Abstract
PURPOSE
To develop a technique for optimizing coverage of brain 3D 1H Magnetic Resonance Spectroscopic Imaging (MRSI) by automatic placement of outer-volume suppression (OVS) saturation bands (sat bands) and to compare the performance for PRESS MRSI protocols with manual and automatic placement of sat bands.
MATERIALS AND METHODS
The automated OVS procedure includes the acquisition of anatomic images from the head, obtaining brain and lipid tissue maps, calculating optimal sat band placement and then using those optimized parameters during the MRSI acquisition. The data were analyzed to quantify brain coverage volume and data quality.
RESULTS
3D PRESS MRSI data were acquired from 3 healthy volunteers and 29 patients using protocols that included either manual or automatic sat band placement. On average, the automatic sat band placement allowed the acquisition of PRESS MRSI data from 2.7 times larger brain volumes than the conventional method, while maintaining data quality.
CONCLUSION
The technique that has been developed helps solve two of the most significant problems with brain PRESS MRSI acquisitions: limited brain coverage and difficulty in prescription. This new method will facilitate routine clinical brain 3D MRSI exams and will be important for performing serial evaluation of response to therapy in patients with brain tumors and other neurological diseases.
Keywords: 3D 1H MRSI, Brain, Outer-volume suppression, Sat Band Optimization
INTRODUCTION
Proton Magnetic Resonance Spectroscopic Imaging (MRSI) is an important tool for diagnosis and evaluation of neurological diseases. MRSI data have been used for characterization of disease state, prediction of disease progression, treatment planning and evaluation of response to therapy (1–8). In particular, it has been shown that MRSI can detect the spatial extent of brain tumor more accurately than conventional T1 and T2-weighted MRI pulse sequences (9).
Excitation of subcutaneous lipids has been a major problem in 3D brain MRSI that has limited the coverage obtained. In the head, metabolites of interest, Choline, Creatine, Lactate and N-Acetyl-Aspartate (NAA) appear in concentrations on the order of 104 smaller than that of water or subcutaneous lipids (fatty tissues under the skin). If subcutaneous lipids are excited, a “ringing” artifact may appear in voxels within brain tissue due to side-lobes of the point-spread function of the excited protons (10).
Current MRSI protocols commonly use Point Resolved Spectroscopy (PRESS) volume selection, where a rectangular excitation volume results from an intersection of 3 infinite slabs, orthogonal to each other. The rectangular shape of the volume of interest requires that the region being excited is kept relatively small in order to avoid artifacts from the subcutaneous fat and sinuses. This limits the areas that can be evaluated and means that some metabolically-abnormal areas may be missed.
Several methods to suppress the subcutaneous lipid signal have been used in the past. Inversion recovery can be used to reduce the lipid signal by discriminating the tissues based on T1 relaxation times (11). This method, however, negatively affects the SNR of the acquisition (12). Other methods (13,14) use frequency-selective pulses to suppress the signal around the lipid resonance frequency. All those methods not only suppress the subcutaneous lipid signal, but also signal from the lipids within the brain. This is not desirable, since lipid in the tumor is a valuable biomarker in diseases, such as glioblastoma multiforme (15).
Manually placed sat bands are used to further reduce the lipid excitation. A sat band can eliminate most of the MR signal from a slab in space by excitation with radio-frequency (RF) pulses, such as the Very Selective Suppression pulses (VSS) (16), in the presence of slice-selection gradients and then de-phasing excited spins using crusher gradients. When PRESS excitation is performed, the area under the sat band cannot be excited any more until its longitudinal magnetization recovers. Manual placement of these sat bands is time-consuming and depends on the skill of the operator. Thus, it is hard to ensure consistently good placement of the sat bands, which is necessary to get useful MRSI data. This is one major reason why MRSI is currently not widely used in a clinical setting.
To improve coverage of the brain and to simplify prescription of 3D MRSI, we have developed a technique for automatic optimization of sat band position and orientation that simultaneously maximizes coverage of subcutaneous fat by the sat bands and minimizes the portion of brain tissue that is suppressed. This allows the prescription of a much larger PRESS box and provides MR spectra from a significantly larger volume of the brain. In this study we examine the performance of this technique.
METHODS
Overview
The technique that was developed included the acquisition of a volume of anatomic images from the head with high lipid/tissue contrast, obtaining brain and lipid tissue maps, calculation of optimal sat band placement to cover the lipids in that image volume and integration of those parameters into the spectroscopy pulse sequence.
To test the performance of the automatic sat band placement and reproducibility of the results in controlled experimental setup, data were acquired from 3 healthy volunteers (Table 2), each receiving two exams on different days, performed by different operators. Each exam contained 2 MRSI acquisitions: one with manually prescribed sat bands (STANDARD protocol, Table 1) and one with automatic sat band placement (AUTOSAT+OCTAGONAL protocol).
Table 2.
Reproducibility of technique, measured by acquiring data from 3 healthy volunteers.
| volunteer 1 | volunteer 2 | volunteer 3 | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 1 | 2 | 1 | 2 | |
| VS, cm3 | 224 | 324 | 324 | 288 | 324 | 324 |
| VA, cm3 | 404 | 515 | 403 | 500 | 505 | 570 |
| Voverlap, cm3 | 427 | 327 | 502 | |||
| Cho/Cre (S) | 1.009 | 1.044 | 1.198 | 1.177 | 1.286 | 1.290 |
| Cho/Cre (A) | 1.067 | 1.09 | 1.211 | 1.162 | 1.287 | 1.291 |
| NAA/Cre (S) | 1.985 | 2.093 | 2.103 | 2.078 | 2.196 | 2.139 |
| NAA/Cre (A) | 2.163 | 2.168 | 2.138 | 2.131 | 2.275 | 2.293 |
Each volunteer received two exams, prescribed by two different operators. Each exam contained STANDARD (S) and AUTOSAT+OCTAGONAL (A) MRSI acquisitions. Covered volumes (VS, VA) were defined as the volumes within the PRESS box, not covered by sat bands. Voverlap was calculated as intersection between the volumes, covered by AUTOSAT+OCTAGONAL acquisitions. Mean metabolite ratios were calculated within volumes, covered by all four acquisitions of the same subject.
Table 1.
3D MRSI acquisition protocols that were considered in this study
| STANDARD | OCTAGONAL | AUTOSAT | AUTOSAT+ OCTAGONAL |
|
|---|---|---|---|---|
| Fixed ROI-edge sat bands | ✓ | ✓ | ||
| Manually prescribed sat bands | ✓ | ✓ | ||
| Octagonal selection | ✓ | ✓ | ||
| Automatically prescribed sat bands | ✓ | ✓ | ||
| FOV (cm) | 16×16×16 | 16×16×16 | 18×18×16 | 18×18×16 |
| Lactate Editing | ✓ | ✓ | ||
| TR | 1104 | 1134 | 1500 | 1500 |
| Tacq (min) | 9 | 9 | 6.5 | 6.5 |
| Slices in PRESS box | 4 | 4 | 6 | 6 |
STANDARD = standard lactate edited MRSI, OCTAGONAL = lactate-edited MRSI with octagonal selection, AUTOSAT = MRSI with automatically generated sat band placement, AUTOSAT+OCTAGONAL = combination of automatic sat band placement and octagonal selection
To evaluate the technique as part of a clinical protocol, data were collected from 29 patients with brain tumors (Table 3), often acquiring two datasets per patient exam: one with automatic sat band placement and one without, for comparison. All subjects provided informed consent as approved by the Institutional Review Board (IRB).
Table 3.
Performance of the protocols in patients
| STANDARD | OCTAGONAL | AUTOSAT | AUTOSAT+ OCTAGONAL |
||
|---|---|---|---|---|---|
| N datasets | 12 | 13 | 10 | 14 | |
| Coverage (cm3) | 227.67±30.86 (224) | 322.62±60.01 (340) | 621.8±121.93 (672.5) | 612.64±88.33 (616.00) | |
| Cho | SNR | 8.27±3.27 (8.16) | 8.04±3.43 (8.14) | 8.41±3.43 (8.58) | 8.30±3.97 (8.06) |
| SNReff | 0.56±0.22 (0.56) | 0.54±0.23 (0.55) | 0.62±0.25 (0.63) | 0.61±0.29 (0.59) | |
| linewidth (Hz) | 6.65±2.21 (6.63) | 6.80±2.16 (6.76) | 6.78±2.15 (6.86) | 6.97±2.10 (7.11) | |
| Cre | SNR | 6.81±3.00 (6.97) | 6.36±3.01 (6.57) | 7.33±3.23 (7.57) | 6.49±3.42 (6.45) |
| SNReff | 0.55±0.24 (0.56) | 0.50±0.24 (0.52) | 0.61±0.27 (0.63) | 0.54±0.29 (0.54) | |
| linewidth (Hz) | 5.96±2.27 (5.80) | 6.09±2.26 (6.20) | 5.59±1.97 (5.69) | 5.70±2.02 (5.76) | |
| NAA | SNR | 13.07±5.33 (13.37) | 11.29±4.98 (11.69) | 14.67±6.05 (14.63) | 12.43±6.09 (13.03) |
| SNReff | 1.05±0.43 (1.07) | 0.89±0.39 (0.92) | 1.23±0.51 (1.23) | 1.04±0.51 (1.09) | |
| linewidth (Hz) | 7.20±2.05 (7.22) | 7.31±2.30 (7.12) | 7.43±2.63 (7.58) | 7.60±2.49 (7.66) | |
| Lipid | SNR | −0.04±1.55 (−0.07) | 0.15±2.48 (0.12) | −0.10±3.79 (−0.26) | −0.35±2.78 (−0.45) |
| SNReff | 0.00±0.07 (0.00) | 0.01±0.11 (0.01) | −0.01±0.21 (−0.01) | −0.02±0.16 (−0.03) | |
| Cho/Cre | 1.22±0.35 (1.24) | 1.26±0.41 (1.26) | 1.16±0.34 (1.13) | 1.28±0.41 (1.29) | |
| NAA/Cre | 1.91±0.56 (1.94) | 1.76±0.61 (1.78) | 1.99±0.57 (2.01) | 1.91±0.59 (1.92) | |
Mean coverage volumes, uncorrected metabolite SNR, SNR efficiency, linewidth and metabolite ratios and their standard deviations for the datasets, acquired from patients using the protocols that were compared (see table 1). Median values are shown in parentheses.
Image Acquisition
T1-weighted spoiled gradient echo axial image series (Fig. 1a) was obtained (flip angle = 20°, TE =2.1 ms, TR = 4.5 ms, acquisition matrix: 256×160, FOV: 240×150 mm slice thickness = 3 mm, 92 slices, Tacq ≈ 1 min) on a 3T MR scanner (GE Healthcare, Waukesha, WI, USA). Acquisition parameters were chosen to achieve maximum contrast between subcutaneous lipids and brain tissues. Images were automatically downloaded from the scanner to a separate computer, running Linux operating system for post-processing. Only the lowest 64 slices were used in the analysis since wrap-around artifacts from the neck were present in the top slices above the head.
Fig. 1.
(a) One slice from acquired image series with high lipid-brain contrast used for sat band placement calculation; (b,c) initial masks of segmented brain and fat; (d,e) brain and fat masks after post-processing.
Automatic Sat Band Placement
Image processing and sat band optimization algorithms were implemented in Matlab (The MathWorks, Natick, MA, USA). Lipid and brain tissue masks were generated using k-means clustering with k=3 (Fig. 1 b,c). K-means clustering is an iterative algorithm that aims to partition a given set of data (in our case, pixel values) into k clusters (e.g., air, brain tissue, lipids) by trying to minimize the distances between data points and cluster centroids (17). The masks were post-processed using morphological closing and opening to remove segmentation artifacts (Fig. 1 d,e). To reduce the computation time their resolution was decreased by a factor of 3 in X and Y directions. While more advanced algorithms could obtain more precise segmentation, this simple approach was chosen to achieve acceptable calculation time. Sat band placement algorithm required only a general shape of the brain and lipid layer to work robustly.
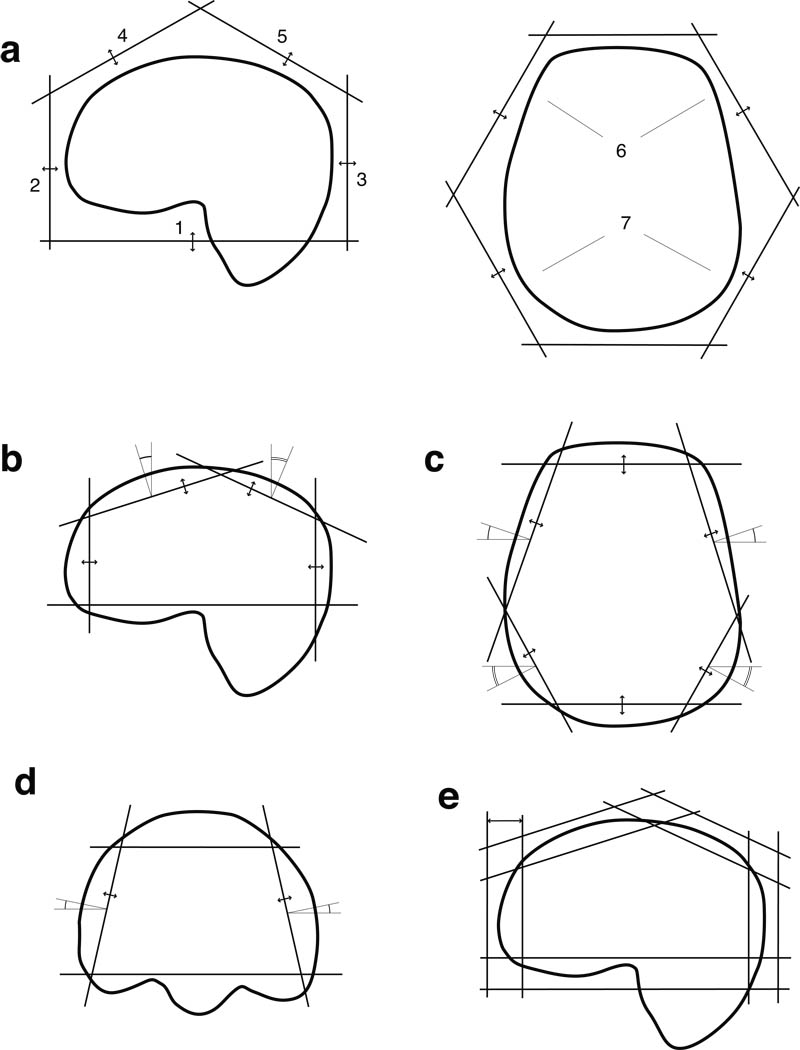
Position, orientation and thickness of 9 sat bands were calculated to achieve optimal lipid coverage. Placement of each sat band was defined by the following parameters: d - distance from origin, t - thickness, α - rotation angle around Z axis of a normal vector to the sat band plane, β - angle between the normal vector and Z axis. The sat bands were initially placed at the same distance from the center of the head in the following configuration: one band horizontally below the head, six bands forming a hexagon around the head and two bands above the head, tilted 30° in the anterior and posterior directions (Fig. 2, a).
Fig. 2.
The sequence of optimizations to obtain the sat band prescription consisted of: (a) the distances to individual sat bands were optimized in the order, indicated by the numbers; (b) optimization in the sagittal plane; (c) optimization in the axial plane; (d) tilt angle and distance optimization for side bands (first anterior ones, then - posterior ones); second sagittal optimization (not shown); (e) optimization of thickness of individual bands
A cost function, similar to that in (18), was defined to measure the quality of any given sat band configuration for a given image dataset:
| [1] |
where: wf, wb, wa, wd – weights, Nf, Nb, Na, - approximate the number of fat, brain, air pixels, covered by the bands, as described below, d – measure of distances of all bands from origin. Intuitively, this function reached its minimum when the sat bands covered most of the lipid, but as little brain tissue and air as possible. The distance term was added for regularization to discourage the configurations where the sat bands were placed far from the origin.
The cost function, as described above, was not continuous; there was a step whenever a sat band started covering a new pixel or stopped covering a pixel that was covered earlier. Non-continuous functions cause problems in optimization when the step size is small. To overcome this, Nf, which approximated the amount of lipid tissue, covered by sat bands, was redefined as:
| [2] |
where F is a set of fat pixels in the image and r(pi,sj) was a piecewise-linear function whose value was defined as follows: 0 if pixel pi is far outside sat band sj, 1 if it was deep inside the sat band; and interpolated linearly between 0 and 1 when the pixel was within distance δ of the sat band edge. Nb and Nf, which approximated the volume of brain tissue and air, covered by sat bands, were redefined similarly.
To find the optimal sat band placement the algorithm iteratively searched for parameters that minimized this cost function. The Nelder-Mead simplex method was chosen since it did not impose a requirement that the cost function be differentiable.
Due to the large number of degrees of freedom, simultaneous optimization of all parameters was impractical. Instead, optimization was done in several iterations on subsets of parameters (Fig. 2). During each step values of some parameters remained fixed, while other parameters were allowed to change to minimize the cost function. The optimum parameters, obtained in each step served as initial values for the next one. The sequence of optimizations included: 1) d (distance from origin) of individual sat bands (Fig. 2, a); 2) d, β of the bands, orthogonal to the sagittal plane (Fig. 2, b); 3) d, α of the bands, orthogonal to the axial plane (Fig. 2, c); 4) d, α, β of the same bands (Fig. 2, d); 5) repeat of step 2; 6) t (thickness) of individual bands, in the same order as in step 1 (Fig. 2, e).
For each stage, the weights were tuned to ensure the fastest convergence. To further reduce the number of parameters to optimize, the assumption was made that the problem was symmetrical in the right-left direction, so that a single parameter could control the distance or orientation of up to two symmetrical sat bands at the same time. The calculated parameters were written to a file and transferred back to the MRI scanner host computer.
MRSI Pulse sequence
This was modified to add up to 16 additional saturation pulses that could be controlled by externally generated prescription file. The parameters for all sat bands were read from the file and used to calculate gradient amplitudes, frequency offsets and rotation matrices for saturation pulses. All sat bands (fixed, manually prescribed and automatically-prescribed) used VSS pulses (16) with the following parameters: nominal B1: 0.116 G, pulse width: 3.0 ms, bandwidth: 5868 Hz. VSS pulse flip angles started at 91.5° for the last VSS pulse and increased by 1.5° for each earlier pulse, as implemented in the vendor pulse sequence, to compensate for T1 relaxation and minimize the longitudinal magnetization of saturated areas at the start of PRESS excitation. Cosine modulation could be turned on for any of the 16 additional pulses to achieve two parallel sat bands from one pulse. This was used to implement octagonal selection within the pulse sequence using 6 sat pulses (4 cosine-modulated, 2 regular) by automatically placing the sat bands (width = 4 cm) at the edges of the PRESS box and over the corners of the PRESS box in the axial plane (Fig. 5, purple) (19).
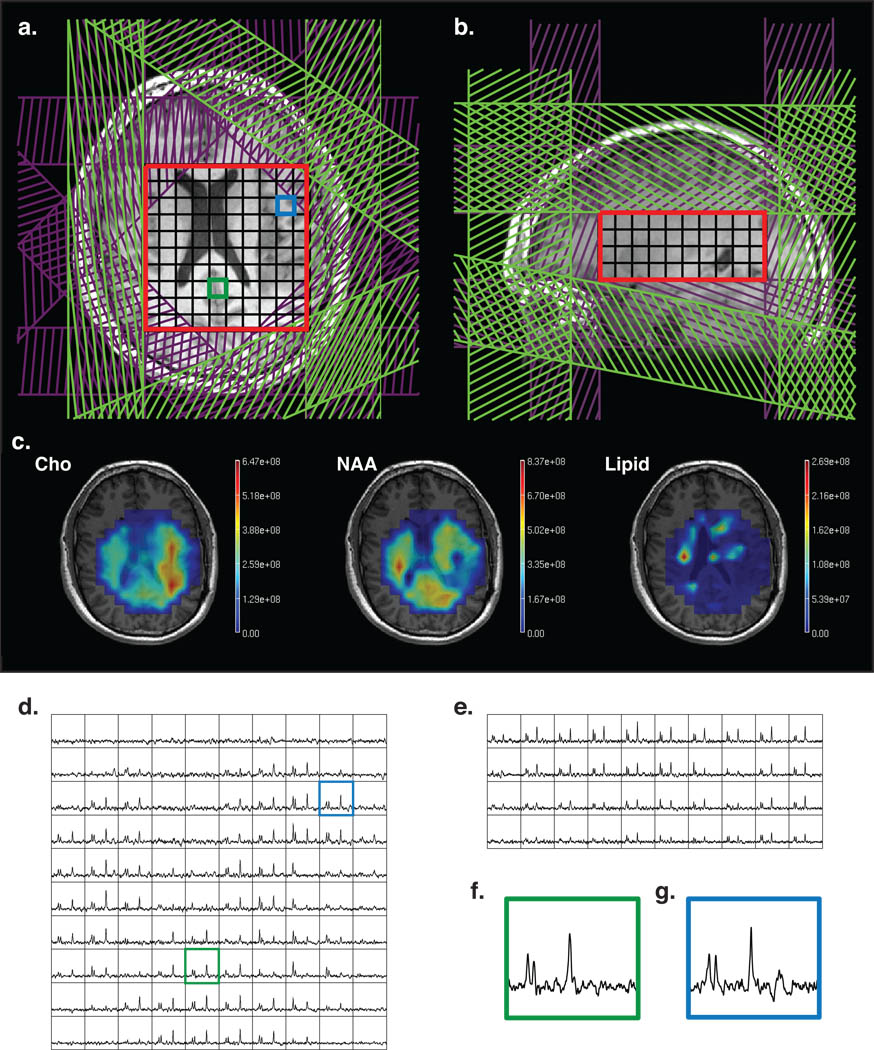
Fig 5.
(a, b) MRSI prescription with octagonal VSS sat bands (purple) and manually prescribed sat bands (green). PRESS volume is shown in red; (c) metabolite maps, overlaid on the anatomical images; (d, e) axial and sagittal slices through 3D MRSI data; (f,g) sample central (green) and periphery (blue) voxels (location is shown on (a) and (d)).
MRSI Acquisition
MRSI data were acquired using a commercially available 8-channel head coil (isotropic nominal voxel size 10 mm, TE = 144 ms, TR = 1100–1500 ms) with an EPSI flyback sequence (20). Raw data were transferred to a Linux workstation, then reconstructed and processed offline using an approach described previously in (9,21). Data from each channel was processed separately with 4Hz apodization, FFT in spatial and frequency dimensions, phase and frequency corrections, residual water and baseline removal. Data from all channels was combined using weights, obtained from low-resolution proton-density images.
The following acquisition protocols were used to acquire patient data (see table 1).
1. Standard MRSI (STANDARD)
This protocol was previously used for obtaining routine clinical and research brain patient exams. Six fixed sat bands with a width of 40 mm were implicitly applied by the pulse sequence at the superior, inferior, left, right, anterior and posterior edges of the PRESS box. PRESS box (SI dimension: 40 mm) was prescribed by the operator to cover the tumor and a portion of healthy brain tissue, while avoiding subcutaneous lipids as much as possible. Six additional sat bands were manually prescribed at the sides of the PRESS box to further suppress the signal from subcutaneous lipids. The sat bands were rotated or rearranged to improve suppression in areas where PRESS box was located close to the lipids, according to operator’s skill and experience. J-difference lactate editing scheme with dual BASING pulses (22) was used to provide summed data for Cho, Cre, NAA, lipids and difference data for lactate (not used in the current study, but needed for the clinical protocol). This scheme required two excitations for each k-space point, making acquisition time 9 min.
MRSI with octagonal selection (OCTAGONAL)
Octagonal selection (19) was used in place of fixed ROI edge sat bands to provide additional suppression of signal at the corners of the PRESS box. The operators were instructed to allow the corners of the PRESS box to extend over the lipid layer, while ensuring that those corners were covered by one or two additional manual sat bands.
MRSI with automatically generated sat band placement (AUTOSAT)
The PRESS box (SI dimension: 60 mm) was prescribed to cover as much of the brain as possible, with corners that often extended outside the brain. Operators used the images that overlaid the automatically placed sat bands over the brain anatomy to put the PRESS box over the area, enclosed by the sat bands. The matrix size was increased to 18×18×16 to accommodate the larger PRESS box. Fixed sat bands were implicitly placed at the edges of the PRESS box by the pulse sequence and there were 9 automatically optimized sat bands applied. Acquisition time was 6.5 minutes with one excitation per k-space point.
MRSI with automatically generated sat band placement and octagonal selection. (AUTOSAT+OCTAGONAL)
This was the same as option 3 but with octagonal selection instead of fixed sat bands.
Data Analysis
The data were analyzed to determine whether the new technique allowed increased coverage of the brain and whether the acquired data had similar quality to the data obtained with the standard protocol.
To determine the coverage volume, masks of PRESS volume and sat bands were generated using the parameters, extracted from the MRSI raw files and sat band prescription files. The coverage volume of an MRSI acquisition was defined as the number of 1cc voxels that lied within the PRESS box, but not in the areas, suppressed by sat bands. A voxel was considered to be suppressed by a sat band if more than 50% of its volume was within a sat band.
To quantify the quality of data and the amount of lipid contamination, peak height, linewidth and metabolite ratios of choline, creatine, NAA and lipid peaks were calculated within the healthy brain tissue (defined by white-matter mask, obtained with FMRIB's Automated Segmentation Tool, FAST (23)) based on predefined peak descriptor files that were adjusted using frequency correction parameters. Noise level was determined from the variance of the signal at the lower ppm end of the spectra. To correct for differences in protocol parameters, SNR efficiency values were calculated using total acquisition times and repetition times for each protocol and metabolite T1 relaxation times, estimated earlier (24,25). Mean, median values and pooled variances were calculated for each of the protocols that were compared.
To measure reproducibility of the results, the two exams from each volunteer were aligned to the same coordinate system using software, developed in or laboratory. For each pair of exams of the same subject, the overlap volume was calculated as the intersection of the volumes, covered by the AUTOSAT+OCTAGONAL acquisitions. Metabolite ratios were calculated within the volume, defined as intersection of the covered volumes of all 4 datasets of the same subject.
RESULTS
Figure 4 shows an example of a standard 3D MRSI dataset. Fixed ROI edge sat bands are shown in gray and manually prescribed sat bands in green. The PRESS box is 7×8×4 cm. Figure 5 shows an example of a dataset with octagonal selection, using cosine modulated sat bands (purple). Manual sat bands are shown in green. The PRESS box was made larger (10×10×4) due to better suppression in the corners. Figure 6 shows a dataset with automatic placement of sat bands (orange) and octagonal selection (purple). The prescribed PRESS box was 10×14×6 cm.
Fig. 4.
(a, b) Standard MRSI prescription with sat bands (gray) at the edge of the PRESS volume (red) and manually prescribed sat bands (green); (c) metabolite maps, overlaid on the anatomical images; (d, e) axial and sagittal slices through 3D MRSI data; (f,g) sample central (green) and periphery (blue) voxels (location is shown on (a) and (d)).
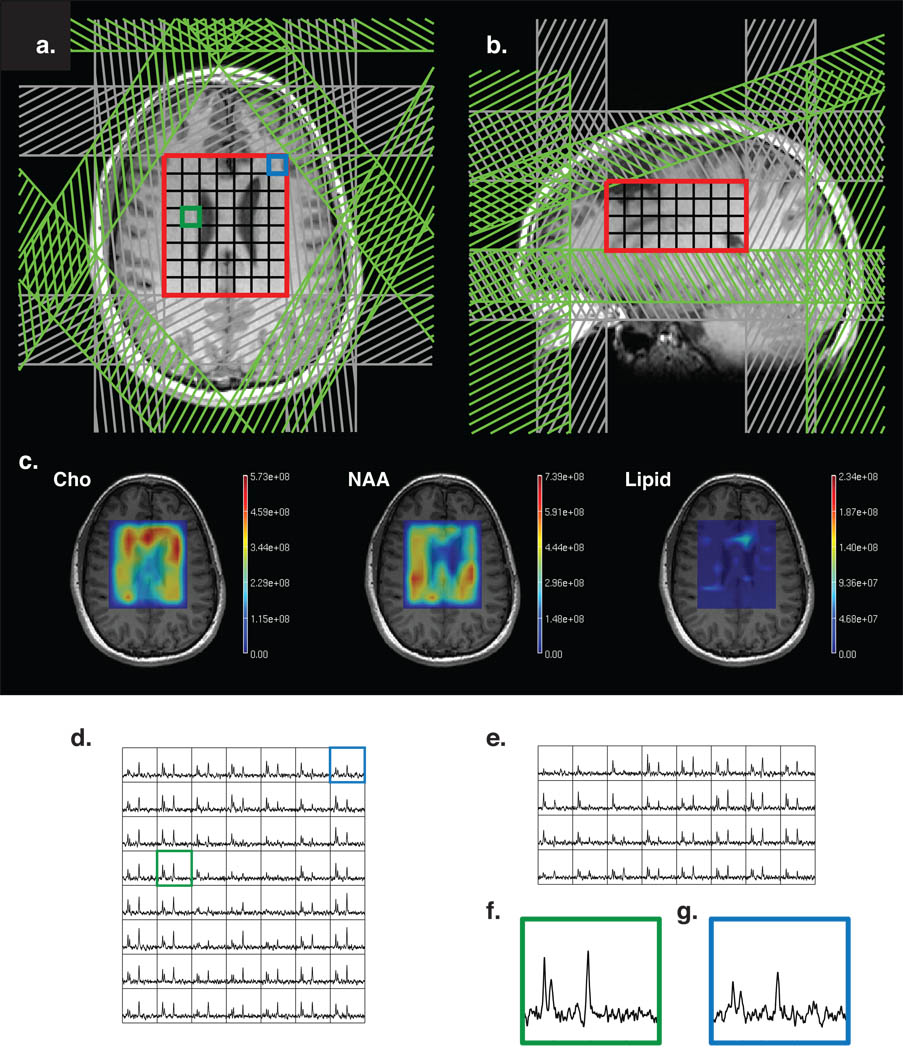
Fig. 6.
(a,b) MRSI dataset with automatically prescribed sat bands (orange), PRESS volume in red and octagonal selection in purple. (c) metabolite maps, overlaid on the anatomical images; (d, e) axial and sagittal slices through 3D MRSI data; (f,g) sample central (green) and periphery (blue) voxels (location is shown on (a) and (d)).
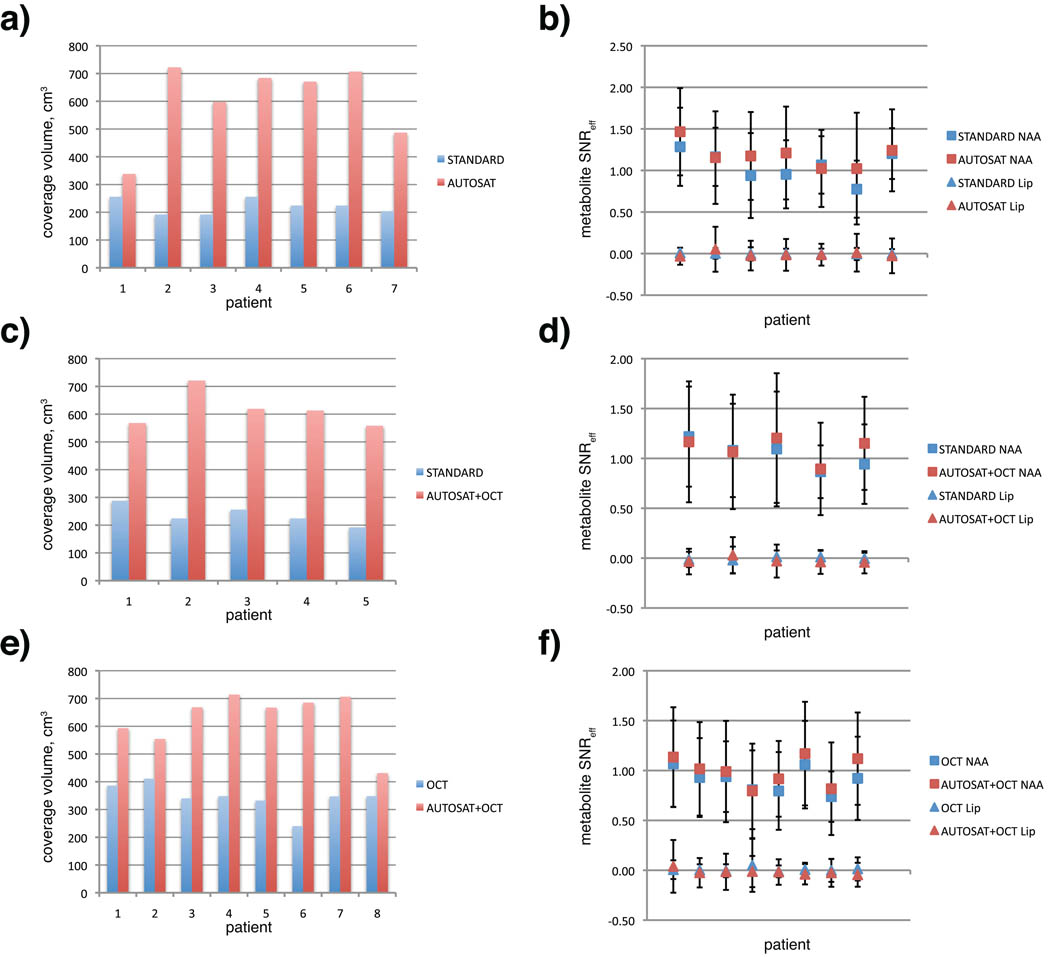
To quantify the observed improvement in coverage in patients, the results from the protocols were compared to those from the standard clinical 3D MRSI protocol. Table 3 shows mean, median and standard deviation for coverage volumes, metabolite signal-to-noise ratios (SNR), linewidth and metabolite ratios. Mean coverage for AUTOSAT protocol was 621.8 cm3 (s.d. = 121.93 cm3), compared to 227.67 cm3 for STANDARD protocol (s.d. = 30.86) - a 2.7 fold improvement. Pairwise coverage and average metabolite SNR of protocols with automatic and manual prescriptions for exams that had two MRSI acquisitions are shown on figure 7. In every case automatic prescription resulted in a larger coverage volume, while the average SNR values were consistent within each exam.
Fig. 7.
Pairwise comparison of coverage volume (cm3, a, c, e) and SNR efficiency of NAA and lipid peaks (b, d, f) for exams with two MRSI acquisitions: (a, b) STANDARD and AUTOSAT, (c, d) STANDARD and AUTOSAT+OCT, (e, f) OCTAGONAL and AUTOSAT+OCTAGONAL
The NAA signal shows no decline in data quality in datasets with bigger coverage. There was no serious increase in lipid contamination, although the variance of the lipid signal was slightly bigger in those datasets. Lipid leakage could be detected in voxels adjacent to the sat bands, but was small enough not to affect the NAA peak (fig 6g). That was expected due to increased lipid excitation, caused by proximity and overlap between the PRESS box and lipids. It also demonstrated satisfactory ability of the sat bands to minimize this excitation. Metabolite linewidth values show no degradation in spectral resolution in protocols with automatic sat band placement. Although the data does not show major difference in terms of coverage and data quality between AUTOSAT and AUTOSAT+OCTAGONAL protocols, the use of octagonal sat band configuration together with automatically-placed sat bands made the protocol more robust to PRESS box placement without any additional drawbacks.
To test the reproducibility of the protocol with automatic sat band placement in a controlled setting, results from the AUTOSAT+OCTAGONAL protocol, as well as STANDARD protocol were compared with those, acquired from the same healthy volunteer on a separate date (Table 2). The high overlap volume for exams with AUTOSAT+OCTAGONAL protocol indicated that the technique was able to cover much of the same volume despite different head position and different operators, running the scan. In all cases very similar metabolite ratios within the same anatomical region were observed.
DISCUSSION
Achieving large coverage volume, while maintaining low lipid contamination of the data by manually placing sat bands is time-consuming and requires a highly-skilled operator. Previously, Osorio et al developed an OVS scheme using octagonal selection with cosine-modulated VSS pulses (19). This allowed an increase in coverage within plane, but the curvature of the skull meant that it was only sufficient for single-slice acquisitions. Nevertheless, we found that using the octagonal sat band configuration together with manual sat bands (OCTAGONAL protocol) in place of a rectangular configuration (STANDARD protocol) improved the lipid suppression in the corners of the volume of interest in multi-slice MRSI acquisitions and allowed the operator to place a larger PRESS box closer to the edges of the brain, improving brain coverage.
Hovdebo and Ryner developed a technique that achieved non-cuboidal excited volume for single-voxel MRS by iteratively simplifying the polyhedral shape and using its faces to prescribe sat bands (26,27). 3D sat band optimization based on image data for spectroscopic imaging was introduced by Li et al in their 2006 ISMRM abstract (18) and later elaborated in a paper by Martinez-Ramon (28).
While initially based on the same idea of maximizing the number of lipid pixel and minimizing the number of brain pixels covered by the sat bands, described in (18), the technique used in our study had several major differences from the one implemented in (28).
One difference of our algorithm was that optimization was performed in several steps on subsets of parameters. Resulting values from one step were used as starting conditions for the next one. This made the algorithm find the optimal solution faster and, more importantly, prevented it from getting stuck in a local minimum. Fig. 3 (red, blue) shows how the algorithm converged on an optimal solution in a reasonable number of iterations. The other two curves (green, purple) show what would happen if all 36 unknowns were optimized simultaneously, using the same cost function and starting with the same initial configuration. While it was able to achieve some initial improvement, the optimization got stuck in a configuration that was likely a local minimum of the cost function and was not able to make much improvement after that. Also the cost function (eq. 1) contained the regularization term that discouraged configurations where no brain or lipid pixels were covered and the sat bands were far away from the origin. This obviated the need for a separate coarse placement of sat bands as in (28), since the initial distances to the sat bands could be calculated as a part of multi-step optimization with the same cost function.
Fig. 3.
Convergence of the optimization algorithm: blue and red - number of lipid and brain pixels, covered by sat bands using a multi-stage algorithm; green and purple - number of covered lipid and brain pixels using an algorithm in which all parameters are optimized simultaneously.
Another difference of this technique was the use of linear interpolation of covered volume to achieve a continuous cost function, since the number of covered pixels was inherently discrete. Even with interpolation, the cost function was not differentiable, so the gradient-based optimization methods could get stuck in areas where the gradient was not continuous. Instead, we chose to use Nelder-Mead simplex method, which did not use the gradient of the cost function.
A major part of the study was development of the pulse sequence that could use additional calculated sat bands. While the technique in (28) calculated the placement of 16 sat bands, only 8 could be used by the pulse sequence. In contrast, the pulse sequence used in the in-vivo experiments, described in this paper, used 9 calculated sat bands in addition to 10 fixed bands in octagonal configuration, implemented using cosine-modulated RF pulses. This allowed to effectively cover the top of of the head, as well as provide additional suppression in the corners of the PRESS box.
The study in (28) presented MRSI data from six healthy volunteers and showed sat band placement placement very similar to that of a highly-trained operator. The techniques developed in our study were able to achieve more optimal placement of sat bands than human operators did. That made possible to acquire good quality 3D MRSI data from a much larger volume of the brain than the current protocols. Optimal placement of the sat bands allowed the acquisition of MRSI data from tissues near the edge of the brain that were previously difficult to cover due to the curvature of the skull. It also ensured effective lipid suppression and allowed the PRESS box to extend beyond the boundary of the brain. Automatic placement of the sat bands was also designed to reduce the variability in the quality of the scan that is inherent for manual prescription. Repeated acquisition of data from the same subject with different head position and different operators (Table 2) shows very good reproducibility in terms of data quality and covered volume.
To verify that the technique works robustly in a clinical setting and allows for larger coverage volumes, we added an additional MRSI acquisition to the routine exams of patients with brain tumors. The data quality parameters (metabolite peak SNR, peak width, metabolite ratios) were calculated within the areas of healthy white matter to measure the effects of the technique itself on data quality as opposed to the variation in the parameters caused by the disease. These data show no compromise in data quality, while achieving significantly larger coverage volume. It was observed that the technique would bring the most benefit to the cases with multi-focal and heterogenous tumors. Fig. 6 shows one such case, which would have required two standard MRSI acquisitions to cover both disease sites, while the AUTOSAT+OCTAGONAL covered both areas of interest at the same time.
One limitation of the study was that the authors had no control over the population of patients, scheduled to receive scans at our facility, so few patients in the study had challenging tumors like this. Also, due to experimental nature of the protocols with automatic sat bands, acquired data was not used by the physicians treating the patients. Future studies will assess the effects of the technique on radiological interpretation of tumor spectra.
The protocols with automatic sat band placement may add time to the patient exam. The optimization algorithm can be made to work with standard T1-weighted images, making it unnecessary to acquire an extra image series for sat band optimization. Calculation for the sat band placement took around 3 minutes (2.8 GHz Intel Xeon computer), which was usually less time consuming than adjusting sat bands manually and was usually run while another image series was being acquired.
The largest limiting factor encountered in our study was signal drop-off in the anterior of the lower slices, presumably due to field inhomogeneity caused by tissue-air interface in sinuses. This meant that tumors below the level of the eye-sockets could not be covered. This effect can be reduced by placing the PRESS box obliquely and performing high-order shimming over an oblique volume.
In conclusion, the technique described in this study has helped solve two of the most significant problems with brain PRESS MRSI acquisitions: limited brain coverage and difficulty in prescription. The improved coverage will be useful for evaluating heterogeneous and infiltrative tumors, as well as tumors at the periphery of the brain, which are difficult to evaluate with current protocols. It should make possible a more accurate assessment of the progression of tumors in serial studies. The use of this technique reduces the need for extensive operator training, thus facilitating wider utilization of MRSI in the clinical setting.
ACKNOWLEDGEMENTS
We would like to thank Adam Kerr and Joseph Osorio for help with pulse sequence development, Jason Crane for help with computing infrastructure and data processing software, Adam Elkhaled, Wei Bian, Trey Jalbert and Emma Essock-Burns for help with data acquisition and processing.
Grant Support:
UC Discovery grant ITL-BIO04-10148 in conjunction with GE Healthcare, NIH R01 CA127612 and NIH R01 CA111291.
REFERENCES
- 1.Di Costanzo A, Scarabino T, Trojsi F, et al. Proton MR spectroscopy of cerebral gliomas at 3 T: spatial heterogeneity, and tumour grade and extent. Eur Radiol. 2008;18(8):1727–1735. doi: 10.1007/s00330-008-0938-5. [DOI] [PubMed] [Google Scholar]
- 2.Xu M, See SJ, Ng WH, et al. Comparison of magnetic resonance spectroscopy and perfusion-weighted imaging in presurgical grading of oligodendroglial tumors. Neurosurgery. 2005;56(5):919–926. discussion 919–926. [PubMed] [Google Scholar]
- 3.Rijpkema M, Schuuring J, van der Meulen Y, et al. Characterization of oligodendrogliomas using short echo time 1H MR spectroscopic imaging. NMR Biomed. 2003;16(1):12–18. doi: 10.1002/nbm.807. [DOI] [PubMed] [Google Scholar]
- 4.Hattingen E, Delic O, Franz K, et al. (1)H MRSI and progression-free survival in patients with WHO grades II and III gliomas. Neurol Res. 2009 doi: 10.1179/016164109X12478302362770. [DOI] [PubMed] [Google Scholar]
- 5.Kuznetsov YE, Caramanos Z, Antel SB, et al. Proton magnetic resonance spectroscopic imaging can predict length of survival in patients with supratentorial gliomas. Neurosurgery. 2003;53(3):565–574. doi: 10.1227/01.neu.0000079331.21178.8e. discussion 574–566. [DOI] [PubMed] [Google Scholar]
- 6.Chang SM, Nelson S, Vandenberg S, et al. Integration of preoperative anatomic and metabolic physiologic imaging of newly diagnosed glioma. J Neurooncol. 2009;92(3):401–415. doi: 10.1007/s11060-009-9845-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narayana A, Chang J, Thakur S, et al. Use of MR spectroscopy and functional imaging in the treatment planning of gliomas. Br J Radiol. 2007;80(953):347–354. doi: 10.1259/bjr/65349468. [DOI] [PubMed] [Google Scholar]
- 8.Sajja BR, Wolinsky JS, Narayana PA. Proton magnetic resonance spectroscopy in multiple sclerosis. Neuroimaging Clin N Am. 2009;19(1):45–58. doi: 10.1016/j.nic.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson SJ. Analysis of volume MRI and MR spectroscopic imaging data for the evaluation of patients with brain tumors. Magn Reson Med. 2001;46(2):228–239. doi: 10.1002/mrm.1183. [DOI] [PubMed] [Google Scholar]
- 10.Patel MS, Hu X. A robust algorithm for reduction of truncation artifact in chemical shift images. IEEE Trans Med Imaging. 1993;12(4):812–818. doi: 10.1109/42.251133. [DOI] [PubMed] [Google Scholar]
- 11.Spielman DM, Pauly JM, Macovski A, Glover GH, Enzmann DR. Lipid-suppressed single- and multisection proton spectroscopic imaging of the human brain. J Magn Reson Imaging. 1992;2(3):253–262. doi: 10.1002/jmri.1880020302. [DOI] [PubMed] [Google Scholar]
- 12.Hetherington HP, Pan JW, Mason GF, et al. 2D 1H spectroscopic imaging of the human brain at 4.1 T. Magn Reson Med. 1994;32(4):530–534. doi: 10.1002/mrm.1910320417. [DOI] [PubMed] [Google Scholar]
- 13.Star-Lack J, Nelson SJ, Kurhanewicz J, Huang LR, Vigneron DB. Improved water and lipid suppression for 3D PRESS CSI using RF band selective inversion with gradient dephasing (BASING) Magn Reson Med. 1997;38(2):311–321. doi: 10.1002/mrm.1910380222. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H, Ouwerkerk R, Barker PB. Dual-band water and lipid suppression for MR spectroscopic imaging at 3 Tesla. Magn Reson Med. 2010;63(6):1486–1492. doi: 10.1002/mrm.22324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford FW, Khayal IS, McGue C, et al. Relationship of pre-surgery metabolic and physiological MR imaging parameters to survival for patients with untreated GBM. J Neurooncol. 2009;91(3):337–351. doi: 10.1007/s11060-008-9719-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran TK, Vigneron DB, Sailasuta N, et al. Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer. Magn Reson Med. 2000;43(1):23–33. doi: 10.1002/(sici)1522-2594(200001)43:1<23::aid-mrm4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 17.Seber GAF. Multivariate observations. New York: Wiley; 1984. p. 686. [Google Scholar]
- 18.Li T, Martinez-Ramon M, Heileman G, Posse S. Automatic Outer Volume Suppression (OVS) Slice Placement for Proton-Echo-Planar-Spectroscopic-Imaging (PEPSI). Proceedings of the 14th Annual Meeting of ISMRM; 2006. (abstract 3086) [Google Scholar]
- 19.Osorio JA, Xu D, Cunningham CH, et al. Design of cosine modulated very selective suppression pulses for MR spectroscopic imaging at 3T. Magn Reson Med. 2009;61(3):533–540. doi: 10.1002/mrm.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham CH, Vigneron DB, Chen AP, et al. Design of flyback echo-planar readout gradients for magnetic resonance spectroscopic imaging. Magn Reson Med. 2005;54(5):1286–1289. doi: 10.1002/mrm.20663. [DOI] [PubMed] [Google Scholar]
- 21.Osorio JA, Ozturk-Isik E, Xu D, et al. 3D 1H MRSI of brain tumors at 3.0 Tesla using an eight-channel phased-array head coil. J Magn Reson Imaging. 2007;26(1):23–30. doi: 10.1002/jmri.20970. [DOI] [PubMed] [Google Scholar]
- 22.Star-Lack J, Spielman D, Adalsteinsson E, Kurhanewicz J, Terris DJ, Vigneron DB. In vivo lactate editing with simultaneous detection of choline, creatine, NAA, and lipid singlets at 1.5 T using PRESS excitation with applications to the study of brain and head and neck tumors. J Magn Reson. 1998;133(2):243–254. doi: 10.1006/jmre.1998.1458. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Srinivasan R, Ratiney H, Lu Y, Chang SM, Nelson SJ. Comparison of T(1) and T(2) metabolite relaxation times in glioma and normal brain at 3T. J Magn Reson Imaging. 2008;28(2):342–350. doi: 10.1002/jmri.21453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han E, Gold G, Stainsby J, Wright G, Beaulieu C, Brittain J. In-Vivo T1 and T2 Measurements of Muskuloskeletal Tissue at 3T and 1.5T. Proceedings 11th Scientific Meeting, International Society for Magnetic Resonance in Medicine; Toronto. 2003. (abstract 450) [Google Scholar]
- 26.Hovdebo J, Ryner L. An Improved Method for Automatic Placement of Spatial Saturation Planes in MR Spectroscopy. Proceedings of the 17th Annual Meeting of ISMRM; Honolulu. 2009. (abstract 2376) [Google Scholar]
- 27.Ryner L, Westmacott G, Davison N, Latta P. Automated Positioning of Multiple Spatial Saturation Planes for Non-Cuboidal Voxel Prescription in MR Spectroscopy. Proceedings of the 13th Annual Meeting of ISMRM; Miami. 2005. (abstract 350) [Google Scholar]
- 28.Martinez-Ramon M, Gallardo-Antolin A, Cid-Sueiro J, et al. Automatic placement of outer volume suppression slices in MR spectroscopic imaging of the human brain. Magn Reson Med. 2010;63(3):592–600. doi: 10.1002/mrm.22275. [DOI] [PubMed] [Google Scholar]