Summary
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) and the associated proteins (Cas) comprise a system of adaptive immunity against viruses and plasmids in prokaryotes. Cas1 is a CRISPR-associated protein that is common to all CRISPR-containing prokaryotes but its function remains obscure. Here we show that the purified Cas1 protein of Escherichia coli (YgbT) exhibits nuclease activity against single-stranded and branched DNAs including Holliday junctions, replication forks, and 5′-flaps. The crystal structure of YgbT and site-directed mutagenesis have revealed the potential active site. Genome-wide screens show that YgbT physically and genetically interacts with key components of DNA repair systems, including recB, recC and ruvB. Consistent with these findings, the ygbT deletion strain showed increased sensitivity to DNA damage and impaired chromosomal segregation. Similar phenotypes were observed in strains with deletion of CRISPR clusters, suggesting that the function of YgbT in repair involves interaction with the CRISPRs. These results show that YgbT belongs to a novel, structurally distinct family of nucleases acting on branched DNAs and suggest that, in addition to antiviral immunity, at least some components of the CRISPR-Cas system have a function in DNA repair.
Keywords: Cas1, CRISPR, DNA recombination, DNA repair, nuclease, YgbT
Introduction
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) represent the most widely distributed family of DNA repeats in prokaryotes (Makarova et al., 2006; Marraffini and Sontheimer, 2010a; Sorek et al., 2008; van der Oost et al., 2009). The CRISPRs and their associated proteins (CRISPR-associated, Cas) appear to comprise a novel microbial defense (immune) system which functions to some extent analogously to the eukaryotic RNA silencing machinery (Makarova et al., 2006; Sorek et al., 2008; van der Oost et al., 2009). CRISPRs are widespread among prokaryotes and are present in approximately 90% of archaeal and approximately 40% of bacterial genomes (Grissa et al., 2007; Horvath and Barrangou, 2010; Karginov and Hannon, 2010; van der Oost et al., 2009).
Most CRISPR-containing prokaryotes possess multiple CRISPR clusters (from 2 to 20 loci), each of which is organized as a tandem array of up to 100 identical repeats of ~25–50 base pairs (Sorek et al., 2008). A unique feature of CRISPRs is the separation of the direct repeats by non-repetitive spacers of similar size (Fig. 1A). The CRISPR clusters are transcribed as multi-unit precursors that are subsequently cleaved into smaller units that consist of one spacer flanked by two partial repeats (Brouns et al., 2008; Hale et al., 2009; Tang et al., 2002; Tang et al., 2005).
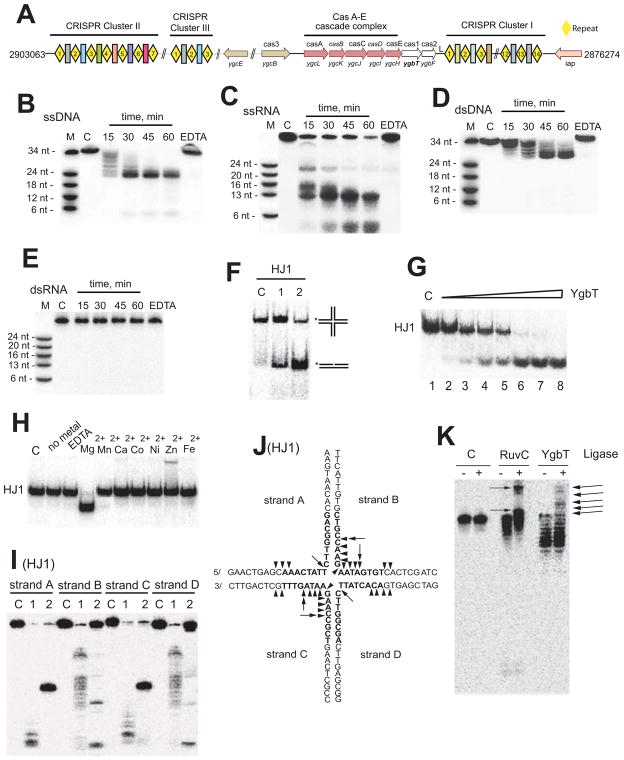
Fig. 1.
E. coli CRISPR system and nuclease activity of YgbT. (A) CRISPR-Cas system of E. coli K12 W3110. Repeats are shown as yellow diamonds, and spacers as colored rectangular boxes. (B, C, D, E) Cleavage of ssDNA (B), ssRNA (C), dsDNA (D) or dsRNA (E) by YgbT (denaturing PAGE/autoradiography). The 5′-[32P]-labeled ssDNA (34 nt), ssRNA (37 nt) or in a complex with complementary oligonucleotides was incubated without protein (C), with YgbT for the indicated times (37 oC) or in the presence of 10 mM EDTA for 60 min (EDTA). (F) Cleavage of HJ1 by YgbT (native PAGE/autoradiography). The 5′-[32P]-labeled (*) HJ1 substrate was incubated at 37 oC for 45 min without protein (C) or with YgbT (1) or RuvC (2). (G) Dependence of HJ1 cleavage on YgbT concentration (native PAGE). The 5′-[32P]-labeled HJ1 substrate (20 nM) was incubated in the absence (1) or in the presence of YgbT (Lanes 2–8: 0.05, 0.1, 0.2, 0.4, 0.8, or 1.0 μg) at 37 oC for 30 min. (H) Effect of divalent metal cations on HJ1 cleavage by YgbT (30 min, 37 °C, 5 mM metals or 10 mM EDTA; native PAGE). (I, J) Denaturing gel analysis of the cleavage of HJ1 by YgbT (triangles) or RuvC (arrows). The HJ1 substrate labeled by [32P] on different strands (A, B, C, or D) was incubated without protein (C) or with YgbT (1) or RuvC (2). (K) Re-ligation assay of the HJ cleavage products generated by YgbT or RuvC. The [32P]-labeled asymmetric HJ2 was incubated (30 min at 37oC) without protein (C) or with RuvC or YgbT, and the reaction products were then incubated with T4 DNA ligase prior to PAGE. Re-ligation products are indicated by arrows.
Most of the CRISPR spacers lack detectable sequence homologues, but in some organisms a varying, often small fraction of the spacers are homologous to sequences from bacteriophage and plasmid genomes. This key finding suggests that the spacer elements of the CRISPRs represent traces of past invasions by phages and plasmids (Bolotin et al., 2005; Marraffini and Sontheimer, 2010a; Mojica et al., 2005; Pourcel et al., 2005). Recently, a role for CRISPR spacers in defense against specific foreign DNA has been demonstrated in two gram-positive bacteria, Streptococcus thermophilus and Staphylococcus epidermidis (Barrangou et al., 2007; Marraffini and Sontheimer, 2008 – 2010b), and in an engineered gram-negative species, Escherichia coli (Brouns et al., 2008). However, in other studies, the presence of phage-specific spacers in CRISPR clusters of various bacteria failed to confer immunity against the cognate phage (Semenova et al., 2009; van der Ploeg, 2009; Zegans et al., 2009), suggesting that at least some of the CRISPRs may perform cellular functions other than phage immunity. Recently, it has been shown that some of the CRISPR-containing genomes also carry self-targeting CRISPR spacers which are homologous to chromosomal genes and might represent a form of autoimmunity or a regulatory mechanism (Aklujkar and Lovley, 2010; Stern et al., 2010).
CRISPR loci are associated with a large number of Cas protein-coding genes, which have been classified into approximately 45 protein families and 8 types of CRISPR/Cas systems (CRISPR sub-types) (Haft et al., 2005; Makarova et al., 2006). Six Cas protein families (Cas1–6) represent the core group of CRISPR-associated proteins with Cas1 and Cas2 proteins found in all CRISPR-bearing organisms (Haft et al., 2005; Makarova et al., 2006). Pseudomonas aeruginosa Cas1 (PaCas1) has a metal-dependent DNase activity but its role in CRIPSR function and cell biology remains unknown (Wiedenheft et al., 2009 ). In contrast, no nuclease activity has been detected in the Cas1 protein SSO1450 from Sulfolobus solfataricus (in the presence of 1 mM Mg2+ or Ca2+) (Han et al., 2009). The small Cas2 proteins possess endoribonuclease activity (Beloglazova et al., 2008; Han and Krauss, 2009), whereas the Cas3 protein is predicted to be a helicase that in many prokaryotes also contains a predicted nuclease domain (Haft et al., 2005; Makarova et al., 2002; Makarova et al., 2006; van der Oost et al., 2009). The biochemical activities of Cas4 and Cas5 are unknown. Cas6 proteins belong to a diverse class of proteins (15 families), named RAMP (Repeat Associated Mysterious Protein) (Haft et al., 2005; Makarova et al., 2006). The RAMP proteins are predicted to function as RNA-binding modules (Makarova et al., 2006). Cas6 from the archaeon Pyrococcus furiosus (PF1131) can cleave long precursor CRISPR transcripts into small guide RNAs (Carte et al., 2008). In P. furiosus, four different RAMP proteins form a complex, which binds to the CRISPR-derived guide RNAs to specifically target and cleave the invader RNA, but not DNA (Hale et al., 2009). By contrast, two studies in S. epidermidis and in the engineered E. coli suggest that the CRISPR/Cas systems destroy invading DNA rather than RNA (Brouns et al., 2008; Marraffini and Sontheimer, 2008 ). Thus, the CRISPR systems of archaea and bacteria show a great diversity in spacer and protein composition and appear to use various molecular mechanisms for protection against alien genetic elements.
The E. coli K12 strain W3110 contains three CRISPR loci with the spacers showing no homology to known phage or plasmid sequences. The CRISPR locus-1 has 13 spacers (14 repeats) and 8 cas genes which encode three core Cas proteins Cas1 (ygbT), Cas2 (ygbF), and Cas3 (ygcB) and five non-core Cas proteins, Cse1 (ygcL), Cse2 (ygcK), Cse3 (ygcH), Cse4 (ygcJ), and Cas5e (ygcI) (Fig. 1A) (Diez-Villasenor et al., 2010). In E. coli, the five non-core Cas proteins have been shown to form a complex, “Cascade”, which processes long CRISPR RNA transcripts into short guide RNAs (~57 nt) (Brouns et al., 2008). The “Cascade” complex and Cas3 (YgcB) can confer phage resistance when the E. coli strain is engineered to incorporate CRISPR spacer sequences homologous to an infecting phage λ (Brouns et al., 2008). By contrast, co-expression of Cas1 (YgbT) with Cascade in this strain had no effect on the sensitivity to phage λ, leaving the role of this universal Cas protein enigmatic (Brouns et al., 2008).
To gain insight into the function(s) of Cas1, we characterized the E. coli Cas1 protein YgbT using biochemical, genetic and structural approaches, and found that it is a multifunctional nuclease that cleaves Holliday junctions (HJs) and other intermediates of DNA repair and recombination. We found that YgbT interacts physically and genetically with multiple proteins involved in DNA recombination and repair, and that strains lacking YgbT show defects in DNA repair and chromosome segregation. Similar defects are caused by the deletion of the CRISPR repeats. Taken together, these results indicate that, in addition to their role in antiviral immunity, at least some CRISPR-Cas systems function in DNA repair.
Results
YgbT is a nuclease that can cleave linear substrates and Holliday junctions
Recently, it has been shown that the P. aeruginosa Cas1 protein is a metal-dependent DNase (Wiedenheft et al., 2009 ); however, no nuclease activity was found in the Cas1 protein SSO1450 from S. solfataricus (in the presence of 1 mM Mg2+ or Ca2+) (Han et al., 2009). To characterize the biochemical activity of the E. coli Cas1 protein YgbT, we purified this protein to homogeneity and tested its activity on a wide range of the 32P-labeled linear DNA and RNA substrates (Suppl. Table 1). We found a prominent divalent metal cation-dependent nuclease activity against single-stranded (ss) DNAs and a lower activity against ssRNAs (Fig. 1B, 1C; Suppl. Fig. 1A, 1B). YgbT also cleaved short linear double-stranded (ds) DNAs (34 nt), but no cleavage activity was observed with dsRNAs (16 – 39 nt) or longer dsDNAs (60 nt) (Fig. 1D, 1E; Suppl. Fig. 1C).
An activity common to DNA integration and recombination that also requires the cleavage of ssDNA is the resolution of Holliday Junctions (HJs), a cruciform-like DNA intermediate produced by reciprocal strand exchange between two dsDNA molecules (Garcia et al., 2000; Sharples, 2001). HJs are formed in all organisms during DNA integration, recombination, and recombinational DNA repair, as well as the regression or restart of a replication fork. The purified YgbT was tested for the HJ resolving activity using the general HJ substrate HJ1, which contains a homologous core formed by four partially complementary oligonucleotides, one of which is 5′-32P-labeled (Lilley and White, 2001). The presence of a homologous core allows the junction point to move freely by branch migration (up/down or left/right) permitting the HJ resolving enzymes to select an optimal sequence or site for cleavage. As shown in Figure 1F, purified YgbT displayed significant activity against the HJ1 substrate, producing a nicked duplex with electrophoretic mobility similar to that of the RuvC product. The cleavage of HJ1 was proportional to the concentration of YgbT (Fig. 1G) with the highest activity at pH 8.5 in the presence of Mg2+ (10 mM) (Figure 1H) and K+ (100 mM) (data not shown).
To characterize the cleavage pattern of YgbT, the products of HJ1 resolution were analyzed by denaturing PAGE and compared with the products of RuvC which cleaves this substrate at one major site in strands A and C and two major sites in strands B and D (Fig. 1I and J). In contrast to RuvC, YgbT did not show any pronounced sequence preference in the cleavage of HJ1 and introduced multiple symmetrical nicks in strands A and C, as well as in B and D (Fig. 1I, 1J). Strands A and C were cleaved preferentially on the 5′-side of the homologous core, whereas a distinct preference for cleavage at the center of the substrate was apparent in strands B and D (Fig. 1I, 1J). The HJ cleavage pattern of YgbT is similar to that of the human HJ resolvases MUS81-EME1 and SLX1 which likewise exhibit low sequence selectivity and introduce multiple nicks close to the 5′-side or at the center of the substrate (Constantinou et al., 2002 ; Svendsen et al., 2009). To determine whether YgbT, similarly to RuvC, resolves HJs through the introduction of ligatable nicks, we used the asymmetric substrate HJ2. The treatment of RuvC cleavage products with T4 DNA ligase yielded two main ligation products, whereas re-ligation of the YgbT cleavage products generated several products (indicated by the arrows) (Fig. 1K). Formation of several re-ligatable products was also observed with human HJ resolvase MUS81-EME1 (Constantinou et al., 2002 ). Hence, analogously to other HJ cleaving nucleases, YgbT cleaves HJs with the formation of ligatable products.
YgbT is a branched DNA nuclease
In addition to HJs, many known HJ cleaving nucleases can cleave other branched DNA substrates such as replication forks, Y-junctions, splayed arms, and 3′- and 5′-flaps (Abraham et al., 2003; Benson and West, 1994 ; Ciccia et al., 2008; Sharples, 2001; Svendsen et al., 2009). The substrate specificity of YgbT was further characterized using a series of branched 5-32P-labeled DNA substrates containing sense and antisense sequences of the E. coli CRISPR repeat, including a static HJ (HJ3), replication fork, 5′-flap, 3′-flap, and splayed arm duplex, structures that mimic various DNA repair and recombination intermediates (Fig. 2A). At pH 8.5, YgbT cleaved all these substrates with variable efficiency, whereas at pH 7.0 it was more specific and showed significant activity only against HJ3 (Fig. 2A). Purified YgbT also bound to 5′-flaps structures, HJs, ssDNAs and ssRNAs, and produced oligomeric complexes, as detected using mobility shift assays (Fig. 2B, only 5′-flap and HJ are shown). Like RuvC (Benson and West, 1994), YgbT cleaved the three-way junction substrate (3wHJ) but showed no activity against a Y-junction or a heterologous loop (data not shown). Analysis of the cleavage products by denaturing PAGE revealed that YgbT introduced a limited number of cuts into the replication fork and 5′-flap structures, but produced multiple cleavage products with the HJ3, 3′-flap, and splayed arm substrates (Fig. 2C, 2D).
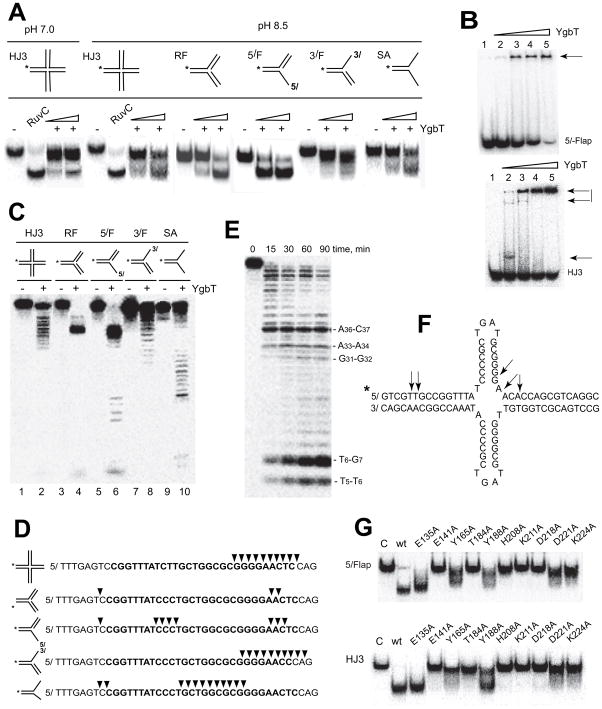
Fig. 2.
Nuclease activity of YgbT against branched DNA substrates containing sequences of the E. coli CRISPR. (A) Cleavage of branched DNA substrates by YgbT (native PAGE). The substrates including static HJ (HJ3), replication fork (RF), 5′-flap (5/F), 3′-flap (3/F), and splayed arm duplex (SA) were 5′-[32P]-labeled on the indicated strand (*) and incubated (30 min at 37 oC) in the absence or in the presence of 0.2 or 0.4 μg of YgbT at pH 7.0 or pH 8.5. (B) Gel-shift assay showing the binding of YgbT to 5′-flap or HJ3 (native PAGE). The 5′-[32P]-labeled substrates were pre-incubated for 15 min at 25 oC without YgbT (1) or with YgbT (Lanes 2–5: 0.2 μg, 0.4 μg, 0.6 μg, and 0.8 μg). Arrows indicate the position of the DNA-YgbT complexes. (C, D) Denaturing PAGE analysis of YgbT cleavage sites. The 5′-[32P]-labeled (*) substrates were incubated without or with YgbT (30 min, 37 oC). Panel D shows the sequences of the labeled strands and major cleavage sites of YgbT (arrowheads). The sequence of the E. coli CRISPR repeat-1 is shown in bold letters. (E, F) Cleavage of the CRISPR cruciform-like substrate CF1 by YgbT (denaturing PAGE analysis). The 5′-labeled CF1 (shown in panel F) was incubated with YgbT (0.2 μg) at 37 oC. (G) Alanine replacement mutagenesis of YgbT: cleavage of 5′-flap and HJ3 substrates by purified mutant proteins (30 min at 37 oC; native PAGE/autoradiography).
In E. coli and many other CRISPR-containing genomes, CRISPRs encompass palindromic repeats that can potentially generate cruciform (CF)-like structures. To test whether these structures represent possible substrates for YgbT, we designed two artificial CF-like substrates (CF-1 and CF-2) using oligonucleotides that correspond to the sequence of the E. coli CRISPR repeat-1 (Suppl. Table 1). YgbT efficiently cleaved both CF-like substrates and generated multiple cleavage products (Fig. 2E, 2F; only CF-1 is shown). With the CF-1 substrate, the five main cleavage products had the lengths of 5 nt, 6 nt, 29 nt, 31 nt, and 34 nt (Fig. 2E, 2F).
Crystal structure and site-directed mutagenesis of YgbT
The recently published crystal structure of PaCas1 revealed the presence of a small N-terminal β-strand domain (residues 1–106) connected by a flexible linker to a larger α-helical domain (residues 113–324) (Wiedenheft et al., 2009 ). We cloned and purified the C-terminal domain of YgbT without the N-terminal domain (residues 96–278) and found that the C-terminal domain retained all the activities observed in the intact YgbT protein (HJ cleavage, ssDNase and ssRNase), indicating that this domain contains the active site of YgbT (Suppl. Fig. 1D, 1E). We then determined the crystal structures of both the YgbT C-terminal domain (residues 96–278; 1.40 Å resolution; PDB code 3NKE) and the full-length protein (1.95 Å resolution; PDB code 3NKD) (Fig. 3, Suppl. Table 3). The structure of the full-length YgbT showed that this protein is a homodimer (Fig. 3A), consistent with our gel-filtration results (Mr = 70 kDa; predicted monomer Mr = 33.2 kDa). Like PaCas1 and the Cas1 protein aq_369 from Aquifex aeolicus (PDB code 2YZS), the YgbT monomer consists of two domains: a small N-terminal domain (amino acids 1–89) with a beta-sandwich-like structure connected by a flexible linker (aa 90–95) to a larger, mostly α-helical, C-terminal domain (aa 96–305) (Fig. 3B). Our gel-filtration experiments showed that the purified C-terminal domain of YgbT (predicted monomer Mr 20 kDa) exists as a mixture of the dimeric (Mr 38 – 46 kDa; 5% to 20%) and monomeric (Mr 22 – 27 kDa; 80% to 95%) forms in solution.
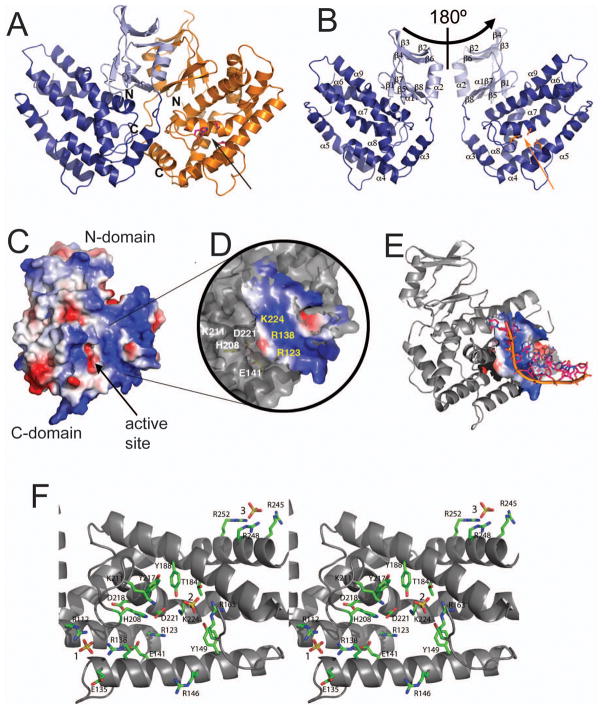
Fig. 3.
Crystal structure of YgbT and the potential active site. (A) Overall structure of the YgbT dimer. (B) Two views of the YgbT monomer (related by a 180° rotation) showing the presence of two domains: the N-terminal β-sandwich-like domain (light-blue) and the C-terminal all-α domain (dark-blue). The position of the potential active site is indicated by the side chains of three conserved residues (shown as sticks). (C) Surface charge presentation of the YgbT monomer showing the presence of several large basic patches representing potential DNA-binding sites (colored in blue). (D) Close-up view of the main basic patch of YgbT located close to the potential active site (E141, H208, and D221) and showing the position of several conserved residues. (E) Superposition of the ssDNA fragment from the E. coli topoisomerase III DNA-binding site (1i7d) onto the potential DNA-binding site of YgbT. The DNA docking was performed using the Surface Screen analysis (Binkowski and Joachimiak, 2008). (F) Close-up stereo view of the YgbT active site from the structure of the C-terminal domain showing the position of three bound sulfate molecules shown as sticks and labelled (1, 2, and 3). The YgbT residues are shown as green sticks along a YgbT ribbon (grey). Two arrows in A and B indicate the protein side and position of the active site shown in C.
A Dali search (Holm and Sander, 1998) for structures similar to that of YgbT identified three other Cas1 proteins as the best hits including PaCas1 (3GOD, Z-score 21.9, rmsd 3.4 Å) and two unpublished structures of Cas1 proteins from A. aeolicus (aq_369, PDB code 2YZS, Z-score 15.3, rmsd 3.4 Å) and Thermotoga maritima (TM1797, PDB code 3LFX, Z-score 16.6, rmsd 4.6). However, these proteins share relatively low overall sequence similarity (16–23% sequence identity) and belong to different CRISPR subtypes (CASS-2: YgbT; CASS-3: PaCas1; CASS-7: aq_369 and TM1797) (Makarova et al., 2006) (Suppl. Fig. 2). Surface charge analysis of YgbT revealed the presence of several large patches of positively charged residues which represent potential DNA-binding sites (Fig. 3C). In PaCas1, several basic residues surround the negatively charged metal-binding site creating a potential nucleic acid binding site in proximity to the catalytic metal cation (Wiedenheft et al., 2009 ). YgbT contains a larger cluster of highly conserved basic residues (R112, R123, R138, R146, R163, K211, and K224) positioned around the potential metal-binding site (E141, H208, D221) (Fig. 3C, 3D). In addition, YgbT contains another cluster of positively charged residues located at the same protein side near the connection of the two domains (K37, R59, R84, R245, R248, and R252); this cluster has no counterpart in PaCas1, suggesting that these proteins might differ in the details of the substrate binding (Fig. 3C). Surface screen analysis (Binkowski and Joachimiak, 2008) showed that the surface of the YgbT main basic patch is similar to that of the ssDNA-binding site of E. coli topoisomerase III (PDB code 1I7D), which binds ssDNA through the direct or water-mediated interactions with the phosphate groups of the DNA phosphodiester backbone (Changela et al., 2001). Fig. 3E shows the possible position of ssDNA modeled on the YgbT surface based on the superposition of the YgbT and topoisomerase III surfaces.
Previous sequence analysis of Cas1 proteins showed that four strictly conserved residues (three carboxylates and one histidine) represent the signature motif of the Cas1 family (Makarova et al., 2002). Two of these residues have been shown to be required for the nuclease activity of PaCas1 (D265 and D268) (Wiedenheft et al., 2009 ). In YgbT, the signature residues (E141, H208, D218, and D221) form a cluster of closely positioned side chains (2.5 to 6.6 Å) located at the bottom of the most prominent cleft of the C-terminal domain (Fig. 3F). These four residues are surrounded by the positively charged side chains of 7 highly conserved residues (R112, R123, R138, R146, R163, K211, and K224) which form the main basic patch of the YgbT surface and are potentially involved in the coordination of the phosphate backbone of bound DNA. In the structure of the YgbT C-terminal domain, three of these residues (R112, R138, and R163) interact with two bound sulfates which can be construed as mimicking the phosphates of the DNA backbone (Fig. 3F). The third sulfate molecule in this structure is bound to three residues from the second basic surface patch (R245, R248, and R252) (Fig. 3F).
Alanine replacement mutagenesis of YgbT showed that its nuclease activity against all substrates was abolished in the purified mutant proteins E141A, H208A, D218A, and D221A (Fig. 2G: 5′-flap and HJ; Suppl. Fig. 1F: ssRNA; Suppl. Fig. 1G), in accord with the prediction that these residues contribute to the active site. Mutations of other residues near the potential active site (T184A, K211A, and K224A) also had strong inhibitory effects on the activity of YgbT, whereas the E135A, Y165A, and Y188A mutant proteins retained significant activity (Fig. 2G), suggesting that the active site of YgbT is located on its C-terminal domain close to the potential ssDNA-binding site.
Taken together, these results indicate that YgbT is a nuclease that can cleave HJs and other branched DNA substrates, as well as linear ss/dsDNAs and ssRNAs. The ability of YgbT to cleave branched DNA substrates potentially could contribute to the addition of new spacers to CRISPR clusters. Moreover, the identified activity of YgbT against branched DNAs suggests that this protein might also participate in some of the DNA repair or recombination pathways. This possibility is consistent with previous studies which proposed a function for the CRISPR system in DNA repair or in chromosomal segregation (DeBoy et al., 2006; Makarova et al., 2002; Mojica et al., 1995).
YgbT is associated with proteins involved in DNA repair
To probe the potential role of YgbT in DNA repair and recombination, we performed genome-wide assays for physical and genetic interactions between YgbT and other E. coli proteins. Endogenous YgbT was C-terminally tagged using a cassette encoding a calmodulin binding peptide and a triple FLAG-tag (Zeghouf et al., 2004). Proteins stably associated with the tagged YgbT were treated with DNase and affinity-purified in two steps on anti-FLAG and calmodulin resins and then identified using tandem mass spectrometry (LC-MS) (Butland et al., 2005). Among the proteins co-purifying with YgbT (Suppl. Table 3) were RecB and RecC, two subunits of the RecBCD nuclease-helicase (recombinase) complex, which is a major player in the recombinational repair of DNA double-strand breaks (DSBs) (Kowalczykowski, 2000). In addition, the DNA repair proteins that co-purified with YgbT included RuvB, a recombinational DNA helicase which, together with RuvA, binds to HJs and catalyzes HJ branch migration to facilitate the cleavage of HJs by the RuvC resolvase (West, 1996). Although no physical interactions between YgbT and other Cas proteins in E. coli have been reported previously (Brouns et al., 2008), we reproducibly detected two subunits of the CRISPR-associated Cascade complex, YgcJ (CasC) and YgcH (CasE) (Fig. 4A, Suppl. Table 3). Interestingly, purified recombinant YgcH (a Cascade subunit) inhibited the HJ cleavage activity of YgbT in a concentration-dependent manner (Fig. 4B), suggesting that YgbT and the Cascade complex might interact and regulate each other’s activities.
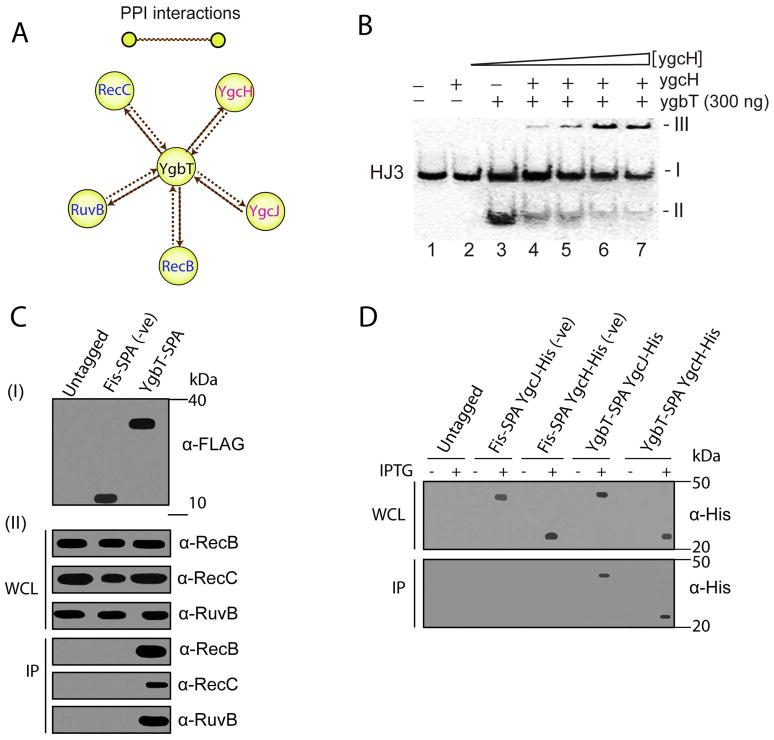
Fig. 4.
Physical interactions of YgbT with other E. coli proteins. (A) Interactions of YgbT with DNA repair/recombination and Cascade proteins validated by reciprocal SPA-tagging and purification. Yellow nodes represent tagged proteins used as “baits”, and brown edges represent protein-protein associations. (B) Purified YgcH (Cse3) inhibits the HJ cleavage activity of YgbT (native gel/autoradiography). The 5′-[32P]-labeled HJ3 substrate was pre-incubated for 2 min at room temperature with various amounts of YgcH (0, 0.1, 0.2, 0.3 and 0.4 μg) before the incubation with YgbT (0.3 μg, 45 min, 37 oC). The band-I represents the HJ3 substrate, the band-II - the HJ3 cleavage product (by YgbT), and the band-III (on the top) – the protein/DNA aggregates which are unable enter the gel. (C, D) Co-immunoprecipitation of YgbT with RecB, RecC, RuvB, YgcH, and YgcJ. Panel C show immunoblot analysis of the whole cell lysates (WCL) and anti-FLAG immunoprecipitates (IP) from the untagged DY330 strain (Panel C) or from strains expressing the SPA-tagged Fis or YgbT probed using RecB, RecC and RuvB antisera. Panels D show immunoblot analysis of the WCL and IP from the E. coli C41 (DE3) strain expressing a SPA-tagged Fis or YgbT with His6-tag YgcJ or YgcH probed using anti-His antibody.
The observed physical interactions of YgbT with DNA repair/recombination proteins (RecB, RecC, RuvB, and UvrC) and with two other Cas proteins (YgcJ and YgcH) were validated by reciprocally purifying C-terminally tagged RecB, RecC, RuvB, UvrC, YgcJ and YgcH proteins. In each case, mass spectrometry analyses of the affinity-purified protein confirmed its association with YgbT (Fig. 4A, Suppl. Table 3). We also confirmed the physical association of YgbT with RecB, RecC, RuvB, YgcH and YgcJ by co-immunoprecipitation using cell lysates from exponential phase cultures of strains expressing C-terminal affinity-tagged YgbT; a strain expressing tagged Fis, a nucleoid associated DNA-binding protein which modulates gyrase (Cho et al., 2008) and topoisomerase I production (Weinstein-Fischer and Altuvia, 2007), was used as a control. The tagged YgbT was immunoprecipitated from the cell lysates, and the presence of RecB, RecC and RuvB was probed by immunoblotting using anti-RecB, -RecC, or - RuvB antisera, or anti-(His6)-tag monoclonal antibody (to detect the (His6)-tagged YgcH or YgcJ) (Fig. 4C, 4D). As expected, all these proteins (RecB, RecC, RuvB, YgcH and YgcJ) co-precipitated from extracts containing tagged YgbT but not from control extracts prepared from an untagged parental E. coli strain or the Fis-tagged strain. Thus, our results reveal a stable physical association of the E. coli Cas1 protein YgbT with some of the key proteins involved in DNA repair pathways and with two other Cas proteins, YgcH and YgcJ.
YgbT genetically interacts with DNA repair-recombination pathways
To identify E. coli genes that genetically interact with ygbT, we used the recently developed E. coli Synthetic Genetic Array (eSGA) approach (Butland et al., 2008). Double mutants were constructed by conjugating a ygbT donor strain (marked with Δ::CmR) with single gene deletions of almost all other non-essential E. coli genes (recipients marked with Δ::KanR) or hypomorphic alleles of selected essential E. coli genes (marked with KanR) (Suppl. Table 4, Suppl. Information). The colony growth and relative fitness of the resulting double mutants that survived double drug selection were examined by digital imaging. Using a statistical interaction score (S) to quantify both the strength and confidence of the interactions between each E. coli mutant gene pair, we detected cases of both synthetic sick or lethal (SSL; i.e. negative or aggravating interactions) and alleviating (i.e. positive or suppressing) growth phenotypes (Suppl. Table 4, Suppl. Information). In congruence with the results of the protein-protein interaction experiments, we observed genetic interactions of ygbT with DNA repair and recombination genes, including SSL interactions with recBC and recN and strong alleviating interactions with the ruvABC genes (Suppl. Table 4). Over-representation analysis (Irizarry et al., 2009) using Gene Ontology annotation terms (Suppl. Information) showed that the interacting genes identified in the ygbT deletion screen were significantly enriched (q-value < 0.005) for genes involved in DNA repair, DNA recombination, cell division, and chromosomal segregation and condensation (Suppl. Table 5).
Because the eSGA method depends on recombination in the recipient cells and genes like recBC are important for recombination, we validated the observed genetic interactions of ygbT with recABCD and ruvABC in reciprocal conjugation experiments using recABCD and ruvABC single gene deletion mutants as donors and ΔygbT as recipient (Fig. 5A, 5B). Consistent with the ygbT genetic interaction results, the recipient ygbT deletion strain exhibited a striking suppression (alleviating) phenotype when combined with individual gene deletions of ΔruvABC donor strains. Conversely, a significant SSL growth defect was consistently observed between the ΔygbT recipient and the ΔrecB and ΔrecC donor mutants, whereas hardly any growth deficiency was seen with recA and recD donor mutants (Fig. 5B). These genetic interactions were not a consequence of recombination suppression resulting from gene proximity because no detectable effects on growth were observed when recBC or ruvABC deletion donors were combined with deletions of functionally unrelated genes flanking ygbT. Moreover, the use as donors of other, functionally unrelated genes from the same genome region, namely csdA and rpoS, did not reveal synthetic genetic interaction with ygbT, recABCD or ruvABC, except in the case of csdA and recB which are too close to one another (Fig. 5A, 5B). Recently, Pougach et al. (Pougach et al., 2010) have demonstrated substantial readthrough transcription from the kanamycin resistance cassette inserted into several cas genes (ygcL, ygcK, or ygcJ) located upstream of ygbT (Fig. 1A) using E. coli strains from the Keio collection, which was also used in our work. However, the readthrough transcription from the kanamycin resistance cassette inserted into ygbT is unlikely to have a substantial effect on the observed genetic interactions, because only one cas gene (ygbF) is located downstream of ygbT (Fig. 1A). Thus, our results suggest that the observed genetic interactions between ygbT and recBC and ruvABC represent bona fide functional relationships. The detection of a SSL association in the first case and an alleviating interaction in the second case indicates that the putative function of YgbT in repair might be redundant with the function of RecBC but cooperates with the function of RuvABC.
Fig. 5.
Genetic interactions of ygbT and DNA damage sensitivity of the ygbT deletion strain. (A) Chromosomal positions of ygbT (green arrow), rec (purple arrows), ruv (yellow arrows), and other gene (red arrows) deletions used as donors (marked with CmR) or recipients (marked with KanR) in eSGA experiments. csdA and rpoS donor deletions (blue arrows) located close to the rec and ygbT genes served as controls for effects of proximity on recombination. The numbers indicate the gene coordinates on the E. coli chromosome (in minutes). OriT is the F origin of transfer in Hfr Cavalli (12.2 minutes). (B) Validation of ygbT-rec and ygbT-ruv synthetic genetic interactions. rec (Panel I), ruv (Panel II), and control (csdA and rpoS, Panel III) Hfr Cavalli donor deletion strains were crossed with the indicated F- recipient deletion strains (deleted for ruv, ygbT, or various genes flanking ruv and ygbT), followed by selection on plates containing chloramphenicol and kanamycin. (C) Serial cell dilution assay showing growth inhibition of ygbT-ruvABC double mutants and their single mutants by MMC (0.15 μM). Growth is classified as sensitive to MMC (+) or not sensitive compared with wild-type (−). Also shown are the effects of complementing ΔygbT with pBAD vectors expressing wild-type or inactive mutant YgbT. (D) Effects of the indicated doses of UV irradiation on the survival of ygbT-ruvABC double mutants and their single mutants. The values plotted are averages from three independent experiments.
In addition, ygbT exhibited a strong SSL interaction with recN and weak SSL interactions with other rec genes (recF, recO and recQ) in the RecF recombination pathway (Suppl. Table S5). In E. coli, the major homologous recombination mechanism is the RecBC pathway, which is involved in both conjugal and transductional recombination, as well as in the repair of DSBs and the degradation of foreign DNA, whereas the RecF recombinational repair pathway is implicated preferentially in the repair of UV-induced daughter strand gaps (Kuzminov, 1999; Tseng et al., 1994). Thus, the SSL interactions of ygbT with recBC and recF genes suggest that YgbT could be involved in both pathways or yet a third parallel pathway of DSB repair.
The ΔygbT screen also identified genetic interactions outside the Rec and Ruv systems, including synthetic lethality following inactivation of the site-specific recombinase XerD, the chromosome partitioning protein MukB, and the essential cell division protein FtsK (Suppl. Table 4). These proteins function in chromosome segregation and the separation of replicated chromosome dimers, which involves the dynamic formation and resolution of HJs and other branched DNA intermediates (Barre et al., 2001; Gordon and Wright, 2000.; Sherratt et al., 2001). Aggravating genetic interactions were also observed with other genes involved in cell division (e.g., dinF, envC, gidA, gidB and dicB) and in methyl-directed mismatch repair (MMR) (e.g., mutH, mutL and mutS) (Suppl. Table 4). Thus, the genetic interaction data appear to support the functional link between YgbT and DNA repair-recombination.
YgbT is required for the resistance of E. coli to DNA damage
Our biochemical results together with physical and genetic interaction data pointed to a role of YgbT in DNA repair-recombination pathways and to a cooperation between YgbT and RuvABC. To investigate the biological implications of these results, we compared the sensitivity of the E. coli ygbT deletion strain to DNA damage induced by either the genotoxic agent mitomycin C (MMC) or UV light with the sensitivities of wild-type or ruvABC deletion strains. Both MMC and UV introduce an array of lesions, including pyrimidine dimers (UV) and inter-strand cross-links (MMC), leading to the formation of DSBs in DNA (De Silva et al., 2000; Garinis et al., 2005). In E. coli, UV- or MMC-induced DNA damage is repaired by a multitude of pathways, including homologous recombination and nucleotide excision repair (Cole, 1973; Keller et al., 2001). Strains with individually deleted ruvABC genes are known to be substantially more sensitive than the wild-type to UV- or MMC-induced DNA damage (Le Masson et al., 2008; Lloyd et al., 1984), and there is little difference in the sensitivity among the strains carrying mutations in each of the ruv genes (Lloyd et al., 1984; Otsuji et al., 1974; Sargentini and Smith, 1989). In our experiments, knockout of ygbT increased the sensitivity of E. coli cells to both MMC and UV to approximately the same extent as the knockout of each of the ruv genes (Fig. 5C, 5D). The sensitivity of double deletion combinations of ygbT with the ruvABC genes to MMC and UV was similar to each of the single knockout mutants (Fig. 5C, 5D). Moreover, the sensitivity of ygbT-ruvABC double mutants to cisplatin (40 μM), another genotoxic agent known to induce DSBs that are repaired by excisional and recombinational repair pathways (Zdraveski et al., 2000), was also comparable with the sensitivities of the single mutants (data not shown). Thus, the lack of synergy in the ygbT-ruv double knockouts suggests, in agreement with the eSGA results, that YgbT functions in the same DNA repair pathway(s) with RuvABC.
To ascertain that the observed phenotype was caused solely by the ygbT deletion, we showed that the MMC and UV resistance of the ygbT null strain could be restored to the wild type level by introducing a pBAD-plasmid expressing the wild-type ygbT gene under the control of an arabinose-inducible promoter but not ygbT mutants with replacements of the predicted catalytic amino acid residues (E141A, H208A and D218A) (Fig. 5C, complementation experiment with UV not shown). Similarly, the resistance of ygbT-ruv double mutants to MMC and UV-irradiation was fully restored to wild-type levels by ectopic expression of both YgbT and the respective Ruv proteins (data not shown). Thus, the nuclease activity of YgbT appears to be critical for the resistance of E. coli to DNA damage.
YgbT is recruited to DNA damage sites in MMC-treated cells
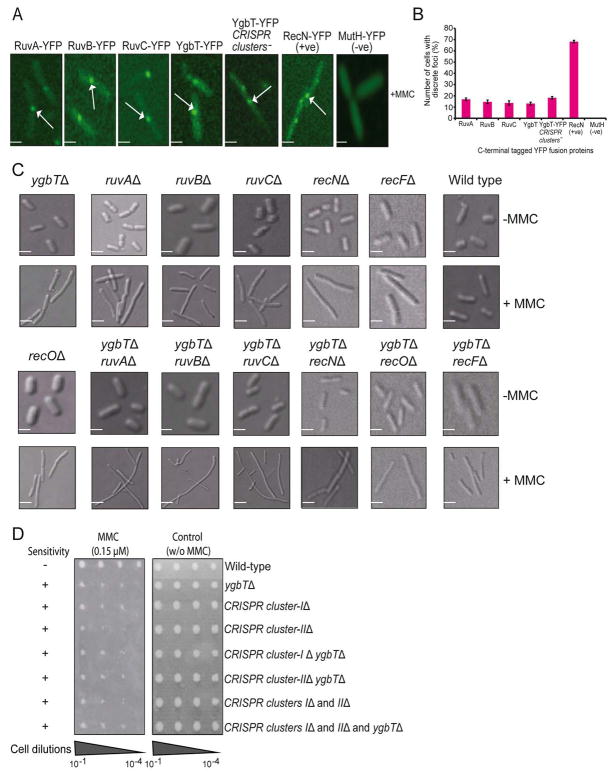
In bacteria, MMC treatment induces the formation of DSBs at replication forks, which then rapidly recruit proteins involved in DNA repair (e.g., RecN) leading to the formation of discrete foci detectable using fusions with appropriate tags (Kidane et al., 2004; Sanchez et al., 2006). To test whether YgbT is likewise recruited to DSBs in E. coli, we constructed chromosomal yellow fluorescent protein (YFP) fusions (Taniguchi et al., 2010) in which YgbT as well as RuvA, RuvB, and RuvC genes were tagged with the YFP coding sequence at their C-terminal ends. A strain expressing a YFP fusion of RecN was used as a positive control. Fluorescence microscopy showed that YFP-labeled YgbT, as well as RuvABC and RecN, formed discrete fluorescent foci on the nucleoids after 120 min of MMC treatment. In YgbT-YFP and RuvABC-YFP strains, 13–17% of the > 250 analyzed cells contained a single focus per nucleoid (Fig. 6A, 6B), whereas in the RecN-YFP strain, the fusion protein localized to a discrete focus in the majority (~70%) of cells. Although it remains unclear why a smaller fraction of MMC-treated cells had YgbT or RuvABC localized to foci compared with RecN, no foci were detected in the absence of MMC in YgbT-YFP or RecN-YFP expressing cells (Suppl. Fig. 3A). Thus, YgbT appears to be specifically recruited to DNA DSBs, in agreement with the other indications of a role of this protein in DNA repair.
Fig. 6.
Cell morphology of the ygbT deletion strain and intracellular localization of YgbT in response to DNA damage. (A) Intracellular localization of RuvA-YFP, RuvB-YFP, RuvC-YFP, YgbT-YFP, RecN-YFP and MutH-YFP fusion proteins in exponentially growing E. coli cells treated with MMC for 120 min. Scale bar, 2 μM. (B) The percentage of E. coli cells with discrete foci of various YFP fusion proteins in their nucleoids after MMC treatment. In each case, 250 different cells from five independent biological replicates (50 cells per replicate) were examined. Error bars indicate standard deviation from mean. (C) Differential interference contrast images of MMC-induced changes of cell morphology in ygbT-ruvABC, ygbT-recN, ygbT-recO, and ygbT-recF double mutants and their respective single mutants in the presence (+) and in the absence (−) of MMC. Scale bar, 2 μM. The average cell length of these cells is shown in Suppl. Fig. 4. (D) E. coli strains with deleted CRISPR clusters exhibit increased sensitivity to MMC. Serial cell dilution assays showing the effects of MMC (0.15 μM) on the growth of E. coli strains with deletions of one or both CRISPR clusters and/or ygbT. Growth is classified as sensitive to MMC (+) or not sensitive (−) in comparison to wild-type.
YgbT is required for cell division in MMC-treated cells
Strains showing hypersensitivity to MMC, such as ruvABC, recN, recF and recO loss-of-function mutants, exhibit impaired cell division and chromosomal segregation in the presence of unrepaired DNA damage, leading to the formation of abnormally long, non-septate, multi-nucleate cells (Ishioka et al., 1998; Kidane et al., 2004; Meddows et al., 2005; Otsuji et al., 1974; Suzuki et al., 1967; Zahradka et al., 1999). This unusual morphology might be a consequence of the inability of the cells to resolve chromosome dimers. The demonstration of the HJ cleavage activity of YgbT and the physical and genetic interactions between YgbT and several repair proteins described above suggest that YgbT could also be involved in resolving chromosomes during cell division. A role of YgbT in chromosome segregation is also suggested by its aggravating genetic interactions with several genes known to be involved in cell division and chromosome segregation factors (e.g., FtsK, MukB, and XerD) (Suppl. Table 4). Should YgbT participate in the resolution of sister chromosomes, the ΔygbT strain would be expected to form abnormal filaments in the presence of MMC. Indeed, we found that the ygbT deletion strain formed greatly elongated cells containing one long nucleoid after 120 min of MMC treatment (Fig. 6C). The average length of ygbT mutant cells (~5.8 ± 0.3 μm) in the presence of MMC was almost twice that of wild type cells (from ~2.5 to 3.9 μm) (Suppl. Fig. 3B). This extent of cell elongation was similar to that seen in recN, recF, or recO deletion strains (from ~4.7 to 6.1 μm) or ruvABC single mutants (from ~5.0 ± 0.6 to 5.9 ± 0.3 μm). Double mutants of ygbT with ruvABC, but not with recN, recF, or recO, produced even longer filamentous cells in the presence of MMC, with average cell lengths ranging from ~8.6 ± 0.8 to 9.7 ± 0.3 μm after 120 min of MMC treatment (Fig. 6C, Suppl. Fig. 3B). Thus, our findings indicate that YgbT might be involved in chromosome segregation following DNA damage. This result is consistent with observations on the archaea Haloferax volcanii and H. mediterranei which implicated the CRISPR repeats in replicon partitioning (Mojica et al., 1995). Surprisingly, when it comes to chromosome segregation, YgbT appears to function in cooperation with the RecF pathway and redundantly with RuvABC.
Association of YgbT with CRISPR clusters
The CRISPR repeats of E. coli and many other organisms contain a 5–7 nt palindromic repeat (Jansen et al., 2002; Kunin et al., 2007 ; Sorek et al., 2008). Such palindromes could potentially form cruciform structures (in dsDNA) or hairpins (in ssDNA), whereas direct tandem repeats can form slipped-strand DNA (S-DNA) with mispaired complementary repeats (Lilley, 1989; Mirkin and Mirkin, 2007). These unusual DNA structures can interfere with DNA replication, resulting in replication fork stalling and genomic instability (Lindsey and Leach, 1989; Mirkin and Mirkin, 2007). As the cas1 gene is present only in bacteria and archaea that also possess CRISPRs, functional coupling between the Cas1 protein and CRISPRs appears most likely (Brouns et al., 2008; Makarova et al., 2006). The E. coli K12 laboratory strain W3110 carries three CRISPR clusters (Cluster-1 with 14 repeats, Cluster-2 with 7 repeats, and Cluster-3 with 3 repeats), with the cas1 (ygbT) gene located close (~300 bp) to CRISPR cluster-1 (Fig. 1A). Taking into account the function of YgbT in DNA recombination-repair that is strongly suggested by the results of this work and the obligate association of cas1 with CRISPR clusters, we speculated that the repair function of YgbT might involve alleviating the potential deleterious effect of CRISPR. Should that be the case, the requirement for YgbT in DNA repair would be lifted by deletion of the CRISPR clusters. To test this possibility, we constructed E. coli strains lacking ygbT and either CRISPR cluster-1, or cluster-2, or both CRISPR clusters, and compared their MMC sensitivities with that of the ygbT mutant strain (Fig. 6D). The results showed that the requirement of ygbT for MMC resistance was unrelated to the prevention of the potential deleterious effect of CRISPRs because the triple deletion of both CRISPR clusters and ygbT exhibited MMC sensitivity similar to that of the ygbT mutant (Fig. 6D).
Interestingly, deletion of one or both CRISPR clusters itself increased the sensitivity of E. coli to MMC (Fig. 6D). One possible explanation of this observation could be that CRISPR clusters are required for the expression of YgbT (Pul et al., 2010). However, this appears not to be the case because the assembly of YgbT at DSBs was independent of the presence of either CRISPR cluster (Fig. 6A, 6B). Thus, CRISPR clusters are apparently required for the function of YgbT in DNA repair but not for YgbT recruitment to MMC-induced DSBs. To elucidate the molecular mechanism of the CRISPR-dependent role of YgbT in repair, additional experiments are obviously required.
Discussion
The results presented here indicate that the E. coli Cas1 protein YgbT is a novel nuclease that can cleave HJs and other branched DNA substrates, as well as linear ss/ds DNAs and ssRNAs. To date, E. coli is known to encode one functional HJ resolvase, RuvC, and one cryptic resolvase, RusA, which is normally not expressed (Benson and West, 1994 ; Sharples et al., 1994). In contrast to RuvC, YgbT shows no apparent target sequence specificity. In addition, YgbT cleaves in vitro a broad range of branched DNA substrates (asymmetrical Holliday junction, replication fork, 5′-flap, 3′-flap and splayed arm substrates) which represent various intermediates of DNA recombination and repair (Fig. 2). Like human MUS81-EME1 and SLX1, YgbT produces multiple HJ cleavage products and can also cleave replication fork and 3′-flap structures; however, in addition, YgbT is active with 5′-flap and splayed arm substrates which are not cleaved by MUS81 (Constantinou et al., 2002 ). The 5′-flap endonucleases, which are required for the removal of RNA primers during replicative and repair DNA synthesis and typically can cleave both RNA and DNA, are encoded as distinct proteins in eukaryotes (FEN-1), archaea and some DNA viruses, whereas their bacterial homologs are N-terminal domains of DNA polymerase I (Shen et al., 1998 ). Given that YgbT cleaves both ssRNA and 5′-flap DNAs but is unrelated to the FEN-1 family it might represent a new group of stand-alone flap endonucleases.
When compared to one another, the three experimentally characterized Cas1 proteins display related but different biochemical properties: SSO1450 binds DNA/RNA but appears not to possess nuclease activity, PaCas1 cleaves ss/dsDNA, whereas YgbT is also active against branched DNA substrates and linear ssRNAs. These proteins belong to different CRISPR subtypes (Makarova et al., 2006) and share rather low overall sequence similarity (18.9 – 21.6 % of sequence identity), suggesting that they might possess genuinely different substrate preferences. This possibility is consistent with the presence of several (2 to 5) distinct paralogous Cas1 proteins in many bacteria and archaea. Further, detailed biochemical studies of Cas1 proteins from diverse organisms are required to delineate the functional versatility of this protein family.
The ability of YgbT to cleave branched DNA substrates in vitro suggests that this activity might contribute to the addition/removal of CRISPR spacers, which is proposed to proceed through DNA recombination (Mojica et al., 2009). Four strictly conserved amino acid residues of YgbT (E141, H208, D218 and D221) are critical for activity and represent its active site, which is located close to the potential DNA-binding site in the C-terminal domain. Further detailed analysis of the role of YgbT in the CRISPR mechanism requires the use of a natural experimental model of CRISPR spacer addition/removal, which has yet to be established in E. coli.
The incorporation of a novel CRISPR spacer and the accompanying repeat apparently involves the recognition of foreign DNA followed by processing steps and insertion into the CRISPR cluster. Given that YgbT showed no obvious sequence selectivity in the cleavage of branched DNAs, we hypothesize that other Cas (and non-Cas) proteins are also involved in the formation of novel CRISPR spacers, whereas YgbT might contribute to one of the final steps in the integration of the spacers into the chromosomal or plasmid DNA (e.g., HJ resolution or flap substrate cleavage). The physical interaction between YgbT and two components of the Cascade complex (Cse4/CasC and Cse3/CasE) reported in this work also suggests that Cascade might contribute to the integration of new spacers in E. coli.
The key conclusion of the present work is that YgbT and CRISPR are involved in one or more repair-recombination pathways and contribute to the resistance of E. coli to at least some types of DNA damage and chromosome segregation. This conclusion is consistently supported by several lines of genetic and biochemical evidence: (1) knockout of the ygbT gene results in a substantial increase in the sensitivity of E. coli to DNA damage caused by UV or MMC; (2) the rescue of the knockout mutants requires catalytically active YgbT, indicating that the apparent role of YgbT in repair-recombination depends on its demonstrated endonuclease activity towards the characteristic intermediates of several DNA repair pathways, including recombinational (HJs and splayed arms), base excision (5′-flaps), and nucleotide excision (3′-flaps) pathways; (3) YgbT physically interacts with several key repair proteins including RecB, RecC and RuvB; (4) the ygbT gene genetically interacts with repair genes, including synthetic-sick interactions with recB, recC and recN, indicative of functional redundancy, and alleviating interactions with ruvABC, indicative of involvement in the same repair-recombination pathway(s); (5) YgbT is recruited to DSBs in MMC-treated cells; (6) YgbT is required for cell division in MMC-treated cells, as evidenced by the unusual morphology of ygbT knockouts that resembles the morphology of knockout mutants of other repair genes upon treatment with DNA-damaging agents. In addition, we found that the function of YgbT in DNA repair apparently requires interaction with CRISPRs because deletion of the CRISPRs or the double deletion of the CRISPRs and ygbT led to the same phenotype as the ygbT knockout. The specific role of CRISPRs in repair remains to be elucidated, but the involvement of their recombinogenic potential seems plausible; previously, it has been suggested that CRISPRs mediate genome rearrangements in Thermotogales via recombination (DeBoy et al., 2006).
Thus, YgbT appears to be a multifunctional nuclease that can cleave various branched DNA intermediates produced by DNA repair pathways, chromosome segregation mechanisms, and (potentially) by the CRISPR system. More generally, our results suggest an intrinsic connection between the function of the CRISPR-Cas system in the acquired antiviral immunity and its emerging role in DNA repair in prokaryotes. These findings reconcile the latest observations on the antiviral functions of this system that depend on unique spacers homologous to viral genes (Horvath and Barrangou, 2010; Karginov and Hannon, 2010) and the earlier hypothesis on a repair function of the Cas proteins (Makarova et al., 2002) that was proposed before the discovery of the virus-specific spacers, solely on the basis of the predicted enzymatic activities of the Cas proteins (a helicase, a polymerase and multiple nucleases). Furthermore, a dual function of the CRISPR-Cas system in defense and repair is compatible with the typically small fraction of virus-specific CRISPR spacers and the inability of some of these spacers to protect the host against infection (Semenova et al., 2009; van der Ploeg, 2009; Zegans et al., 2009). It is conceivable that the array of diverse enzymes and nucleic acid-binding proteins associated with the CRISPRs evolved at early stages of the evolution of bacteria and archaea within the intrinsically interlinked contexts of DNA damage repair and antiviral defense. Further research into the functions of various CRISPR-Cas systems should reveal the relationships between its apparently diverse functions and more specific roles of the individual components.
Experimental procedures
Bacterial strains, gene cloning, and protein purification
All parental and mutant bacterial strains are listed in Suppl. Table 6 and are described in Suppl. Methods. For enzymatic assays and crystallization, the proteins were over-expressed in the E. coli BL21 (DE3) strain (Novagen) and affinity purified as previously described (Proudfoot et al., 2008; Zhang et al., 2001) (Suppl. Methods). This E. coli strain has been shown to contain no cas genes (Brouns et al., 2008).
Nuclease assays
DNA substrates used in this work and nuclease assays are described in Suppl. Table 1 and Suppl. Methods. The reaction products were analyzed by native (8% PAAG) or denaturing (15% polyacrylamide/8M urea PAAG) electrophoresis and visualized by autoradiography.
Crystallization and structure determination
The Se-Met-substituted C-terminal domain of YgbT (aa 92 – 281) and native two-domain YgbT (aa 7–281) were crystallized by the hanging drop vapor diffusion method as described in Suppl. Methods. The structure of the YgbT C-terminal domain was determined using the single anomalous scattering dispersion (SAD) method; whereas the structure of the two-domain YgbT was solved by molecular replacement using the structure of the YgbT C-terminal domain as a model (Suppl. Table 2 and Suppl. Methods).
SPA-tagging, purification and immunoprecipitation
YgbT, Ruv, Rec and Cas proteins were C-terminally tagged and purified essentially as previously described (Butland et al., 2005). The co-purifying proteins were identified using SDS-PAGE fractionation followed by peptide mass fingerprinting or using gel-free liquid chromatography/tandem mass spectrometry (LCMS) essentially as previously described (Babu et al., 2009). The physical association of YgbT with RecB, RecC, RuvB, YgcH and YgcJ proteins that were confirmed by co-immunoprecipitation are described in detail in Supplementary Methods.
Genome-wide eSGA screen and gene set enrichment analysis
A genome-wide eSGA screen using ygbTΔ::CmR in Hfr Cavalli as donor was carried out and analyzed as previously described (Butland et al., 2008), as were various mini-array crosses. Gene set enrichment analysis (Irizarry et al., 2009) was performed on the interaction S scores to identify the significantly enriched GO terms (Suppl. Table S6) spanning various biological processes (see Suppl. Methods).
DNA damage sensitivity assays
UV irradiation experiments were performed as described previously (Nair and Finkel, 2004). Diluted cultures were plated onto LB plates and colonies formed after overnight incubation at 32 °C were counted. Cell survival results were derived from the mean of three independent experiments. For the MMC and cisplatin sensitivity assays, exponentially growing cells were serially diluted in LB medium and pinned onto LB plates in the absence or presence of the indicated concentration of the DNA damaging agent. Sensitivity to DNA damage was assessed after 36 to 48 hrs of incubation at 32 °C.
Cell morphology and protein localization analysis
The C-terminal fusions of chromosomal YgbT, RuvA, RuvB, RuvC, RecN, and MutH to YFP proteins were constructed in the DY330 background by converting the SPA tags and kanamycin resistance cassette in these strains to YFP tags via the λ-RED recombination system (Yu et al., 2000). PCR amplification of a YFP-chloramphenicol resistance cassette was performed using ~40 base primers with homology to the insertion site (SPA) and the kanamycin resistance cassette. The deletion mutants or YFP-fusion strains were grown exponentially in LB medium at 32 °C. Prior to imaging, the cultures (A600=0.5) were incubated for 120 min with MMC (0.15 μM), pelleted, and 1.5 μl of suspension was spotted onto a glass slide for image analysis. The images were captured using the Quorum WaveFX Spinning Disc Confocal System, and the cell length was measured using the Volocity program (Improvision Ltd) as previously described (Sydorskyy et al., 2010).
Coordinates
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (http://www.rcsb.org) with accession codes 3NKD and 3NKE for the structures of YgbT and its C-terminal domain, respectively.
Supplementary Material
Acknowledgments
We thank members of the Emili and Greenblatt laboratories and of the Structural Proteomics in Toronto (SPiT) Centre for technical assistance. We are grateful to Paul Choi and Sunney Xie from Harvard University for providing us with the YFP-chloramphenicol resistance cassette. We thank Andrew Taylor and Gerry Smith from the Fred Hutchinson Cancer Research Center, Seattle WA for generous gifts of anti-RecBCD monoclonal antibodies. This work was supported by the Government of Canada through Genome Canada and the Ontario Genomics Institute (JG, AE and AFY; 2009-OGI-ABC-1405), by the Canadian Institutes of Health Research grant CIHR 82852 (to JG and AE), by the National Institutes of Health grant GM074942 (to AJ), and by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357 (to AJ). EVK is supported by the intramural funds of the US Department of Health and Human services (NIH, National Library of Medicine).
Footnotes
Supporting information is available in the online version of this article.
References
- 1.Abraham J, Lemmers B, Hande MP, Moynahan ME, Chahwan C, Ciccia A, Essers J, Hanada K, Chahwan R, Khaw AK, McPherson P, Shehabeldin A, Laister R, Arrowsmith C, Kanaar R, West SC, Jasin M, Hakem R. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. Embo J. 2003;22:6137–6147. doi: 10.1093/emboj/cdg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aklujkar M, Lovley DR. Interference with histidyl-tRNA synthetase by a CRISPR spacer sequence as a factor in the evolution of Pelobacter carbinolicus. BMC Evol Biol. 2010;10:230. doi: 10.1186/1471-2148-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babu M, Butland G, Pogoutse O, Li J, Greenblatt JF, Emili A. Sequential Peptide Affinity Purification System for the Systematic Isolation and Identification of Protein Complexes from Escherichia coli. Methods in Molecular Biology. 2009;564:373–400. doi: 10.1007/978-1-60761-157-8_22. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 5.Barre FX, Søballe B, Michel B, Aroyo M, Robertson M, Sherratt D. Circles: the replication-recombination-chromosome segregation connection. Proc Natl Acad Sci USA. 2001;98:8189–8195. doi: 10.1073/pnas.111008998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W, Koonin EV, Edwards AM, Savchenko A, Yakunin AF. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson FE, West SC. Substrate specificity of the Escherichia coli RuvC protein. Resolution of three- and four-stranded recombination intermediates. Journal of Biological Chemistry. 1994;269:5195–5201. [PubMed] [Google Scholar]
- 8.Binkowski TA, Joachimiak A. Protein functional surfaces: global shape matching and local spatial alignments of ligand binding sites. BMC Struct Biol. 2008;8:45. doi: 10.1186/1472-6807-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 10.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butland G, Peregrín-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 12.Butland G, Babu M, Díaz-Mejía JJ, Bohdana F, Phanse S, Gold B, Yang W, Li J, Gagarinova AG, Pogoutse O, Mori H, Wanner BL, Lo H, Wasniewski J, Christopolous C, Ali M, Venn P, Safavi-Naini A, Sourour N, Caron S, Choi JY, Laigle L, Nazarians-Armavil A, Deshpande A, Joe S, Datsenko KA, Yamamoto N, Andrews BJ, Boone C, Ding H, Sheikh B, Moreno-Hagelseib G, Greenblatt JF, Emili A. eSGA: E. coli synthetic genetic array analysis. Nat Methods. 2008;5:789–795. doi: 10.1038/nmeth.1239. [DOI] [PubMed] [Google Scholar]
- 13.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 15.Cho BK, Knight EM, Barrett CL, Palsson BO. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/ATtracts. Genome Res. 2008;18:900–910. doi: 10.1101/gr.070276.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 17.Cole RS. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc Natl Acad Sci U S A. 1973;70:1064–1068. doi: 10.1073/pnas.70.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Constantinou A, Chen X-B, McGowan CH, West SC. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO Journal. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeBoy RT, Mongodin EF, Emerson JB, Nelson KE. Chromosome evolution in the Thermotogales: large-scale inversions and strain diversification of CRISPR sequences. J Bacteriol. 2006;188:2364–2374. doi: 10.1128/JB.188.7.2364-2374.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diez-Villasenor C, Almendros C, Garcia-Martinez J, Mojica FJ. Diversity of CRISPR loci in Escherichia coli. Microbiology. 2010;156:1351–1361. doi: 10.1099/mic.0.036046-0. [DOI] [PubMed] [Google Scholar]
- 22.Garcia AD, Aravind L, Koonin EV, Moss B. Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc Natl Acad Sci USA. 2000;97:8926–8931. doi: 10.1073/pnas.150238697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garinis GA, Mitchell JR, Moorhouse MJ, Hanada K, de Waard H, Vandeputte D, Jans J, Brand K, Smid M, van der Spek PJ, Hoeijmakers JH, Kanaar R, van der Horst GT. Transcriptome analysis reveals cyclobutane pyrimidine dimers as a major source of UV-induced DNA breaks. Embo J. 2005;24:3952–3962. doi: 10.1038/sj.emboj.7600849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon GS, Wright A. DNA segragation in bacteria. Annual Review Microbiology. 2000;54:681–708. doi: 10.1146/annurev.micro.54.1.681. [DOI] [PubMed] [Google Scholar]
- 25.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han D, Krauss G. Characterization of the endonuclease SSO2001 from Sulfolobus solfataricus P2. FEBS Lett. 2009;583:771–776. doi: 10.1016/j.febslet.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 29.Han D, Lehmann K, Krauss G. SSO1450--a CAS1 protein from Sulfolobus solfataricus P2 with high affinity for RNA and DNA. FEBS Lett. 2009;583:1928–1932. doi: 10.1016/j.febslet.2009.04.047. [DOI] [PubMed] [Google Scholar]
- 30.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 32.Irizarry RA, Wang C, Zhou Y, Speed TP. Gene Set Enrichment Analysis Made Simple. John Hopkins University, Dept. of Biostatistics; 2009. Working Papers. : Working Paper 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishioka K, Fukuoh A, Iwasaki H, Nakata A, Shinagawa H. Abortive recombination in Escherichia coli ruv mutants blocks chromosome partitioning. Genes Cells. 1998;3:209–220. doi: 10.1046/j.1365-2443.1998.00185.x. [DOI] [PubMed] [Google Scholar]
- 34.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 35.Karginov FV, Hannon GJ. The CRISPR System: Small RNA-Guided Defense in Bacteria and Archaea. Mol Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller KL, Overbeck-Carrick TL, Beck DJ. Survival and induction of SOS in Escherichia coli treated with cisplatin, UV-irradiation, or mitomycin C are dependent on the function of the RecBC and RecFOR pathways of homologous recombination. Mutat Res. 2001;486:21–29. doi: 10.1016/s0921-8777(01)00077-5. [DOI] [PubMed] [Google Scholar]
- 37.Kidane D, Sanchez H, Alonso JC, Graumann PL. Visualization of DNA double strand breaks repair in live bacteria reveals dynamic recruitment of Bacillus subtilis RecF, RecO and RecN protein to distinct sites on the nucleoids. Molecular Microbiology. 2004;52:1627–1639. doi: 10.1111/j.1365-2958.2004.04102.x. [DOI] [PubMed] [Google Scholar]
- 38.Kowalczykowski SC. Initiation of recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 39.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Masson M, Baharoglu Z, Michel B. ruvA and ruvB mutants specifically impaired for replication fork reversal. Mol Microbiol. 2008;70:537–548. doi: 10.1111/j.1365-2958.2008.06431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lilley DMJ. Structural isomerization in DNA: the formation of cruciform structures in supercoiled DNA molecules. Chem Soc Rev. 1989;18:53–82. [Google Scholar]
- 43.Lilley DMJ, White MF. The junction-resolving enzymes. Nature Reviews Molecular Cell Biology. 2001;2:433–443. doi: 10.1038/35073057. [DOI] [PubMed] [Google Scholar]
- 44.Lindsey JC, Leach DR. Slow replication of palindrome-containing DNA. J Mol Biol. 1989;206:779–782. doi: 10.1016/0022-2836(89)90584-6. [DOI] [PubMed] [Google Scholar]
- 45.Lloyd RG, Benson FE, Shurvinton CE. Effect of ruv mutations on recombination and DNA repair in Escherichia coli K12. Mol Gen Genet. 1984;194:303–309. doi: 10.1007/BF00383532. [DOI] [PubMed] [Google Scholar]
- 46.Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010a;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010b;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meddows TR, Savory AP, Grove JI, Moore T, Lloyd RG. RecN protein and transcription factor DksA combine to promote faithful recombinational repair of DNA double-strand breaks. Mol Microbiol. 2005;57:97–110. doi: 10.1111/j.1365-2958.2005.04677.x. [DOI] [PubMed] [Google Scholar]
- 52.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mojica FJ, Diez-Villasenor C, Garcia-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 54.Mojica FJM, Ferrer C, Juez G, Rodríguez-Valera F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol Microbiol. 1995;17:85–93. doi: 10.1111/j.1365-2958.1995.mmi_17010085.x. [DOI] [PubMed] [Google Scholar]
- 55.Mojica FJM, Díez-Villaseñor C, García-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 56.Nair S, Finkel SE. Dps Protects Cells against Multiple Stresses during Stationary Phase. Journal of Bacteriology. 2004;186:4192–4198. doi: 10.1128/JB.186.13.4192-4198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Otsuji N, Iyehara H, Hideshima Y. Isolation and characterization of an Escherichia coli ruv mutant which forms nonseptate filaments after low doses of ultraviolet light irradiation. J Bacteriol. 1974;117:337–344. doi: 10.1128/jb.117.2.337-344.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, Severinov K. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol. 2010;77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 60.Proudfoot M, Sanders SA, Singer A, Zhang R, Brown G, Binkowski A, Xu L, Lukin JA, Murzin AG, Joachimiak A, Arrowsmith CH, Edwards AM, Savchenko AV, Yakunin AF. Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain. J Mol Biol. 2008;375:301–315. doi: 10.1016/j.jmb.2007.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 62.Sanchez H, Kidane D, Castillo Cozar M, Graumann PL, Alonso JC. Recruitment of Bacillus subtilis RecN to DNA Double-Strand Breaks in the Absence of DNA End Processing. Journal of Bacteriology. 2006;188:353–360. doi: 10.1128/JB.188.2.353-360.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sargentini NJ, Smith KC. Role of ruvAB genes in UV- and gamma-radiation and chemical mutagenesis in Escherichia coli. Mutat Res. 1989;215:115–129. doi: 10.1016/0027-5107(89)90224-8. [DOI] [PubMed] [Google Scholar]
- 64.Semenova E, Nagornykh M, Pyatnitskiy M, Artamonova II, Severinov K. Analysis of CRISPR system function in plant pathogen Xanthomonas oryzae. FEMS Microbiol Lett. 2009;296:110–116. doi: 10.1111/j.1574-6968.2009.01626.x. [DOI] [PubMed] [Google Scholar]
- 65.Sharples GJ, Chan SN, Mahdi AA, Whitby MC, Lloyd RG. Processing of intermediates in recombination and DNA repair: identification of a new endonuclease that specifically cleaves Holliday junctions. Embo J. 1994;13:6133–6142. doi: 10.1002/j.1460-2075.1994.tb06960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharples GJ. The X philes: structure-specific endonucleases that resolve Holliday junctions. Mol Microbiol. 2001;39:823–834. doi: 10.1046/j.1365-2958.2001.02284.x. [DOI] [PubMed] [Google Scholar]
- 67.Shen B, Qiu J, Hosfield D, Tainer JA. Flap endonuclease homologs in archaebacteria exist as independent proteins. Trends Biochem Sci. 1998;23:171–173. doi: 10.1016/s0968-0004(98)01199-2. [DOI] [PubMed] [Google Scholar]
- 68.Sherratt DJ, Lau IF, Barre F-Xa. Chromosome segregation. Current Opinion in Microbiology. 2001;4:653–659. doi: 10.1016/s1369-5274(01)00265-x. [DOI] [PubMed] [Google Scholar]
- 69.Sorek R, Kunin V, Hugenholtz P. CRISPR a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 70.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzuki H, Pangborn J, Kilgore WW. Filamentous cells of Escherichia coli formed in the presence of mitomycin. J Bacteriol. 1967;93:683–688. doi: 10.1128/jb.93.2.683-688.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sydorskyy Y, Srikumar T, Jeram SM, Wheaton S, Vizeacoumar FJ, Makhnevych T, Chong YT, Gingras AC, Raught B. A novel mechanism for SUMO system control: regulated Ulp1 nucleolar sequestration. Mol Cell Biol. 2010;30:4452–4462. doi: 10.1128/MCB.00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci U S A. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Huttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 76.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tseng YC, Hung JL, Wang TC. Involvement of RecF pathway recombination genes in postreplication repair in UV-irradiated Escherichia coli cells. Mutat Res. 1994;315:1–9. doi: 10.1016/0921-8777(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 78.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 79.van der Ploeg JR. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology. 2009;155:1966–1976. doi: 10.1099/mic.0.027508-0. [DOI] [PubMed] [Google Scholar]
- 80.Weinstein-Fischer D, Altuvia S. Differential regulation of Escherichia coli topoisomerase I by Fis. Mol Microbiol. 2007;63:1131–1144. doi: 10.1111/j.1365-2958.2006.05569.x. [DOI] [PubMed] [Google Scholar]
- 81.West SC. The RuvABC proteins and Holliday junction processing in Escherichia coli. J Bacteriol. 1996;178:1237–1241. doi: 10.1128/jb.178.5.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 83.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zahradka D, Vlahovic K, Petranovic M, Petranovic D. Chromosome segregation and cell division defects in recBC sbcBC ruvC mutants of Escherichia coli. J Bacteriol. 1999;181:6179–6183. doi: 10.1128/jb.181.19.6179-6183.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zdraveski ZZ, Mello JA, Marinus MG, Essigmann JM. Multiple pathways of recombination define cellular responses to cisplatin. Chem Biol. 2000;7:39–50. doi: 10.1016/s1074-5521(00)00064-8. [DOI] [PubMed] [Google Scholar]
- 86.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O’Toole GA. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol. 2009;191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zeghouf M, Li J, Butland G, Borkowska A, Canadien V, Richards D, Beattie B, Emili A, Greenblatt JF. Sequential Peptide Affinity (SPA) System for the Identification of Mammalian and Bacterial Protein Complexes. Journal of proteome research. 2004;3:463–468. doi: 10.1021/pr034084x. [DOI] [PubMed] [Google Scholar]
- 88.Zhang RG, Skarina T, Katz JE, Beasley S, Khachatryan A, Vyas S, Arrowsmith CH, Clarke S, Edwards A, Joachimiak A, Savchenko A. Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase. Structure. 2001;9:1095–1106. doi: 10.1016/s0969-2126(01)00675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.