. Low outer membrane permeability in combination with multidrug efflux systems account for its intrinsic mechanisms of resistance. The resistance-nodulation-cell division (RND) family of transporters is responsible for a significant portion of clinically relevant drug resistance among Gram-negative bacteria and facilitates active efflux of multiple antimicrobial substrates [18]. The broad specificity of the RND-type family allows many structurally unrelated molecules, such as biocides, detergents, dyes and organic solvents to act as substrates [19]. The majority of RND efflux systems are chromosomally encoded and are overexpressed following mutations in their respective negative regulator genes. At least seven RND efflux systems (e.g., MexAB-OprM, MexCD-OprJ, MexEF-OprN and MexXY-OprM) have been described in P. aeruginosa [20,21]. Broad antimicrobial resistance can result from overexpression of these pumps. For example, the MexAB-OprM system has the broadest substrate specificity and contributes to resistance to macrolides, aminoglycosides, sulfonamides, fluoroquinolones, tetracyclines and many β-lactams [20]. The loss of the outer membrane protein (porin) OprD, is associated with imipenem resistance and reduced susceptibility to meropenem.
Table 1.
Common mechanisms of antibiotic resistance in Pseudomonas aeruginosa. Isolates may express and/or harbor multiple mechanisms leading to a multidrug-resistant phenotype
| Type | Examples | Antibiotic classes/agents affected |
|---|---|---|
| Drug access | ||
| Overexpression of RND-type multidrug efflux pumps |
MexAB-OprM, MexCD-OprJ, MexEF-OprN | Macrolides, aminoglycosides, sulfonamides, fluoroquinolones, tetracyclines, β-lactams |
| Porin deletions | OprD | Imipenem, meropenem |
| Enzyme-based | ||
| β-lactamases | PSE-1, PSE-4 AmpC Metallo-β-lactamases |
Penicillins Third-generation cephalosporins, piperacillin Cephalosporins, carbapenems |
| Aminoglycoside-modifying enzymes | Acetyltransferases Nucleotidyltransferases Phosphotransferases |
Aminoglycosides |
| 16S rRNA methylases | rmtA, rmtB, armA genes | Aminoglycosides (high-level resistance) |
| Target mutations | ||
| Quinolone resistance-determining region | Encoded by gyrA, gyrB, parC, parE | Fluoroquinolones |
RND: Resistance-nodulation-cell division.
Additional resistance mechanisms in P. aeruginosa include enzyme production and target mutations. Expression of aminoglycoside-modifying enzymes (acetyltransferases, nucleotidyltransferases and phosphotransferases), mediating aminoglycoside resistance are common [22]. Most recently, methylation of 16S rRNA has emerged as a mechanism conferring high-level resistance against all commercially available aminoglycosides in P. aeruginosa [23]. Methylation of specific nucleotides within the A-site of 16S rRNA prevents binding of aminoglycosides to their target site. At least six distinct genes (rmtA, rmtB, rmtC, rmtD, armA and npmA) encoding their respective methylase enzymes have been reported to date [24]. The carriage of these genes on plasmids is probably responsible for their worldwide dissemination and transfer amongst various genera and species [25,26]. Furthermore, a wide variety of β-lactamases are produced by P. aeruginosa. PSE-1 and PSE-4 are those most frequently acquired but confer resistance only to the penicillins; activity of the antipseudomonal cephalosporins, carbapenems and monobactam is retained. The overexpression of chromosomally encoded cephalosporinase, AmpC, is prevalent in P. aeruginosa [27]. Antibiotic therapy may induce expression or select for stably derepressed mutants, resulting in resistance to ticarcillin, piperacillin and third-generation cephalosporins. Broad spectrum β-lactam resistance attributed to β-lactamases is generally due to the metallo-β-lactamases (MBLs): IMP, VIM, SPM and GIM [17]. These MBLs hydrolyze antipseudomonal cephalosporins and carbapenems effectively and their activity is not suppressed by the β-lactamase inhibitors that are currently commercially available. Lastly, mutations in DNA gyrase and topoisomerase IV (encoded by the gyrA and parC genes), mediate resistance to the fluoroquinolones [28].
Confounded by the inconsistent definition of multidrug resistance in P. aeruginosa, data regarding its impact on bacterial fitness are somewhat controversial. One generally accepted school of thought regarding antibiotic resistance and virulence is that overexpression/acquisition of resistance determinants comes at a physiological cost to the bacteria, resulting in a reduction in their fitness. This decrease in fitness, however, may be restored by compensatory mutations without the loss of resistance [29]. Conversely, ‘no-cost’ mutations in several bacterial strains have been reported [30,31]. Several in vitro studies in P. aeruginosa suggest virulence impairment in MDR strains. In one study comparing virulence factors of clinical strains, MDR strains had reduced expression of exoenzymes and pyocyanin, and slower growth rates were observed when compared with multidrug-susceptible (MDS) strains [32]. In a second study comparing MDR mutants overexpressing MexAB-OprM and MexCD-OprJ efflux systems to their isogenic parent strain, the mutants displayed impaired survival in water and on dry surfaces, and reduced virulence in a Caenorhabditis elegans nematode model [33]. It is poorly understood how these in vitro data relate to humans, as in vitro results seem to be highly dependent on growth conditions that can be standardized in the laboratory. Ongoing in vitro and in vivo investigations in our laboratory are also generating rather conflicting results [Abdelraouf K et al., Unpublished Data]. Consequently, inferring the in vivo fitness of bacteria based on in vitro studies may not always be accurate.
Risk factors for MDR P. aeruginosa infection
As the prevalence of MDR P. aeruginosa has increased over the past few decades, many studies have focused on the specific risk factors associated with the isolation of MDR P. aeruginosa. Again, direct comparison of these papers is difficult owing to the varied definitions used for multidrug resistance. When focusing on case–control studies using more stringent definitions of multidrug resistance (i.e., resistance to multiple classes of antipseudomonal antimicrobials), prior to use of antibiotics (duration and number of classes) [34-38], history of P. aeruginosa infection or colonization within the previous year [34], length of hospital stay [38], being bedridden or in the ICU [35], mechanical ventilation [38], malignant disease [35], and history of chronic obstructive pulmonary disease [34] have all been identified as independent risk factors for MDR P. aeruginosa infection. When looking specifically at antibiotics associated with the isolation of MDR P. aeruginosa, prior to receipt of carbapenems, broad-spectrum cephalosporins, aminoglycosides and fluoroquinolones have been identified as independent risk factors [34-37]. Hence, patients with MDR infections tend to be critically ill or have prolonged hospital stays and receive multiple antimicrobials (typically with antipseudomonal activity) prior to the isolation of MDR P. aeruginosa.
Impact of antimicrobial therapy
Importance of appropriate empirical therapy
Multiple studies have demonstrated that inappropriate empirical antimicrobial therapy is associated with poorer outcomes in patients with varied types of infections. Specifically, delays in appropriate therapy are linked to increased mortality, morbidity and hospital length of stay [39-43]. One rational strategy is to initiate therapy with broad-spectrum antimicrobials until full culture and susceptibility results are available [44,45]. Once the pathogen(s) and susceptibility profile are known, therapy may be de-escalated or streamlined to the most appropriate patient-specific regimen. However, treatment of infections caused by P. aeruginosa may be particularly difficult owing to the limited number of antipseudomonal agents available.
Four retrospective cohort studies have shown trends toward increased mortality after inappropriate empirical therapy for P. aeruginosa bloodstream infections (Table 2). In a retrospective cohort study of 136 patients, both inappropriate empirical and inappropriate definitive antibiotic treatment were independent risk factors for mortality (odds ratio [OR]: 4.61; 95% confidence interval [CI]: 1.18–18.09; p = 0.028 and OR: 11.68; 95% CI: 2.51–54.38; p = 0.002, respectively) [46]. Appropriate empirical therapy was defined as agent(s) administered within 24 h after blood cultures were obtained and found subsequently to be active in in vitro susceptibility testing. The mean duration of delay in appropriate antimicrobial therapy was 3.5 ± 1.28 days. After excluding those who did not receive appropriate definitive therapy, there was a trend toward a higher 30-day mortality rate compared with the appropriate empirical treatment group (43.4 vs 27.7%; p = 0.079). Similarly, a second retrospective cohort study of 305 patients also identified administration of inappropriate empirical antimicrobial treatment (defined as absence of an agent given with in vitro activity as determined by susceptibility testing) as an independent determinant of hospital mortality [47]. Hospital mortality rate was statistically higher for patients given inappropriate empirical antimicrobial treatment compared with appropriate empirical treatment (30.7 vs 17.8%; p = 0.018). Lastly, a retrospective cohort study of 167 patients evaluated the impact of empirical therapy in three distinct time windows: 8 h before the time the culture was obtained to 24 h afterward, between 24 and 48 h after the culture was obtained; and from 48 h after the culture was obtained to 4 h after antibiotic susceptibility results were available [48]. Empirical antibiotic therapy was considered appropriate if it included antibiotics to which the specific bacterial isolate exhibited in vitro susceptibility (with the exclusion of aztreonam and aminoglycoside monotherapy). The authors found a trend toward a protective effect of appropriate antibiotics at 24 h (OR: 0.93; 95% CI: 0.45–1.92), at 48 h (OR: 0.66; 95% CI: 0.29–1.49), and at the time susceptibility results were available (OR: 0.96; 95% CI: 0.31–2.93), after adjustment for mean modified acute physiology score at time of culture and age in the multivariable logistic regression analysis.
Table 2.
Studies demonstrating increased mortality after inappropriate empirical therapy for Pseudomonas aeruginosa infections
| Study design | Setting/location | Infection type |
Cases/controls (n) |
Outcome | Main fndings | Signifcance (p-value or 95% CI) |
Ref. |
|---|---|---|---|---|---|---|---|
| Retrospective cohort |
Tertiary care; Korea | BSI | 76/47 | 30-day mortality |
Cases: 43.4%; Controls: 27.7% |
p = 0.079 | [46] |
| Retrospective cohort |
Tertiary care; USA | BSI | 75/230 | Hospital mortality |
Cases: 30.7%; Controls: 17.8% |
p = 0.018 | [47] |
| Retrospective cohort |
Tertiary care; USA | BSI | 35/64† 35/79‡ 34/89§ |
Hospital mortality |
OR: 0.93† OR: 0.66‡ OR: 0.96§ |
0.45–1.92 0.29–1.49 0.31–2.93 |
[48] |
| Retrospective cohort |
Tertiary care; USA | BSI | 48/52 | 30-day mortality |
Cases: 43.8%; Controls: 19.2% |
p = 0.008 | [49] |
Appropriate antibiotics at 24 h.
Appropriate antibiotics at 48 h.
Appropriate antibiotics at time of susceptibility results: nonsurvivors compared with survivors.
BSI: Bloodstream infection; OR: Odds ratio.
In an effort to further investigate the length of delay in appropriate therapy associated with increased mortality, Lodise and colleagues analyzed a cohort of 100 patients with P. aeruginosa bloodstream infections [49]. Using classification and regression tree analysis, the most significant delay breakpoint to define the risk of 30-day mortality was 52 h. Patients with delay in appropriate therapy of over 52 h had a greater than twofold increase in 30-day mortality compared with those receiving timely appropriate therapy within 52 h (43.8 vs 19.2%; p = 0.008). Delayed appropriate therapy for 52 h was also independently associated with a 30-day mortality in the multivariate analysis. Antibiotic resistance to multiple (≥3) drug classes was the most important determinant of delayed appropriate therapy (adjusted odds ratio [aOR]: 4.6; 95% CI: 1.9–11.2; p = 0.001).
Collectively, these studies demonstrate the importance of timely initiation of appropriate empirical therapy in relation to mortality associated with P. aeruginosa bloodstream infections. In MDR infections, the likelihood of patients receiving appropriate empirical antibiotics is decreased owing to resistance to multiple antibiotic classes. Therefore, these data provide an additional mechanistic framework as to why MDR infections would be associated with worse clinical outcomes, regardless of their intrinsic biofitness/virulence.
Combination therapy versus monotherapy
Resistance to multiple antimicrobial classes further limits treatment options and decreases the likelihood of appropriate empirical therapy. It is not clearly established whether or not combination therapy for P. aeruginosa bacteremia is associated with more favorable outcomes than monotherapy [43,50]. A potential advantage of combination therapy is the higher probability that a MDR isolate will be susceptible to at least one agent in the combination regimen. In addition, synergistic combination of antimicrobials could also result in enhanced bacterial killing, which may be important for the reduction in bacterial burden at the onset of infection and limiting disease progression [51].
A retrospective cohort analysis of 115 patients who received empirical therapy for P. aeruginosa bacteremia sought to determine whether combination antipseudomonal therapy was superior to monotherapy [52]. Patients were categorized into six treatment groups: appropriate empirical combination therapy, appropriate empirical monotherapy, inappropriate empirical therapy, appropriate definitive combination therapy, appropriate definitive monotherapy and inappropriate definitive therapy. Empirical therapy was defined as treatment that included at least one antipseudomonal agent that was initiated no later than 24 h after the index positive blood culture. Monotherapy consisted of treatment with one antipseudomonal agent (excluding aminoglycoside monotherapy) while combination therapy consisted of piperacillin, ceftazidime, imipenem, or cefepime together with either an aminoglycoside (gentamicin or amikacin) or ciprofloxacin or an aminoglycoside with ciprofloxacin. The risk of death before receipt of the final susceptibility results was similar for the appropriate empirical monotherapy (adjusted hazard ratio [aHR]: 0.81; 95% CI: 0.31–2.1; p = 0.66) and inappropriate empirical therapy (aHR: 1.2; 95% CI: 0.29–5.2; p = 0.79) groups compared with that for the appropriate empirical combination therapy group. However, the risk of death after receipt of final susceptibility results varied according to empirical therapy. The results of a stratified Cox proportional hazard model revealed that patients in the appropriate empirical monotherapy group were 3.7-times more likely (95% CI: 1.0–14.1; p = 0.05) and patients in the inappropriate empirical therapy group were five-times more likely (95% CI: 1.2–20.4; p = 0.02) to have died within 30 days after receipt of the final susceptibility results than patients who received appropriate empirical combination therapy. Likewise, Micek et al. reported that inappropriate empirical antimicrobial therapy (prior to receipt of final susceptibility results) was statistically more likely to occur among patients receiving monotherapy compared with those receiving combination therapy (34.5 vs 20.6%; p = 0.011) [47]. However, there was no statistical difference in hospital mortality (p = 0.214) among patients who received treatment with a single β-lactam (n = 95; mortality: 12.5%), a single aminoglycoside (n = 29; mortality: 10.3%), the combination of a β-lactam and an aminoglycoside (n = 59; mortality: 22%) or ciprofloxacin alone (n = 15; mortality: 6.7%).
Several meta-analyses evaluating combination therapy versus monotherapy for Gram-negative infections have shown conflicting results [53-55]. These pooled results, often derived from subgroup analyses of pseudomonal infections, may be difficult to interpret owing to varied definitions used for ‘combination therapy’ patient populations, and varied study designs (i.e., retrospective versus prospective). The meta-analysis by Safdar and colleagues included five studies of patients with P. aeruginosa bacteremia. A significant mortality benefit was seen with the use of combination therapy (summary OR: 0.5; 95% CI: 0.3–0.79). However, the underlying patient populations in the five studies varied considerably [53]. Three studies included general medical patients (with a large proportion of immunocompromised patients), one study primarily included HIV-infected patients and one study consisted of patients with underlying malignancies. Conversely, a mortality benefit was not seen with the use of combination therapy in septic patients in the meta-analysis by Paul and colleagues [55]. They analyzed patients with P. aeruginosa infections from all anatomic locations and noted a lack of data to specifically study bacteremias. While meta-analyses can provide large numbers of patients with rare conditions/infections on which to draw conclusions, they are not without limitations and should not be solely relied on. Hence, it appears that more clinical data are needed to make an accurate assessment on the use of combination therapy.
Impact on patient outcomes
Mortality
As mentioned earlier, mortality rates following P. aeruginosa bloodstream infections are high, particularly in patients given inappropriate empirical therapy. Several studies have found increased mortality rates with MDR in comparison with MDS P. aeruginosa infections (Table 3). Cao et al. found a significantly higher crude mortality rate in patients with MDR infections (n = 44) compared with MDS infections (n = 68; 54.5 vs 16.2%; p < 0.05) [37]. The study patients had nosocomial infections caused by P. aeruginosa isolated from any clinical culture. The definition of multidrug resistance in this study included resistance to ceftazidime, ciprofloxacin, piperacillin and imipenem. Similarly, Aloush et al. conducted a retrospective matched-cohort study to examine the clinical outcomes of patients with P. aeruginosa isolated from any clinical culture during a 10-month period. They defined multidrug resistance as an organism resistant to all of the following agents: ceftazidime, cefepime, aztreonam, ciprofloxacin, piperacillin and gentamicin. They found a trend toward in-hospital mortality with MDR infection compared with matched control patients (21 vs 12%; OR: 2.3; p = 0.08), and isolation of MDR P. aeruginosa infection was significantly associated with mortality in a multivariate model (OR: 4.4; p = 0.04) [35].
Table 3.
Studies demonstrating negative clinical outcomes in drug-resistant Pseudomonas aeruginosa infections
| Study design | Setting/location | Infection type | Cases/ controls (n) |
Definition of resistance | Outcome | Main finding | Significance (p-value or 95% CI) |
Ref. |
|---|---|---|---|---|---|---|---|---|
| Retrospective cohort |
Tertiary; China | Nosocomial | 44/68 | R to ceftazidime, ciprofloxacin, piperacillin, imipenem |
Crude mortality |
Cases: 54.5%; Controls: 16.2% |
p < 0.05 | [37] |
| Retrospective matched cohort |
Tertiary; Israel | Isolation from any clinical culture |
82/82 | R to all: ceftazidime, cefepime, aztreonam, ciprofloxacin, piperacillin, gentamicin |
In-hospital mortality |
Cases: 21%; Controls: 12%; OR: 2.3 |
p = 0.08 | [35] |
| Retrospective cohort |
Tertiary; USA | FQRPA vs FQSPA; any clinical culture |
320/527 | Imipenem-R isolates were R to median of 4 (2–6) unique drug classes |
Mortality | aOR†: 1.79 | 1.13–2.84 p = 0.01 |
[56] |
| Prospective | Calgary Health Region; Canada |
MBL+ vs MBL− P aeruginosa from any clinical culture |
93/80 | MBL-positive strains R to ≥3 classes of antibiotics |
Crude case–fatality |
Cases: 25%; Controls: 13% RR: 1.98 |
1.00–3.9 p = 0.05 |
[57] |
| Retrospective cohort |
Tertiary; USA | All clinical cultures | 135/719 | Imipenem-R isolates significantly more R to 7 other antimicrobials |
In-hospital mortality |
Cases: 31.3%; Controls: 16.7% RR: 1.86 |
1.38–2.51 p< 0.001 |
[59] |
| Retrospective cohort |
Tertiary and community; USA |
All clinical cultures | 253/2289 | Imipenem-R isolates | In-hospital mortality |
Cases: 17.4%; Controls: 13.4% RR: 1.39 |
1.09–1.76 p = 0.01 |
[60] |
| Case–control | Tertiary; Japan | All clinical cultures | 69/247 |
b/aIMP‡-positive isolates R to 3 classes of antibiotics |
Infection- related mortality |
Cases: 5.8%; Controls: 1.2% OR: 5.00 |
1.09–22.9 p = 0.02 |
[61] |
Imipenem-resistant isolates, in a multivariable analysis.
b/aIMP, IMP-type metallo-β-lactamase.
aOR: Adjusted odds ratio; CI: Confidence interval: FQRPA: Fluoroquinolone-resistant P. aeruginosa; FQSPA: Fluoroquinolone-susceptible P. aeruginosa; MBL: Metallo-β-lactamase; OR: Odds ratio; R: Resistant; RR: Relative risk.
While the following studies do not specifically address patients having MDR infections, the recovered isolates were commonly resistant to multiple antipseudomonal agents. As a result, these isolates could be inferred to exhibit a MDR phenotype. In a study comparing patients with fluoroquinolone-susceptible (n = 527) P. aeruginosa infections to those with fluoroquinolone-resistant (n = 320) infections, Gasink et al. found imipenem resistance to be the only factor significantly associated with mortality (aOR: 1.79; 95% CI: 1.13–2.84; p = 0.01) [56]. Interestingly, 42.6% of these imipenem-resistant isolates were resistant to five or more unique classes (i.e., aminoglycosides, carbapenems, piperacillin, aztreonam and antipseudomonal cephalosporins) of drugs, implying a MDR phenotype. A limitation of this study was that the isolates were not determined to be true pathogens versus colonizers; this could impact outcomes as critically ill patients who have been hospitalized (and/or ventilated) for extended periods of time are often colonized with resistant organisms. In a second study, Laupland and colleagues studied patients treated within the Calgary Health Region with imipenem-resistant P. aeruginosa infections [57]. PCR was used to identify isolates producing MBLs, which are responsible for resistance to the carbapenems and a variety of penicillins and cephalosporins [58]. They found a significantly higher crude case–fatality rate for patients infected with MBL-producing strains compared with that in non-MBL-producing strains (23 out of 93 [25%] vs 10 out of 80 [13%] deaths; relative risk [RR]: 1.98; 95% CI: 1.00–3.90; p = 0.05). Again, the majority (87 out of 98 [89%]) of these MBL-producing strains were resistant to three classes or more of antibiotics in comparison to only 7% in non-MBL-producing strains (p < 0.0001). Two similar case–control studies conducted by the same group found mortality to be significantly higher among patients with imipenem-resistant P. aeruginosa infection/colonization than patients with imipenem-susceptible infection/colonization. In the first single-center study, patients with any clinical isolate positive for P. aeruginosa were evaluated. Mortality was higher among case patients (31.1 vs 16.7%; RR: 1.86; 95% CI: 1.38–2.51; p < 0.001) compared with control patients [59]. Infection with an imipenem-resistant isolate remained significantly associated with mortality in a multivariable analysis adjusted for confounders (aOR: 1.94; 95% CI: 1.22–3.10; p = 0.005). Similar to the previous studies, the imipenem-resistant isolates had significantly higher resistance rates to other tested antimicrobials, including amikacin, aztreonam, cefepime, ciprofloxacin, gentamicin, levofloxacin and piperacillin (p < 0.001 for all comparisons). In the second two-center study by the same group, mortality was found to be significantly higher in patients with imipenem-resistant isolates compared with imipenem-susceptible isolates in the bivariate analysis (17.4 vs 13.4%; RR: 1.39; 95% CI: 1.09–1.76; p = 0.01) [60]. However, after controlling for confounders in the multi variable analysis, an independent association with mortality remained only for patients with imipenem-resistant isolates collected from the bloodstream (aOR: 5.43; 95% CI: 1.72–17.10; p = 0.004). This second study, however, did not address resistance to other tested antimicrobials. Lastly, Hirakata and colleagues conducted a case–control study of 69 inpatients with blaIMP-positive P. aeruginosa and 247 control patients with blaIMP-negative P. aeruginosa [61]. They found more frequent infection-related death among patients with blaIMP-positive P. aeruginosa isolates compared with the control patients (5.8 vs 1.2%; OR: 5.00; 95% CI: 1.09–22.9; p = 0.02) [61]. Similar to the previous studies, blaIMP-positive isolates were more likely to be resistant to three classes of antibiotics than blaIMP-negative isolates (p = 0.01), also implying a MDR phenotype was exhibited.
It should be noted that common limitations in these studies include retrospective study design, possible bias from unadjusted confounding variables, such as baseline severity of illness/comorbidities, and different timing of therapy initiation. The clonality of the isolates recovered was also not routinely investigated. Addressing as many of these limitations as possible, a clinical investigation is currently being undertaken by our research group.
Morbidity
Multidrug-resistant P. aeruginosa infection not only could increase mortality, but it may also be associated with increased patient morbidity. Aloush et al. demonstrated that isolation of MDR P. aeruginosa was associated with a higher incidence of surgery as compared with controls (27 vs 16%; OR: 2.5; p = 0.05) [35]. The surgery objective was often to remove the source of infection (i.e., debridement, amputation and graft removal), when otherwise not controlled with medical therapy. In addition, patients with MDR infection in this study also had an increased number of invasive procedures (i.e., bronchoscopy, tracheostomy or catheter implantation; 38 vs 11%; OR: 5.4; p = 0.001). When patients were classified into groups based upon their destination after discharge (home, rehabilitation center or chronic care facility), more patients with MDR infection were discharged to a chronic care facility (55 vs 24%; OR: 6; p = 0.01) and were lacking full activity at discharge compared with matched controls (59 vs 34%; OR: 6.7; p = 0.002).
Length of hospital stay is another marker frequently used for morbidity [62]. Aloush et al. found isolation of MDR P. aeruginosa to be associated with increased length of stay (p = 0.001) when compared with matched controls [35]. Similarly, Micek et al. found that patients treated with inappropriate empirical therapy had a statistically longer length of stay than those who received appropriate empirical treatment (41.4 ± 47.4 [mean ± standard deviation] days vs 23.9 ± 25.2 days; p = 0.006) [47].
Microbiological outcomes, such as persistence of infection and emergence of resistance during antibiotic therapy, have been shown to negatively affect patient outcomes. In a study of bacteremic patients with P. aeruginosa, patients whose isolates overexpressed AmpC (n = 21), a class C β-lactamase mediating resistance to multiple antipseudomonal penicillins and cephalosporins, were compared with pan-susceptible (wild-type) control patients (n = 33) [63]. In a multivariate analysis, AmpC overexpression was a significant predictor of persistent disease (OR: 12.2; 95% CI: 1.7–87.7; p = 0.013). Persistent disease was defined as a new positive culture for P. aeruginosa at least 24 h after the collection of the index culture. While these patients were not specifically infected with a MDR strain, resistance to multiple antimicrobials was common in the AmpC overexpressed group. Carmeli and colleagues demonstrated that emergence of resistance (defined as subsequent detection of P. aeruginosa with at least a fourfold increase in minimum inhibitory concentration [MIC] relative to baseline) had significant effects on both mortality (RR: 3.3; p = 0.01) and length of stay (multiplicative effect: 1.5; p = 0.005) [64]. Their study population of patients with confirmed Pseudomonas infection using the CDC definitions for infection, had P. aeruginosa recovered from any clinical culture and were hospitalized for at least 2 days. They also estimated that emergence of resistance was associated with an average adjusted increase of 5.7 days in length of hospital stay.
In summary, the current literature demonstrates that patients infected with resistant P. aeruginosa commonly have increased mortality and morbidity. While direct comparison of these studies may be difficult owing to varied study designs and definitions of resistance, the overall significance of these data is apparent.
Economic impact
Literature investigating the economic impact of HAIs and attributable costs of antibiotic-resistant infections appears to be on the rise [65]. This may be due, in part, to the current era of increasing resistance rates and lack of novel antimicrobials in the development stages [66]. Economic data reporting on the burden of Gram-positive resistance are more extensive than Gram-negative resistance; however, the association with increased hospital costs and lengths of stay is evident [67-71].
Few studies have attempted to quantify the direct costs attributed to MDR P. aeruginosa infections and increased costs are not consistently demonstrated. Again, the difficulties in direct comparison of these studies include varied definitions of resistance and economic evaluation methods. Harris and colleagues published a case series of 22 patients with MDR P. aeruginosa recovered from any clinical culture hospitalized from August 1994 to December 1997 [72]. They classified a MDR isolate as one with intermediate susceptibility or resistance to ceftazidime, ciprofloxacin, imipenem and piperacillin. The mean cost of hospitalization during which the first MDR isolate was cultured was US$54,081 (mean length of stay = 18 days) compared with a mean of US$22,116 cost of hospitalization for patients with susceptible P. aeruginosa infections. In total, 17 out of 22 patients required surgery to treat their infection (five required amputations), which could increase both hospital charges and length of hospital stay. In the cohort of patients with fluoroquinolone-resistant P. aeruginosa infections, median hospital charges were significantly higher compared with patients with fluoroquinolone-susceptible infection (US$62,325 [inter-quartile range: US$22,129–188,979] and US$48,733 [US$18,760–124,829], respectively; p = 0.008) [56]. Although these patients had fluoroquinolone-resistant isolates, cross-resistance in P. aeruginosa to other antimicrobial classes was common [73]. In the third study of patients with imipenem-resistant P. aeruginosa isolates, median hospital costs (inter-quartile range) were US$81,330 (US$28,549–228,174) for case patients compared with US$48,381 (US$19,148–131,144) for control patients (p < 0.001) [59]. Increased hospitalization costs were at least partly owing to increased lengths of stay for those patients with imipenem-resistant isolates (15.5 median days vs 9 days; p = 0.02). The last study published in 1999 was a cohort of 489 hospitalized patients with clinical cultures of P. aeruginosa [64]. Hospital charges were studied in 309 of these patients admitted between April 1995 and July 1996. A total of 217 patients had a baseline isolate susceptible to all four study agents (piperacillin, ceftazidime, ciprofloxacin and imipenem), 92 patients had a baseline isolate resistant to at least one study agent, and 17 had emergence of resistance (defined as subsequent detection of P. aeruginosa with at least a fourfold increase in MIC relative to baseline). Neither resistance at baseline nor emergence of resistance was associated with a significant increase in daily hospital charges (multiplicative effects, 1.04 and 1.1, respectively; p = 0.41 and p = 0.43). The adjusted effects of resistance at baseline and emergence of resistance were similar for daily charges in a multivariate model (RR: 1.0 and 1.1, respectively; p = 0.41 and p = 0.43). The authors also estimated the effect of emergence of resistance by combining daily charge and length of stay analyses. Using a matched cohort study design of 15 patients in whom resistance emerged, the cumulative hospital charge was US$7340 higher than that for matched controls (p = 0.14). Although these authors did not find a significant difference in hospital charges, their definition of a resistant isolate was resistance to only one agent. Had they used a more stringent definition of resistance (i.e., to all four antipseudomonal agents), a more significant difference in charges might have been found.
Although the existing literature demonstrates increased costs owing to resistant Gram-negative infections, data focusing on P. aeruginosa are limited and somewhat inconsistent. Well-designed studies are needed to fully appreciate the direct cost burden associated with MDR P. aeruginosa infections.
Expert commentary
The true impact of MDR P. aeruginosa infection on patient outcomes remains controversial. The conventional notion that MDR pathogens are associated with reduced biofitness and/or virulence (and thus unlikely to negatively influence patient outcomes) has been challenged. Available clinical data suggest that MDR P. aeruginosa infections may be associated with worse outcomes (mortality, morbidity, requirement for surgical inter vention, prolonged length of hospital stay and so on). However, these clinical investigations are often confounded by inconsistency in the definition of MDR pathogens, variable mechanism(s) of resistance, retrospective study design and publication bias. In addition to intrinsic biofitness and/or virulence, an additional critical factor to be considered is the appropriateness of empirical therapy, which may be difficult to adjust clinically. Despite these limitations, we may have to make an educated assessment based on such ‘best-available’ data. Well-designed prospective clinical studies addressing these drawbacks are unlikely to be performed in view of ethical concerns. Molecular investigations may be useful to delineate/de-couple the association of various mechanism(s) of resistance and expression of virulence factors. Appropriately designed animal studies (in experimental therapeutics) may also be helpful in bridging the knowledge gap in clinical scenarios where there could be ethical concerns (i.e., exact infection time before treatment, identical baseline inoculum and so on).
Five-year view
As the prevalence rate of multidrug resistance continues to rise and spread worldwide, more frequent outbreaks of MDR infection are anticipated. Crude hospital mortality of patients with infections (whether they can be attributed to multidrug resistance to a scientific certainty or not) is expected to increase. Unless new and effective antimicrobial agents are developed for clinical use in time, we are at risk of returning to the preantibiotic era. In such difficult times, there is an urgent need for the scientific/medical community to unite and devise effective treatment strategies, preferably via interdisciplinary collaborations.
Key issues.
Currently, no international consensus for the definition of multidrug resistance exists, making direct comparison of literature very difficult.
Multidrug efflux systems, enzyme production, outer membrane protein loss and target mutations all play a role in conferring multidrug resistance in Pseudomonas aeruginosa.
Risk factors for multidrug-resistant (MDR) infections include prior use of antibiotics, history of P. aeruginosa infection or colonization within the previous year, length of hospital stay, being bedridden or in the intensive care unit, mechanical ventilation, malignant disease and history of chronic obstructive pulmonary disease.
Inappropriate empirical therapy has been shown to increase mortality in P. aeruginosa infection. Worse clinical outcomes may be associated with MDR infections owing to limited effective antimicrobial options. These agents (polymyxins/aminoglycosides) may be associated with severe adverse effects.
Current studies are somewhat inconsistent in demonstrating increased costs caused by resistant P. aeruginosa infections. More well-designed studies are needed to assess the direct cost burden associated with MDR infections.
Acknowledgments
Financial & competing interest disclosure
Vincent H Tam has received unrestricted research grants from AstraZeneca, Merck, Achaogen and is on the speakers’ bureau of Merck. Vincent Tam has also received funding from the NIH (grant number 1R15AI089671-01) The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Bodey GP, Jadeja L, Elting L. Pseudomonas bacteremia. Retrospective analysis of 410 episodes. Arch. Intern. Med. 1985;145(9):1621–1629. doi: 10.1001/archinte.145.9.1621. [DOI] [PubMed] [Google Scholar]
- 2.Chatzinikolaou I, Abi-Said D, Bodey GP, Rolston KV, Tarrand JJ, Samonis G. Recent experience with Pseudomonas aeruginosa bacteremia in patients with cancer: retrospective analysis of 245 episodes. Arch. Intern. Med. 2000;160(4):501–509. doi: 10.1001/archinte.160.4.501. [DOI] [PubMed] [Google Scholar]
- 3.Gaynes R, Edwards JR. Overview of nosocomial infections caused by Gram-negative bacilli. Clin. Infect. Dis. 2005;41(6):848–854. doi: 10.1086/432803. •• Data from National Nosocomial Infections Surveillance System (1986–2003) showing trends in nosocomial infections and resistance rates in Gram-negative organisms.
- 4.National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control. 2004;32(8):470–485. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- 5.Obritsch MD, Fish DN, MacLaren R, Jung R. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob. Agents Chemother. 2004;48(12):4606–4610. doi: 10.1128/AAC.48.12.4606-4610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falagas ME, Koletsi PK, Bliziotis IA. The diversity of definitions of multidrug-resistant (MDR) and pandrug-resistant (PDR) Acinetobacter baumannii and Pseudomonas aeruginosa. J. Med. Microbiol. 2006;55(Pt 12):1619–1629. doi: 10.1099/jmm.0.46747-0. • Review of various definitions of multidrug resistance used throughout the literature.
- 7.Gales AC, Jones RN, Turnidge J, Rennie R, Ramphal R. Characterization of Pseudomonas aeruginosa isolates: occurrence rates, antimicrobial susceptibility patterns, and molecular typing in the global SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin. Infect. Dis. 2001;32(Suppl. 2):S146–S155. doi: 10.1086/320186. [DOI] [PubMed] [Google Scholar]
- 8.Tam VH, Chang KT, Abdelraouf K, et al. Prevalence, resistance mechanisms, and susceptibility of multidrug-resistant bloodstream isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2010;54(3):1160–1164. doi: 10.1128/AAC.01446-09. • Explores the trend and molecular mechanisms responsible for the multidrug-resistant phenotype among bloodstream isolates.
- 9.Hartzell JD, Neff R, Ake J, et al. Nephrotoxicity associated with intravenous colistin (colistimethate sodium) treatment at a tertiary care medical center. Clin. Infect. Dis. 2009;48(12):1724–1728. doi: 10.1086/599225. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Lee KH, Yoo S, Pai H. Clinical characteristics and risk factors of colistin-induced nephrotoxicity. Int. J. Antimicrob. Agents. 2009;34(5):434–438. doi: 10.1016/j.ijantimicag.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 11.Falagas ME, Kasiakou SK. Toxicity of polymyxins: a systematic review of the evidence from old and recent studies. Crit. Care. 2006;10(1):R27. doi: 10.1186/cc3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosgrove SE, Vigliani GA, Fowler VG, Jr, et al. Initial low-dose gentamicin for Staphylococcus aureus bacteremia and endocarditis is nephrotoxic. Clin. Infect. Dis. 2009;48(6):713–721. doi: 10.1086/597031. [DOI] [PubMed] [Google Scholar]
- 13.Zavascki AP, Goldani LZ, Li J, Nation RL. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 2007;60(6):1206–1215. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 14.Yuan Z, Tam VH. Polymyxin B: a new strategy for multidrug-resistant Gram-negative organisms. Expert Opin. Investig. Drugs. 2008;17(5):661–668. doi: 10.1517/13543784.17.5.661. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Nation RL, Turnidge JD, et al. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 2006;6(9):589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 16.Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 2002;34(5):634–640. doi: 10.1086/338782. •• Review highlighting resistance mechanisms in Pseudomonas aeruginosa.
- 17.Bonomo RA, Szabo D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 2006;43(Suppl. 2):S49–S56. doi: 10.1086/504477. [DOI] [PubMed] [Google Scholar]
- 18.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64(2):159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 19.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009;69(12):1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li XZ, Barre N, Poole K. Influence of the MexA-MexB-oprM multidrug efflux system on expression of the MexC-MexD-oprJ and MexE-MexF-oprN multidrug efflux systems in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2000;46(6):885–893. doi: 10.1093/jac/46.6.885. [DOI] [PubMed] [Google Scholar]
- 21.Poole K. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 2004;10(1):12–26. doi: 10.1111/j.1469-0691.2004.00763.x. [DOI] [PubMed] [Google Scholar]
- 22.Kotra LP, Haddad J, Mobashery S. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 2000;44(12):3249–3256. doi: 10.1128/aac.44.12.3249-3256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 2007;45(1):88–94. doi: 10.1086/518605. [DOI] [PubMed] [Google Scholar]
- 24.Doi Y, Wachino J, Arakawa Y. Nomenclature of plasmid-mediated 16S rRNA methylases responsible for panaminoglycoside resistance. Antimicrob. Agents Chemother. 2008;52(6):2287–2288. doi: 10.1128/AAC.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galimand M, Sabtcheva S, Courvalin P, Lambert T. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob. Agents Chemother. 2005;49(7):2949–2953. doi: 10.1128/AAC.49.7.2949-2953.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamane K, Wachino J, Doi Y, Kurokawa H, Arakawa Y. Global spread of multiple aminoglycoside resistance genes. Emerg. Infect. Dis. 2005;11(6):951–953. doi: 10.3201/eid1106.040924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tam VH, Schilling AN, LaRocco MT, et al. Prevalence of AmpC over-expression in bloodstream isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 2007;13(4):413–418. doi: 10.1111/j.1469-0691.2006.01674.x. [DOI] [PubMed] [Google Scholar]
- 28.Jalal S, Ciofu O, Hoiby N, Gotoh N, Wretlind B. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 2000;44(3):710–712. doi: 10.1128/aac.44.3.710-712.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersson DI. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr. Opin. Microbiol. 2006;9(5):461–465. doi: 10.1016/j.mib.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Ramadhan AA, Hegedus E. Survivability of vancomycin resistant enterococci and fitness cost of vancomycin resistance acquisition. J. Clin. Pathol. 2005;58(7):744–746. doi: 10.1136/jcp.2004.024091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Criswell D, Tobiason VL, Lodmell JS, Samuels DS. Mutations conferring aminoglycoside and spectinomycin resistance in Borrelia burgdorferi. Antimicrob. Agents Chemother. 2006;50(2):445–452. doi: 10.1128/AAC.50.2.445-452.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deptula A, Gospodarek E. Reduced expression of virulence factors in multidrug-resistant Pseudomonas aeruginosa strains. Arch. Microbiol. 2009;192(1):79–84. doi: 10.1007/s00203-009-0528-1. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez P, Linares JF, Ruiz-Diez B, et al. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 2002;50(5):657–664. doi: 10.1093/jac/dkf185. [DOI] [PubMed] [Google Scholar]
- 34.Ohmagari N, Hanna H, Graviss L, et al. Risk factors for infections with multidrug-resistant Pseudomonas aeruginosa in patients with cancer. Cancer. 2005;104(1):205–212. doi: 10.1002/cncr.21115. [DOI] [PubMed] [Google Scholar]
- 35.Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob. Agents Chemother. 2006;50(1):43–48. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paramythiotou E, Lucet JC, Timsit JF, et al. Acquisition of multidrug-resistant Pseudomonas aeruginosa in patients in intensive care units: role of antibiotics with antipseudomonal activity. Clin. Infect. Dis. 2004;38(5):670–677. doi: 10.1086/381550. [DOI] [PubMed] [Google Scholar]
- 37.Cao B, Wang H, Sun H, Zhu Y, Chen M. Risk factors and clinical outcomes of nosocomial multi-drug resistant Pseudomonas aeruginosa infections. J. Hosp. Infect. 2004;57(2):112–118. doi: 10.1016/j.jhin.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 38.Arruda EA, Marinho IS, Boulos M, et al. Nosocomial infections caused by multiresistant Pseudomonas aeruginosa. Infect. Control Hosp. Epidemiol. 1999;20(9):620–623. doi: 10.1086/501683. [DOI] [PubMed] [Google Scholar]
- 39.Kumar A, Ellis P, Arabi Y, et al. Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. Chest. 2009;136(5):1237–1248. doi: 10.1378/chest.09-0087. [DOI] [PubMed] [Google Scholar]
- 40.Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006;34(6):1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 41.Kang CI, Kim SH, Park WB, et al. Bloodstream infections caused by antibiotic-resistant Gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob. Agents Chemother. 2005;49(2):760–766. doi: 10.1128/AAC.49.2.760-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fraser A, Paul M, Almanasreh N, et al. Benefit of appropriate empirical antibiotic treatment: thirty-day mortality and duration of hospital stay. Am. J. Med. 2006;119(11):970–976. doi: 10.1016/j.amjmed.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 43.Leibovici L, Shraga I, Drucker M, Konigsberger H, Samra Z, Pitlik SD. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J. Intern. Med. 1998;244(5):379–386. doi: 10.1046/j.1365-2796.1998.00379.x. [DOI] [PubMed] [Google Scholar]
- 44.Kollef MH. Broad-spectrum antimicrobials and the treatment of serious bacterial infections: getting it right up front. Clin. Infect. Dis. 2008;47(Suppl. 1):S3–S13. doi: 10.1086/590061. [DOI] [PubMed] [Google Scholar]
- 45.Bassetti M, Righi E, Viscoli C. Pseudomonas aeruginosa serious infections: mono or combination antimicrobial therapy? Curr. Med. Chem. 2008;15(5):517–522. doi: 10.2174/092986708783503186. [DOI] [PubMed] [Google Scholar]
- 46.Kang CI, Kim SH, Kim HB, et al. Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin. Infect. Dis. 2003;37(6):745–751. doi: 10.1086/377200. [DOI] [PubMed] [Google Scholar]
- 47.Micek ST, Lloyd AE, Ritchie DJ, Reichley RM, Fraser VJ, Kollef MH. Pseudomonas aeruginosa bloodstream infection: importance of appropriate initial antimicrobial treatment. Antimicrob. Agents Chemother. 2005;49(4):1306–1311. doi: 10.1128/AAC.49.4.1306-1311.2005. •• Article demonstrating statistically significant mortality increases following inappropriate empirical therapy.
- 48.Osih RB, McGregor JC, Rich SE, et al. Impact of empiric antibiotic therapy on outcomes in patients with Pseudomonas aeruginosa bacteremia. Antimicrob. Agents Chemother. 2007;51(3):839–844. doi: 10.1128/AAC.00901-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lodise TP, Jr, Patel N, Kwa A, et al. Predictors of 30-day mortality among patients with Pseudomonas aeruginosa bloodstream infections: impact of delayed appropriate antibiotic selection. Antimicrob. Agents Chemother. 2007;51(10):3510–3515. doi: 10.1128/AAC.00338-07. • Article delineating time breakpoint for increased risk of mortality following delayed appropriate treatment.
- 50.Vidal F, Mensa J, Almela M, et al. Epidemiology and outcome of Pseudomonas aeruginosa bacteremia, with special emphasis on the influence of antibiotic treatment. Analysis of 189 episodes. Arch. Intern. Med. 1996;156(18):2121–2126. [PubMed] [Google Scholar]
- 51.Yuan Z, Ledesma KR, Singh R, Hou J, Prince RA, Tam VH. Quantitative assessment of combination antimicrobial therapy against multidrug-resistant bacteria in a murine pneumonia model. J. Infect. Dis. 2010;201(6):889–897. doi: 10.1086/651024. [DOI] [PubMed] [Google Scholar]
- 52.Chamot E, Boffi El, Amari E, Rohner P, Van Delden C. Effectiveness of combination antimicrobial therapy for Pseudomonas aeruginosa bacteremia. Antimicrob. Agents Chemother. 2003;47(9):2756–2764. doi: 10.1128/AAC.47.9.2756-2764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Safdar N, Handelsman J, Maki DG. Does combination antimicrobial therapy reduce mortality in Gram-negative bacteraemia? A meta-analysis. Lancet Infect. Dis. 2004;4(8):519–527. doi: 10.1016/S1473-3099(04)01108-9. [DOI] [PubMed] [Google Scholar]
- 54.Paul M, Leibovici L. Combination antimicrobial treatment versus monotherapy: the contribution of meta-analyses. Infect. Dis. Clin. North Am. 2009;23(2):277–293. doi: 10.1016/j.idc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 55.Paul M, Silbiger I, Grozinsky S, Soares-Weiser K, Leibovici L. β lactam antibiotic monotherapy versus β lactam-aminoglycoside antibiotic combination therapy for sepsis. Cochrane Database Syst. Rev. 2006;(1):CD003344. doi: 10.1002/14651858.CD003344.pub2. [DOI] [PubMed] [Google Scholar]
- 56.Gasink LB, Fishman NO, Weiner MG, Nachamkin I, Bilker WB, Lautenbach E. Fluoroquinolone-resistant Pseudomonas aeruginosa: assessment of risk factors and clinical impact. Am. J. Med. 2006;119(6):526, e519–525. doi: 10.1016/j.amjmed.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 57.Laupland KB, Parkins MD, Church DL, et al. Population-based epidemiological study of infections caused by carbapenem-resistant Pseudomonas aeruginosa in the Calgary Health Region: importance of metallo-β-lactamase (MBL)-producing strains. J. Infect. Dis. 2005;192(9):1606–1612. doi: 10.1086/444469. [DOI] [PubMed] [Google Scholar]
- 58.Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lautenbach E, Weiner MG, Nachamkin I, Bilker WB, Sheridan A, Fishman NO. Imipenem resistance among Pseudomonas aeruginosa isolates: risk factors for infection and impact of resistance on clinical and economic outcomes. Infect. Control Hosp. Epidemiol. 2006;27(9):893–900. doi: 10.1086/507274. [DOI] [PubMed] [Google Scholar]
- 60.Lautenbach E, Synnestvedt M, Weiner MG, et al. Imipenem resistance in Pseudomonas aeruginosa: emergence, epidemiology, and impact on clinical and economic outcomes. Infect. Control Hosp. Epidemiol. 2010;31(1):47–53. doi: 10.1086/649021. [DOI] [PubMed] [Google Scholar]
- 61.Hirakata Y, Yamaguchi T, Nakano M, et al. Clinical and bacteriological characteristics of IMP-type metallo-β-lactamase-producing Pseudomonas aeruginosa. Clin. Infect. Dis. 2003;37(1):26–32. doi: 10.1086/375594. [DOI] [PubMed] [Google Scholar]
- 62.Cosgrove SE, Carmeli Y. The impact of antimicrobial resistance on health and economic outcomes. Clin. Infect. Dis. 2003;36(11):1433–1437. doi: 10.1086/375081. [DOI] [PubMed] [Google Scholar]
- 63.Tam VH, Chang KT, Schilling AN, LaRocco MT, Genty LO, Garey KW. Impact of AmpC overexpression on outcomes of patients with Pseudomonas aeruginosa bacteremia. Diagn. Microbiol. Infect. Dis. 2009;63(3):279–285. doi: 10.1016/j.diagmicrobio.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 64.Carmeli Y, Troillet N, Karchmer AW, Samore MH. Health and economic outcomes of antibiotic resistance in Pseudomonas aeruginosa. Arch. Intern. Med. 1999;159(10):1127–1132. doi: 10.1001/archinte.159.10.1127. [DOI] [PubMed] [Google Scholar]
- 65.Stone PW, Braccia D, Larson E. Systematic review of economic analyses of health care-associated infections. Am. J. Infect. Control. 2005;33(9):501–509. doi: 10.1016/j.ajic.2005.04.246. [DOI] [PubMed] [Google Scholar]
- 66.Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 67.Roberts RR, Hota B, Ahmad I, et al. Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship. Clin. Infect. Dis. 2009;49(8):1175–1184. doi: 10.1086/605630. [DOI] [PubMed] [Google Scholar]
- 68.Mauldin PD, Salgado CD, Hansen IS, Durup DT, Bosso JA. Attributable hospital cost and length of stay associated with health care-associated infections caused by antibiotic-resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 2010;54(1):109–115. doi: 10.1128/AAC.01041-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Evans HL, Lefrak SN, Lyman J, et al. Cost of Gram-negative resistance. Crit. Care Med. 2007;35(1):89–95. doi: 10.1097/01.CCM.0000251496.61520.75. [DOI] [PubMed] [Google Scholar]
- 70.Cosgrove SE, Kaye KS, Eliopoulous GM, Carmeli Y. Health and economic outcomes of the emergence of third-generation cephalosporin resistance in Enterobacter species. Arch. Intern. Med. 2002;162(2):185–190. doi: 10.1001/archinte.162.2.185. [DOI] [PubMed] [Google Scholar]
- 71.Schwaber MJ, Navon-Venezia S, Kaye KS, Ben-Ami R, Schwartz D, Carmeli Y. Clinical and economic impact of bacteremia with extended-spectrum-β-lactamase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2006;50(4):1257–1262. doi: 10.1128/AAC.50.4.1257-1262.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harris A, Torres-Viera C, Venkataraman L, DeGirolami P, Samore M, Carmeli Y. Epidemiology and clinical outcomes of patients with multiresistant Pseudomonas aeruginosa. Clin. Infect. Dis. 1999;28(5):1128–1133. doi: 10.1086/514760. [DOI] [PubMed] [Google Scholar]
- 73.Neuhauser MM, Weinstein RA, Rydman R, Danziger LH, Karam G, Quinn JP. Antibiotic resistance among Gram-negative bacilli in US intensive care units: implications for fluoroquinolone use. JAMA. 2003;289(7):885–888. doi: 10.1001/jama.289.7.885. [DOI] [PubMed] [Google Scholar]