Abstract
A tRNA gene-like sequence has been identified near the 3’ end of HIV-1. Two segments of this sequence (motif 9 and segment 1) promoted minus strand transfer in vitro. The segments are complementary to the tRNA3Lys primer, and apparently act by binding the tRNA, thereby bringing the 3’ and 5’ ends of viral RNA into proximity for strand transfer. In this report, we used full-length HIV-1 to demonstrate biological relevance of these segments. We constructed HIV-1 genomes capable of single cycle infection and altered in one or both of two segments. We devised a real time PCR method for quantifying the amount of (-)ssDNA that completes transfer. Results showed that depending on the mutation the efficiency of transfer decreased 9% to 26%. Alteration of segment 1 had the greatest effect. Alteration of motif 9 or both sequences also caused a reduction, but smaller than alteration of segment 1 alone.
Keywords: HIV-1, minus strand DNA transfer, tRNA3Lys
Introduction
Upon human immunodeficiency virus 1 (HIV-1) infection, the single stranded viral RNA genome is converted by reverse transcription to double stranded DNA and integrated into host nuclear DNA (Fig. 1A). Conversion to DNA requires the synthesis of long terminal repeats (LTRs) that flank the protein-coding region. The need to make these regions requires two transfers of the growing DNA strands. The first (minus) strand transfer occurs shortly after reverse transcription starts at the primer binding site (PBS) near the 5’ end of viral RNA. HIV-1 uses human tRNA3Lys as the primer and the viral reverse transcriptase (RT) extends it 180 nucleotides to the 5’ end of the RNA creating an intermediate called the minus-strand strong-stop DNA ((-)ssDNA). The RT ribonuclease H (RNase H) degrades the RNA template, rendering portions of the DNA single stranded. That DNA segment is subsequently transferred to the 3’ end of the viral RNA to continue reverse transcription (Basu et al., 2008).
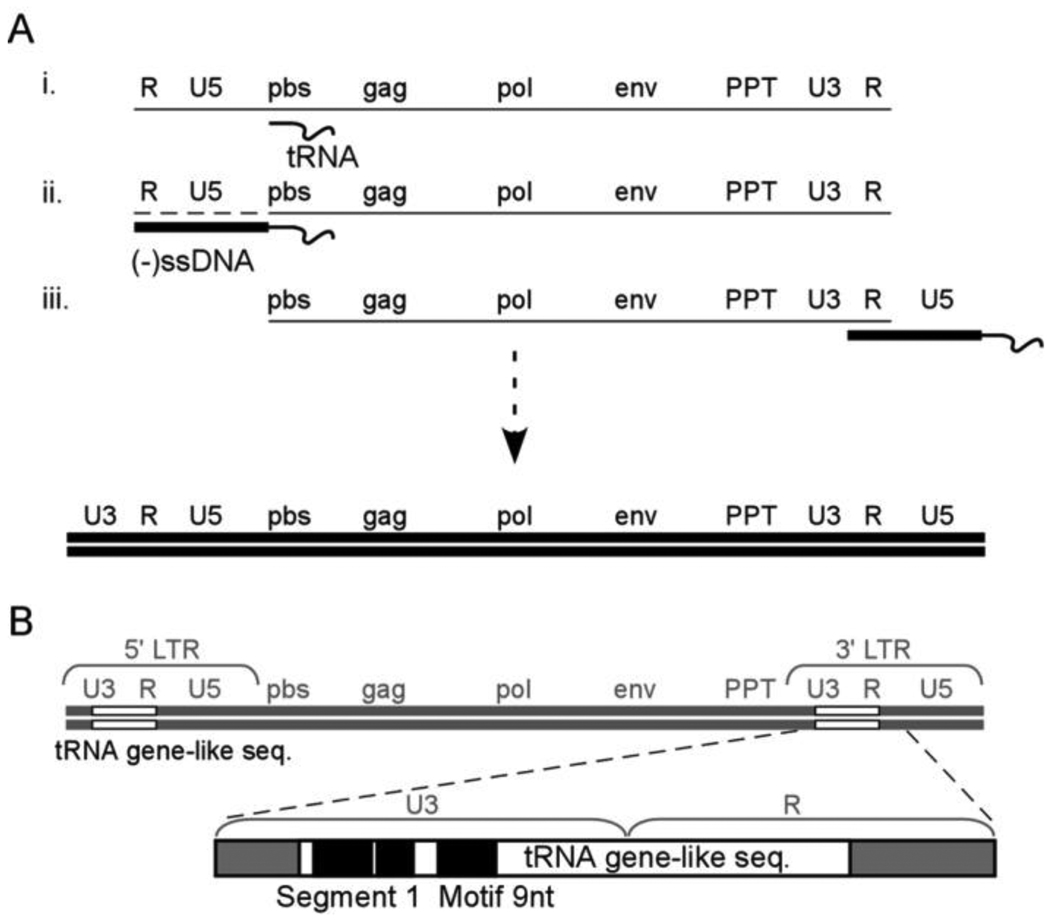
Fig. 1.
(A) Reverse transcription of the HIV-1 RNA genome. (i) Conversion of the RNA genome (thin line) into DNA starts by synthesis of minus strand DNA initiated from a tRNA3Lys primer annealed to the PBS. (ii) The U5 and R elements are copied into cDNA, generating (-)ssDNA (thick line). The 5’ end of the genomic RNA is degraded (dashed line) by the RNase H activity of RT. (iii) Interaction between the R element at 3’ end of the RNA genome and the R sequence in (-)ssDNA allows first (minus) strand DNA transfer. The following steps (not indicated in the drawing) take place to complete synthesis of the proviral DNA genome. Minus strand DNA synthesis is continued until the PBS region. The RNase H activity of RT degrades the RNA except the PPT fragment, which is subsequently used as a primer to start plus strand DNA synthesis. The 3’ end region of tRNA3Lys is copied into plus strand DNA. Complementarity between the PBS region of plus and minus strand DNAs allows second (plus) strand DNA transfer and completion of viral DNA synthesis.
(B) A sequence similar to tRNA3Lys gene is embedded within the U3–R regions of the HIV-1 genome and the LTRs of the double stranded viral DNA. Segment 1 and motif 9nt in U3, which stimulate minus strand DNA transfer in vitro, are complementary to the 3’ region of tRNA3Lys and nucleotides at positions 38-46, respectively.
RNA structure, RNase H activity of RT, nucleocapsid protein (NC), and primer tRNA3Lys, were all indicated by reconstitution experiments to be important factors in efficient minus strand DNA transfer (Brule et al., 2000; Chen et al., 2003a; Negroni and Buc, 2000; Peliska and Benkovic, 1992; Piekna-Przybylska et al., 2010; Rodriguez-Rodriguez et al., 1995; Song et al., 2009; You and McHenry, 1994). Since this is a crucial step of viral replication, we assume that it has evolved to occur with the maximum efficiency attainable by the virus. Terminal repeat (R) elements at both ends of the genomic RNA provide the sites for initial synthesis and then transfer of the (-)ssDNA (Gilboa et al., 1979). Complementarity within the R elements between (-)ssDNA and 3’ end of HIV-1 genomic RNA allows interaction during invasion-driven minus strand transfer, which could lead to transient circularization of the very long (~10kb) RNA to bring the two R regions into proximity for transfer.
The primer tRNA3Lys was proposed to aid this process, serving as a bridging factor holding RNA genome ends together (Brule et al., 2000). The mechanism was proposed to involve complementary interactions between the primer tRNA and a 9 nt sequence in the U3 region at the 3’ end of the viral RNA. Moreover, the presence of the 9 nt sequence was shown to promote minus strand transfer modeled in vitro (Brule et al., 2000). Our recent studies revealed the striking result that the 9 nt sequence is part of a much larger sequence with strong homology to the entire tRNA3Lys (Piekna-Przybylska et al., 2010) (Fig. 1B). This sequence stretches over regions surrounding the U3 and R border. We also used strand transfer assays to assess the influence of the newly discovered sequence. These traditional assays involved measuring the transfer efficiency of a tRNA3Lys primed (-)ssDNA from a donor RNA template to an acceptor template having or lacking regions of the tRNA-like sequence (Piekna-Przybylska et al., 2010). Analyses in vitro revealed that the original 9 nt segment “motif 9nt” and a nearby sequence “segment 1”, both promote transfer, and work most effectively when present together (Basu et al., 2008; Piekna-Przybylska et al., 2010). Motif 9nt in U3 is adjacent to R element complementary to nucleotides at positions 38–46 in the tRNA3Lys. Segment 1 is located just upstream of motif 9nt and resembles the PBS, which interacts with 3’ end of tRNA3Lys. Since the efficiency of minus strand transfer depends on many factors we were interested to know whether these tRNA-like sequences add to other factors to produce a significant incremental increase in transfer efficiency.
In the present studies we analyzed HIV-1 minus strand transfer in cell culture making use of plasmids carrying full-length, infectious HIV-1 sequences. These were designed to produce single-cycle, recombinant-virus infections. We introduced mutations in U3 within motif 9nt and segment 1. The wild-type and mutant viruses were generated and subsequently used for infection. Minus strand transfer was monitored using real-time PCR analysis. The experimental design and the quantitative results are presented, interpreted, and compared to results obtained in vitro.
Results
Experimental design
In order to quantify the role of U3 sequences in strong-stop minus strand DNA transfer in vivo, we introduced mutations in the 3’ LTR of a full-length HIV-1 genome. We adapted the plasmid vector pAT1, which has been used in the previous studies and contains the sequence of the pNL4-3 HIV-1 strain (Dykes et al., 2006). Since our altered sequences in U3 overlap with sequences of HIV-1 promoter in the 5’ LTR, the vector was modified to allow only one round of infection and replication. This was accomplished by deleting 700 nucleotides of the ENV gene in pAT1, producing pDAT1. A single cycle of virus replication prevents creation of next generation of viruses with additional mutations within the promoter of 5’ LTR, inherited during retroviral replication from 3’ LTR.
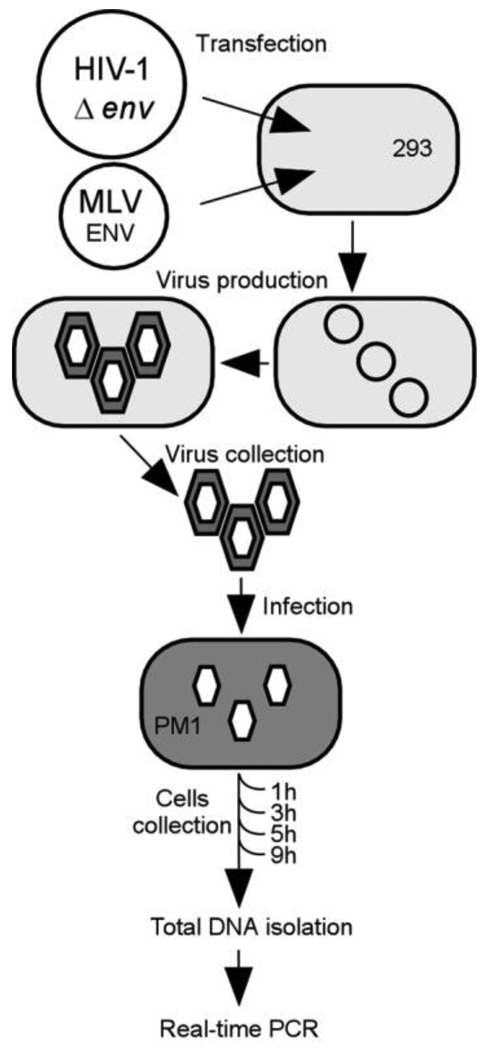
In order to produce infectious virus, the 293 cells were co-transfected with virus DNA and a vector harboring a gene for the MLV envelope protein (Fig. 2). The wild-type and recombinant viruses were harvested after 72 h and analyzed for concentration by using an HIV-1 virus capsid protein p24 ELISA assay. PM1 cells were used for separate viral infection with each mutant virus stock and a wild-type virus as a reference strain. The infected cells were collected after 1 h, 3 h, 5 h and 9 h and total DNA was isolated to be used in quantitative PCR (qPCR) to measure minus strand transfer (Fig. 2).
Fig. 2.
Experimental approach diagram. For virus production virus DNA plasmid and producing ENV protein (MLV) plasmid were co-transfected into 293 cells. After 72 h, viruses were collected and titrated by ELISA with anti-p24 capture antibody. Before infection of PM1 cells, viral stocks were treated with TURBO DNase. Cells were collected 1 h, 3 h, 5 h and 9 h after infection and total DNA was extracted for real-time PCR analysis.
In our experiments, we employed two approaches to reduce transfected plasmid DNA contamination, which can contribute to the signal in real-time PCR. First, to avoid carryover of residual virus plasmid left over from transfection, the virus stocks were treated with Turbo DNase before the infection of PM1 cells (Lu et al., 2004). Second, the total DNA, isolated from samples collected after infection was treated with DpnI endonuclease, which selectively destroys the hemi-methylated DNA (Mbisa et al., 2009). In addition, the DNA samples were tested by PCR reaction with primers specific for the pUC vector to determine a level of plasmid contamination.
Mutagenesis of 3’ LTR U3 in recombinant viruses
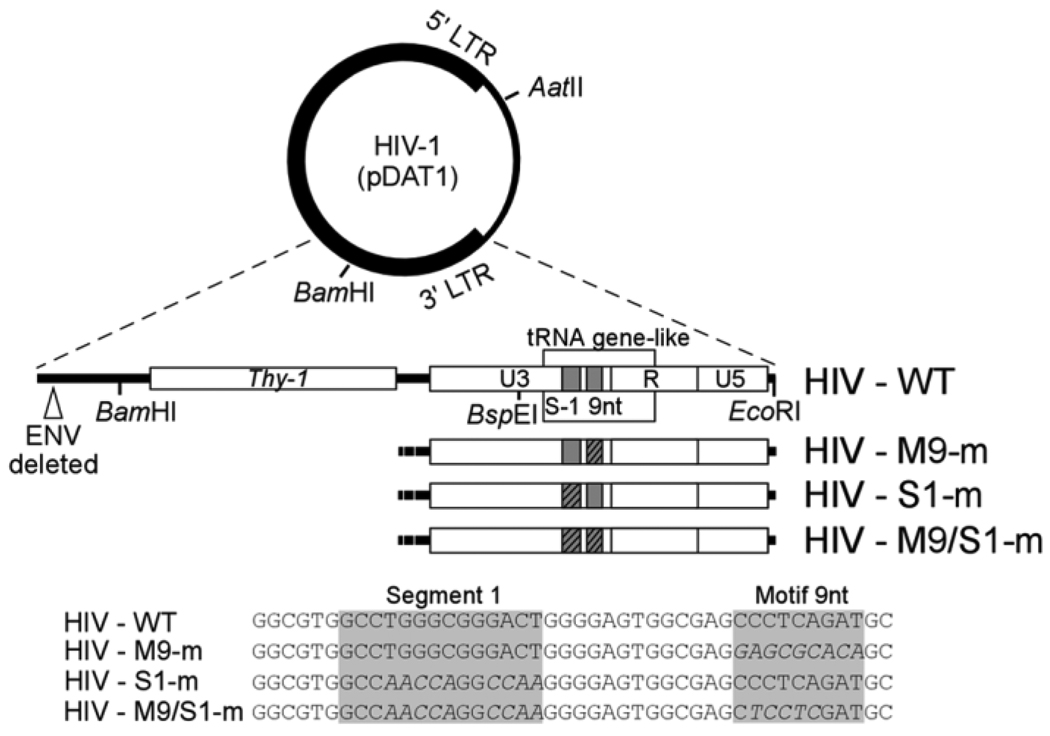
The tRNA gene-like sequence similar to tRNA3Lys is contained within the U3 and R elements of the HIV-1 LTR (Piekna-Przybylska et al., 2010). Based on our previous analyses in vitro two regions of tRNA gene-like sequence in U3, motif 9nt and segment 1, promoted minus strand DNA transfer. We constructed three mutant HIV-1 viruses, two with alterations in a single segment in U3 of 3’ LTR, and one with mutations in both segments (Fig. 3). The viruses were named M9-m and S1-m for mutants with altered sequences in motif 9nt and segment 1, respectively. Virus M9/S1-m has mutations in both segments. The unique BamHI and AatII sites of pDAT1 were used to subclone the fragment of HIV-1 with the 3’ LTR into pUC18 plasmid to generate pD1 for subsequent steps in producing recombinant HIV viruses. We began this preparation by using a PCR approach with application of oligonucleotides harboring altered sequences within the U3 region. Two overlapping DNA fragments were generated and used together to amplify one longer product for subsequent subcloning into unique BspEI/EcoRI sites of pD1. From new constructs of pD1 with the desired mutations in U3, we transferred the BamHI/AatII fragment back to the rest of HIV-1 genome in pDAT1. We produced separate virus stocks for each mutant virus and wild-type virus by transfection in 293 cells. The viral stocks were analyzed by quantifying p24 capsid protein by ELISA. In order to exclude the possibility that mutations in the genome might have effects in the encapsidation of viral RNA, a quantitative measurement of HIV nucleic acid (Viral Load Assay) was performed on viruses treated with TURBO DNase. The similar level of detected p24 capsid protein and RNA genome for wild-type and mutant viruses (Table 1) indicated that mutations in U3 of 3’ LTR did not have an effect on virus production.
Fig. 3.
Plasmid constructs used to analyze the importance of segments in 3’ U3 complementary to tRNA3Lys. The pDAT1 construct was used to create three recombinant viruses, M9-m, S1-m and M9/S1-m by mutation of U3 sequences in the 3’ LTR. Gray boxes show the positions of segment 1 and motif 9nt. Dash lines in boxes indicate the mutated segments of U3. The sequences of segment 1 and motif 9nt for wild-type and mutant strains are indicated below.
Table 1.
Concentration of WT and mutant viruses determined by p24 ELISA and Viral Load Assay (VLA).
| ELISA (p24 ng/ml) |
VLA (number of HIV copies/ml) |
|
|---|---|---|
| WT | 108 | 4.62 × 105 |
| M9-m | 159 | 6.70 × 105 |
| S1-m | 102 | 5.15 × 105 |
| M9/S1-m | 85 | 3.87 × 105 |
Monitoring minus strand DNA transfer in vivo using real-time PCR
Reverse transcription of HIV is a multi-step process in which two template switches occur to generate full-length genomic DNA. A real-time PCR assay can be applied to measure the time-dependent increase in the amounts of specific DNA intermediates synthesized in the cell at different steps during reverse transcription. The approach we took was to measure the production of (-)ssDNA, the starting substrate for minus strand transfer, and the amount of extended (-)ssDNA that could only be created after transfer. The ratio of the shorter over the longer product is the transfer efficiency. The advantage of obtaining results as a ratio is that the approach corrects for virtually all envisioned experimental inconsistencies of conditions and concentrations.
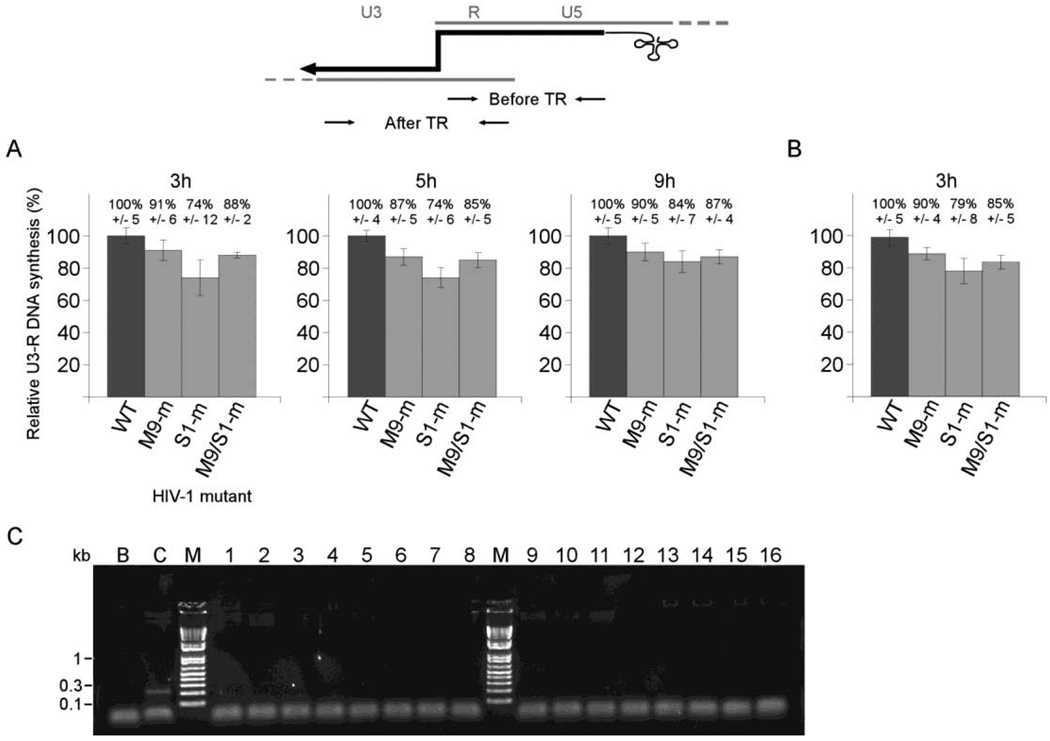
In order to quantify minus strong stop DNA transfer, PCR primers had to be designed for use within U3, R and U5 (Fig. 4). During the first step of reverse transcription, the U5 and R element sequences of viral RNA are synthesized as minus-strand DNA. To monitor the step before transfer by real-time PCR, we designed primers to bind within this region and designated the PCR product as R-U5 DNA. To amplify the longer transfer product, a primer was designed to bind in U3, beyond motif 9nt and segment 1, which are copied into extended minus-strand DNA after first strand transfer occurs. This was matched with a primer that bound the R region, so that a sequence spanning the site of transfer would be amplified. This PCR product was designated U3-R DNA.
Fig. 4.
Comparison of efficiency of minus strand transfer in vivo for U3 HIV-1 mutants. Graphs in (A) and (B) represent results from two independent infection experiments. Total DNA was extracted at different times post-infection from PM1 cells infected with wild-type and three U3 mutant HIV-1 viruses. Viral DNA intermediates were monitored by real-time PCR, as described in the text and in Materials and Methods. The approximate binding positions of the real-time PCR primers used to monitor DNA before and after transfer (TR) are indicated. The amount of the U3-R DNA amplified from the minus strand transfer cDNA by HIV-1 mutants relative to wild-type is shown on the y axis. The virus used to infect cells is indicated on the×axis. The bar graphs represent the mean values of results of PCR experiments performed at least five times, and the error bars represent standard deviations. All differences between wild type and mutant sequence results are statistically significant as determined by Student's t test (Microsoft Excel). For mutants M9-m, S1-m and M9/S1-m the respective p values in (A) are 0.0052, 0.00037, 0.00038, for the 3 h time samples; 0.00076, 0.000019, 0.00019 for the 5 h time samples; 0.0038, 0.00052, 0.00076 for the 9 h time samples; and in (B) are 0.0025, 0.0003, 0.001 for the 3 h time samples. (C) Primers specific for the pUC vector were used in the PCR reaction to determine the level of plasmid contamination in analyzed DNA samples. The picture shows an ethidium bromide-stained gel electrophoresis of the pUC fragment amplified by PCR from a pDAT1 plasmid (control), but not in DNA samples used for analysis of minus strand DNA transfer. M, DNA marker; B, water (negative control); C, pDAT1 (positive control); Lanes 1–4, 1h time point; Lanes 5–8, 9h time point; Lanes 9–12, 3h time point; Lanes 13–16, 5h time point; Lanes 1,5,9,13, WT virus; Lanes 2,6,10,14, M9-m mutant; Lanes 3,7,11,15, S1-m mutant; Lanes 4,8,12,16, M9/S1-m mutant.
To measure the effects of the tRNA-like sequences on transfer, the level of R-U5 DNA was compared with the level of U3-R DNA for each mutant and wild-type virus. If the minus strand transfer were unaffected by altered sequences in U3, the ratio of U3-R to R-U5 DNA should be similar for wild-type and mutant HIV-1 strains. However, if mutations in U3 affected the dynamic of minus strand transfer, a drop in the level of U3-R DNA should be observed when compared to the level of R-U5 DNA. To make this comparison most accurately, we initially tested different sets of primers designed for amplification of R-U5 and U3-R DNA so that we would choose sets that gave a similar level of amplification of both products when using pDAT1, as a template. As the real-time PCR assay can quantify in the cell in real time the viral DNA increase, samples were collected at different time points after infection.
To detect PCR product as it accumulates during PCR cycles, we used reagents with SYBR Green I dye, which is a highly specific, double-stranded DNA binding dye. Since the SYBR Green I dye will bind to all amplified double-stranded DNA, including non-specific reaction products, some background will be observed, but can be monitored by analyzing the Tm profile of all amplified DNA. The quantity of amplified DNA products was calculated from standard curves, generated by using pDAT1 as a template. For samples collected 1h after infection, R-U5 DNA was detectable and above the background level. However, the amount of U3-R DNA was very low and the background contributed significantly to the signal. Consequently, we could not calculate the ratio between U3-R and R-U5 DNA products at that time. The samples collected 3h, 5h, and 9h after infection had more product DNA, and so had proportionally less signal from background. PCR reactions with primers specific for the pUC vector showed that contamination of the samples with the virus DNA plasmid used in the transfection experiments is below a detectable level, and does not contribute to the signal in the real-time PCR experiments (Fig 4C).
Figure 4 presents results from the amplification of U3-R DNA product for wild-type and mutant strains. The ratio of U3-R to R-U5 DNA was used to assess efficiency of minus strand transfer. The ratio for wild-type is defined as 100%. The efficiency of minus strand transfer was diminished in all three strains with altered sequences in U3. The decreases in transfer efficiency were similar over time (A) and similar for two independent infections (B). In (A) the greatest effect, about a 26% drop in transfer, was seen for mutant virus S1-m at time points 3h and 5h post-infection. The drop was about 16% at the 9h time-point. For viruses M9-m and M9/S1-m the efficiency of transfer was reduced 9–13% and 12–15%, respectively. Statistical analysis revealed that the differences between the mutants and wild-type are statistically significant. The probability that they are different is more than 95%.
Discussion
Using a computational approach recently we identified a sequence embedded in the U3-R region of HIV-1 RNA that is highly complementary to human tRNA3Lys (Piekna-Przybylska et al., 2010). By reconstitution in vitro, we showed that segments of this complementary sequence called motif 9 and segment 1 in U3 promote minus strand transfer. These sequences were proposed to bind the priming tRNA so as to circularize the RNA genome to bring the two R regions into proximity for transfer. Our experiences with modeling strand transfer made it clear that many properties of the participating proteins and RNA template structures collaborate to produce the highest transfer efficiency. To simulate HIV-1 minus strand transfer, our systems in vitro contained RT, NC, human tRNA3Lys, and two RNA templates. The donor RNA template for transfer represented the 5’ end of the HIV-1 RNA genome. The acceptor template represented the 3’ end. NC protein and the RT RNase H activity have been shown to be critical for efficient strand transfer, and so were necessary components. However, RNA systems were modified to exclude RNA structural features that promote transfer, other than motif 9 and segment 1. This was done to improve the ability of our assays to measure the effects of those specific sequences. The 3’ end of the RNA template was shortened to eliminate the stimulating effect of the invasion-driven mechanism of transfer and allow only the terminal transfer (Chen et al., 2003b; Song et al., 2009). The RNA template representing the 5’ end of HIV-1 RNA included only the first 199nt, as sequences beyond the PBS in the 3’ direction also stimulated transfer (Piekna-Przybylska et al., 2010; Song et al., 2009). Alteration or deletion of motif 9 and segment 1 reduced the efficiency of minus strand transfer from about 70% to just about 10%, supporting their proposed role in transfer (Piekna-Przybylska et al., 2010).
Our current goal was to determine whether motif 9 and segment 1 had the expected biological function. The approach was to set up a cell culture assay that could detect the efficiency of minus strand transfer in viral systems with normal or altered motif 9 and segment 1. At the outset we realized that the system in vivo could not be as sensitive as the assay in vitro, since in vivo we could not eliminate all of the additional RNA structural factors that promote transfer.
Our approach was to generate three HIV-1 mutant viruses with altered sequences in motif 9 and segment 1. In transfected cells, these mutations do not affect the synthesis and stability of viral RNA, and normal level of virions are produced. However, if the U3 mutations affect the efficiency of minus strand DNA transfer, they would influence expression of the next generation of viruses after first round of reverse transcription in infected cells. The sequence of the promoter for synthesis of the RNA genome is also inherited in the U3 region at the 3’ end of the viral RNA, and overlaps with the tRNA gene-like sequence. Alteration of the promoter and related sequences will result in mutation of binding sites for transcription factor Sp1 and Sp3 (Pereira et al., 2000). By using the single cycle of recombinant virus we were able to exclude these issues and limit the effect of mutations in U3 solely to the minus strand transfer reaction.
The major implication of our results is that tRNA-like U3 sequences have an influence on minus strand DNA transfer in the cell. While the qualitative interpretation of the results seems clear, the quantitative outcomes deserve comment. The observed reductions of 9% to 26% in DNA transfer are generally much smaller than the differences observed in vitro. However, as we mentioned above, our reconstituted system eliminated other RNA structural factors contributing to minus strand transfer. In addition, unknown factors may also help to circularize the template RNA to facilitate transfer. If the interactions of motif 9 and segment 1 with the tRNA were part of a group of circularizing mechanisms, the effect of the two segments would be moderate in terms of percentage change. However, since the minus strand transfer step is likely to affect the overall growth kinetics of the virus, there could be substantial evolutionary pressure supporting any mechanism that even moderately improves transfer efficiency.
The results showed that alteration of either motif 9 or segment 1 reduced transfer. Surprisingly, alteration of both had a lesser than expected effect. Unfortunately, the effects of any mutation can be pleiotropic. For example, strand transfer has been shown to be influenced by the folding structure of the acceptor template, since some structures can interfere with annealing to the (-)ssDNA. Folding analysis of the effects of our mutations does not predict substantial structural changes, but it is not possible to rule out subtle differences among the wild-type and mutant acceptor folding structures.
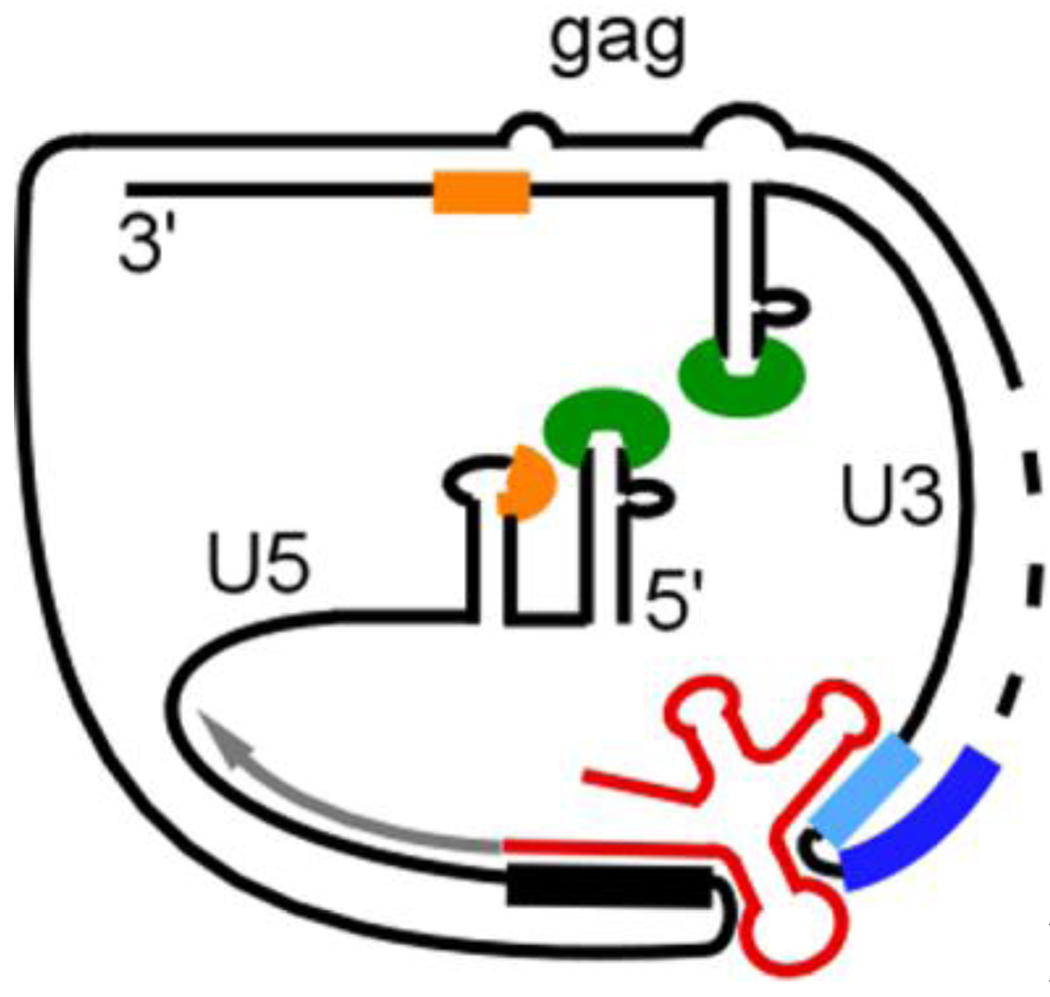
What other factors could bring the 5’ and 3’ ends of the viral RNA template into proximity? It was shown in vitro that the invasion-driven mechanism of minus strand transfer would still support efficient transfer when branch migration of the acceptor template from the invasion site to the terminus transfer site was not allowed (Song et al., 2008). We interpreted this result to mean that interaction of the acceptor RNA at the invasion site held the donor and acceptor RNAs together. This means that the invasion mechanism itself is a mechanism for template circularization, although differing from the U3 interactions in that it occurs after some (-)ssDNA synthesis. A specific interaction was also demonstrated in vitro between the R element at the 3’ end of HIV-1 and a gag sequence, leading to circularization of the genome (Gee et al., 2006; Ooms et al., 2007) (Fig. 5). Analysis in vitro revealed that these interactions enhance minus strand transfer when a DNA primer is used to start reverse transcription (Beerens and Kjems, 2010). As the R element is involved in minus strand transfer the interactions with gag during genome circularization are disrupted by (-)ssDNA synthesis. However, the interactions of tRNA3Lys with sequences in U3 could still maintain circularization until reverse transcription reaches the U3 region after transfer.
Fig. 5.
Proposed RNA-RNA contacts between the 5’ and 3’ ends of the RNA genome in HIV-1. The model shows the interactions between gag sequences and 3’ U3/PolyA sequences, and between tRNA3Lys (red), bound to the PBS region (black thick line), and motif 9nt (light blue) in U3. Sequences of loops of the hairpins TAR (green) and polyA (orange) are indicated. Sequences of segment 1 (dark blue) and the synthesized minus strand DNA (grey) are also shown. The rest of the HIV-1 genome is designated by a dotted line.
The sequences of motif 9 are complementary to nucleotides in the 3' part of the anticodon stem and part of the variable loop of tRNA3Lys (Brule et al., 2000). This region of the tRNA has been proposed to interact with genomic RNA sequences located upstream of the PBS during formation of the initiation complex with RT (Isel et al., 1995). Marquet and coworkers proposed that interaction between U3 and the anticodon stem of tRNA3Lys could be established after initiation of DNA synthesis, when the base pairings within U5 are unwound (Brule et al., 2000). However, segment 1 resembles PBS, which interacts with 18nt at the 3’ end of tRNA3Lys. Soon after minus strand transfer takes place and the PPT sequence is copied, the synthesis of plus strand DNA could start and proceed until tRNA3Lys. It is unknown when tRNA3Lys is displaced from PBS, but displacement is necessary to allow copying of the 3’ end of the tRNA into DNA in preparation for second strand transfer. The interaction with segment 1 of U3 could facilitate this process. Considering the time-dependent alternative structures, the putative interactions between U3 and tRNA3Lys appear to be part of a series of dynamic events that hold the viral ends together as the steps of (-)ssDNA, transfer and subsequent synthesis into U3 proceed.
Overall, U3-tRNA3Lys interactions are suggested by our results to be part of a complex series of changing RNA-DNA structures that promote minus strand transfer. A central theme of structure formation and breakage is to transiently circularize the template genomic RNA to bring its ends into proximity. Disruption of the U3-tRNA3Lys interaction reduces the efficiency of transfer, but only moderately. This observation supports the conclusion that other proposed mechanisms that promote transfer are also important and effective. Significantly, it also suggests that the complex multi-step stimulation process has evolved because the most rapid and efficient minus strand transfer possible is strongly selected in evolution and necessary for viral fitness and long-term survival.
Materials And Methods
Reagents
pDAT1 is a derivative of pNL4-3. The PM1 cell line was obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institute of Health, from Marvin Reitz (Lusso et al., 1995). The 293 cell line was obtained from American Type Culture Collection (ATCC, Rockville, MD). Restriction enzymes were obtained from New England Biolabs (Beverly, MA).
Cell culture
293 cells were grown in Dulbecco modified Eagle medium (DMEM) with 10 % FBS, penicillin (100 U/ml), and streptomycin (100 U/ml). PM1 cells were grown in the presence RPMI (Cellgro, Herndon, VA), supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 U/ml).
Construction of wild-type and mutant HIV-1 vectors
pDAT1 is derivative of NL4-3 with deletion of 700 base pairs in env and the mouse Thy1.1 gene cloned in place of nef. The vector was created by replacing the SalI/BstEII fragment spanning env in pAT1 (Dykes et al., 2006) with the same fragment from pDHIV.3.Thy1.1 (Dykes et al., 2004). Construction of M9-m, S1-m and M9/S1-m vectors shown in Fig. 3 was carried out as follows: First, the BamHI/AatII fragment of pDAT1 with the 3’ LTR and fragment of pUC19 was subcloned into pUC18, generating pD1. To introduce mutations into the U3 region, PCR products were produced from pD1 using pairs of oligomers with altered sequences in the desired region. For mutant M9-m, two PCR products were produced using pairs of oligomers 1/3 and 4/2 and VENT polymerase (NewEngland Biolabs). These fragments were used together with flanking oligomers 1 and 2 to generate a longer product. Following the BspEI/EcoRI digestion, the product fragment was inserted into pD1, to yield pD3. The same approach was used to introduce mutations into segment 1 and to make the mutant with altered sequences in both segments. Two PCR products were amplified using pairs of oligomers 5/6 and 7/2 for mutant S1-m, and oligomers 5/8 and 9/2 for mutant M9/S1-m. Since oligomer 5 overlaps with oligomers 6 and 8, no template was used in the reaction. Next, the longer products were generated from shorter fragments and flanking oligomers 5 and 2. Following the BspEI/EcoRI digestion, the longer PCR products were inserted into pD1, to yield pD15 and pD14 for mutants S1-m and M9/S1-m, respectively. From constructs of pD3, pD14 and pD15 with desired mutations in U3 we transferred the BamHI/AatII fragments to the rest of the HIV-1 genome in pDAT1, producing pD5, pD27 and pD28, respectively.
Oligonucleotides
5’-CTGCATCCGGAGTACTTCAAGAACTGCTGACATCGA-3’
5’-AAGCTTGATATCGAATTCCTGCAGC-3’
5’-GGCGAGGAGCGCACAGCTGCATATAAGCAGCTGCTTTTTGC-3’
5’-GCTTATATGCAGCTGTGCGCTCCTCGCCACTCCCCAGTCCCG-3’
5’-CTGCATCCGGAGTACTTCAAGAACTGCTGACATCGAGCTTGCTACAAGGGACTTTCCGCTGGGGACTTTCC-3’
5’-CTGAGGGCTCGCCACTCCCCTTGGCCTGGTTGGCCACGCCTCCCTGGAAAGTCCCCAGCGGAAAGTCC-3’
5’-GGCGTGGCCAACCAGGCCAAGGGGAGTGGCGAGCCCTCAGATGCTGCATATAAGCAGCTG-3’
5’-CGAGGAGCTCGCCACTCCCCTTGGCCTGGTTGGCCACGCCTCCCTGGAAAGTCCCCAGCGGAAAGTCC-3’
5’-GGCGTGGCCAACCAGGCCAAGGGGAGTGGCGAGCTCCTCGATGCTGCATATAAGCAGCTGCTTTTT-3’
Generation of virus stock
Two days before transfection, 4 × 106 293 cells were seeded in 10 cm plates for each transfection. In order to produce viruses, ten micrograms of pDAT1 or mutant HIV-1 vectors pD5 (M9-m), pD27 (S1-m) and pD28 (M9/S1-m) were used together with ten micrograms of pSV-A8 MLV-ENV (expressing MLV envelope) to transfect 293 cells using 25 µL of Lipofectamine LTX (Invitrogen). First, DNA and Lipofectamine LTX were incubated for 30 min in 2 ml of Opti-Mem (Invitrogen). Next, 8 ml of DMEM with 10 % FBS, penicillin (100 U/ml) and streptomycin (100 U/ml) were added to each tube with DNA/Lipofectamine before it was added to the 293 cells. The transfection medium was removed after 5 h, and then 5 ml of fresh medium was added. Virus-containing supernatant fractions were harvested 72 h later and clarified by centrifugation at 500 × g. To determine concentration of viruses, the amount of HIV-1 virus capsid protein (p24) in each virus stock was measured using an ELISA assay (Perkin-Elmer, Norwalk, CT). In addition, to determine RNA genome concentration, RNA was extracted from viral stocks treated with TURBO DNase. The number of copies of RNA per ml of virus was quantified using the COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test (Roche).
Infection for real-time PCR
The viral stocks were treated with 40 U of Turbo DNase (Ambion, Austin, Tex.) per ml for 1h at 37°C to degrade residual plasmid DNA left over from transfection. Infections with HIV-1 viruses (400 ng p24) were performed in PM1 cells (4 × 106). The viruses were allowed to adsorb to the cells for 1h at 37°C. To remove unbound viruses, cells were washed twice with PBS and sedimented at 500 × g. Cells were then seeded in 4 ml of fresh medium and 1 ml was collected for a 1 h-time point. The rest of the cells were cultured at 37°C. After 2 h, 4 h and 8 h (time-points 3 h, 5 h, 9 h, respectively) 1 ml of cells was removed, washed with PBS and saved for further total DNA extraction.
Determination of DNA copy number by real-time PCR
Genomic DNA was extracted from infected cells by using a QIAGEN DNA Blood Minikit (QIAGEN). DNA was eluted with 200 µl of elution buffer provided in the kit. DNA samples were digested with DpnI restriction enzyme (0.1 U/ l for 4 h at 37°C), which digest only plasmid DNA left over from transfection. The enzyme was deactivated by heating samples at 80°C for 20 minutes. Real-time PCR reactions were set in four repeats for each sample with an ABI7500 apparatus (Applied Biosystems). A 20 µl PCR was performed with 2 µl of genomic DNA, 100 nM of each primer and 10 µl of 2 × Power SYBR Green PCR Master Mix (Applied Biosystems). The pair of primers 5’-GATCTGAGCCTGGGAGCTCTCT-3’, 5’-CTAAAAGGGTCTGAGGGATCT-3’ was used for amplification of cDNA before minus strand DNA transfer, and 5’-ACTTTCCAGGGAGGCG-3’, 5’-TTGAAGCACTCAAGGCAAGC-3’ for amplification of cDNA after minus strand transfer. The pair of primers 5’-TCCGGGAGCTGCATGTGTCAG-3’, 5’-GGGTTATTGTCTCATGAGCGGATACAT-3’, specific for a fragment of the pUC vector, was used to determine the level of viral DNA plasmid contamination in the analyzed DNA samples.
The cycling conditions were 2 min at 50°C and 10 min at 95°C, then 40 cycles of 95°C for 15 s and 60°C for 1 min. To calculate the amount of amplified product the standard curve was generated with serial dilutions of pDAT1 used as a template. The data were averaged from at least five real-time PCR measurements.
Acknowledgments
This work was supported by the US National Institutes of Health research grant GM049573 (to R.A.B.). We thank students of the Bambara laboratory group for helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008;134(1–2):19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Beerens N, Kjems J. Circularization of the HIV-1 genome facilitates strand transfer during reverse transcription. RNA. 2010;16(6):1226–1235. doi: 10.1261/rna.2039610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brule F, Bec G, Keith G, Le Grice SF, Roques BP, Ehresmann B, Ehresmann C, Marquet R. In vitro evidence for the interaction of tRNA(3)(Lys) with U3 during the first strand transfer of HIV-1 reverse transcription. Nucleic Acids Res. 2000;28(2):634–640. doi: 10.1093/nar/28.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Balakrishnan M, Roques BP, Bambara RA. Steps of the acceptor invasion mechanism for HIV-1 minus strand strong stop transfer. J. Biol. Chem. 2003a;278(40):38368–38375. doi: 10.1074/jbc.M305700200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Balakrishnan M, Roques BP, Fay PJ, Bambara RA. Mechanism of minus strand strong stop transfer in HIV-1 reverse transcription. J. Biol. Chem. 2003b;278(10):8006–8017. doi: 10.1074/jbc.M210959200. [DOI] [PubMed] [Google Scholar]
- Dykes C, Balakrishnan M, Planelles V, Zhu Y, Bambara RA, Demeter LM. Identification of a preferred region for recombination and mutation in HIV-1 gag. Virology. 2004;326(2):262–279. doi: 10.1016/j.virol.2004.02.033. [DOI] [PubMed] [Google Scholar]
- Dykes C, Wang J, Jin X, Planelles V, An DS, Tallo A, Huang Y, Wu H, Demeter LM. Evaluation of a multiple-cycle, recombinant virus, growth competition assay that uses flow cytometry to measure replication efficiency of human immunodeficiency virus type 1 in cell culture. J. Clin. Microbiol. 2006;44(6):1930–1943. doi: 10.1128/JCM.02415-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee AH, Kasprzak W, Shapiro BA. Structural differentiation of the HIV-1 polyA signals. J. Biomol. Struct. Dyn. 2006;23(4):417–428. doi: 10.1080/07391102.2006.10531236. [DOI] [PubMed] [Google Scholar]
- Gilboa E, Mitra SW, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer) J. Mol. Biol. 1995;247(2):236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- Lu R, Limon A, Devroe E, Silver PA, Cherepanov P, Engelman A. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 2004;78(23):12735–12746. doi: 10.1128/JVI.78.23.12735-12746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusso P, Cocchi F, Balotta C, Markham PD, Louie A, Farci P, Pal R, Gallo RC, Reitz MS., Jr Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 1995;69(6):3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbisa JL, Delviks-Frankenberry KA, Thomas JA, Gorelick RJ, Pathak VK. Real-time PCR analysis of HIV-1 replication post-entry events. Methods Mol. Biol. 2009;485:55–72. doi: 10.1007/978-1-59745-170-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negroni M, Buc H. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc. Natl. Acad. Sci. U. S. A. 2000;97(12):6385–6390. doi: 10.1073/pnas.120520497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms M, Abbink TE, Pham C, Berkhout B. Circularization of the HIV-1 RNA genome. Nucleic Acids Res. 2007;35(15):5253–5261. doi: 10.1093/nar/gkm564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peliska JA, Benkovic SJ. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258(5085):1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- Pereira LA, Bentley K, Peeters A, Churchill MJ, Deacon NJ. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28(3):663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D, DiChiacchio L, Mathews DH, Bambara RA. A sequence similar to tRNA 3 Lys gene is embedded in HIV-1 U3-R and promotes minus-strand transfer. Nat. Struct. Mol. Biol. 2010;17(1):83–89. doi: 10.1038/nsmb.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez L, Tsuchihashi Z, Fuentes GM, Bambara RA, Fay PJ. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J. Biol. Chem. 1995;270(25):15005–15011. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- Song M, Balakrishnan M, Gorelick RJ, Bambara RA. A Succession of Mechanisms Stimulate Efficient Reconstituted HIV-1 Minus Strand Strong Stop DNA Transfer. Biochemistry. 2009;48(8):1810–1819. doi: 10.1021/bi802149j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Basu VP, Hanson MN, Roques BP, Bambara RA. Proximity and branch migration mechanisms in HIV-1 minus strand strong stop DNA transfer. J. Biol. Chem. 2008;283(6):3141–3150. doi: 10.1074/jbc.M707343200. [DOI] [PubMed] [Google Scholar]
- You JC, McHenry CS. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 1994;269(50):31491–31495. [PubMed] [Google Scholar]