Abstract
We developed a live imaging system enabling dynamic visualization of single cell alignment induced by external mechanical force in a 3-D collagen matrix. The alignment dynamics and migration of smooth muscle cells (SMCs) were studied by time lapse differential interference contrast and /or phase contrast microscopy. Fluorescent and reflection confocal microcopy were used to study the SMC morphology and the microscale collagen matrix remodeling induced by SMCs. A custom developed program was used to quantify the cell migration and matrix remodeling. Our system enables cell concentration-independent alignment eliminating cell-to-cell interference and enables dynamic cell tracking, high magnification observation and rapid cell alignment accomplished in a few hours compared to days in traditional models. We observed that cells sense and response to the mechanical signal before cell spreading. Under mechanical stretch the migration directionality index of SMCs is 46.3% more than those cells without external stretch; the dynamic direction of cell protrusion is aligned to that of the mechanical force; SMCs showed directional matrix remodeling and the alignment index calculated from the matrix in front of cell protrusions is about 3 fold of that adjacent to cell bodies. Our results indicate that the mechanism of cell alignment is directional cell protrusion. Mechano-sensing, directionality in cell protrusion dynamics, cell migration and matrix remodeling are highly integrated. Our system provides a platform for studying the role of mechanical force on the cell matrix interactions and thus find strategies to optimize selected properties of engineered tissues.
Keywords: Molecular imaging, Cell signaling, ECM, Confocal microscopy, Collagen, Smooth muscle cell
1. Introduction
Many types of cells live in a stretch-rich condition in vivo and mechanical factors also control stem cell fate[1], tissue morphogenesis[2] and system development[3]. Mechanical stretch, which is a major form of mechanical force, plays an important role in maintaining cell activity and phenotype[4]. There are also many fundamental biological process which are controlled by mechanical stretch, among which cell alignment parallel or perpendicular to the direction of the mechanical stretch attracts the attention of many scientists[5, 6]. In the past, various in vitro models have been used to study the relationship between mechanical stress and cell alignment, one of which was to seed cells on an elastic silicone sheet and the stress was applied by stretching the sheet[7, 8]. Live imaging was also applied to this 2-D system, which provided scientists direct observation of the process of cell alignment [9]. However, cells live in a three dimensional environment in vivo, in which cells show different morphology and phenotypic characteristics compared with those in 2-D culture [10] [11, 12]. Therefore, a 3-D in vitro model is needed to better understand the in vivo phenomena. In the past, a hydrogel matrix (such as collagen) model using cell-generated force has been widely used[13]. In 1982, a two-blocker assay was introduced by Bellows et. al[14], in which cells compact the gel and the compaction force generates uniaxial stretch to the gel because the gel can only shrink in the direction without the blockers. This model is easy to setup and has been widely used with various cell types [15–17]. However, there are instinct drawbacks to this model. First, it is impossible to use live imaging microscopy to track a single cell within the gel because of the large amount of displacement in X,Y and Z axes[14]. In the past, because cells are commonly out of the working distance of high magnification lenses, it was considered impossible to perform high resolution microscopy, such as reflection confocal microscopy, to study cell-induced matrix remodeling even with static microscopy[18]. And dynamic imaging would obviously be even more challenging.
Another drawback of the model is the high cell density requirement and the cell concentration needs to be at least 105cells/ml (hydrogel) to generate enough cell force to induce cell alignment. The cell contact at high concentration makes it much more complex to study the specific mechanism of the external mechanical effect at the single cell level. It is a further dilemma that low cell concentrations (such as 20,000/ml) generally require 42 days to generate visible alignment[15].
Because of the limitation of the traditional blocker cell alignment assay and the lack of dynamic information, several fundamental questions regarding 3-D mechanical force-induced cell alignment remain unanswered: 1) the mechanism of the cell alignment, 2) at what time point a cell is able to sense the mechanical signal in a 3-D environment, 3) the dynamics of the cell protrusions, and 4) real time interactions in cell migration and matrix remodeling. In order to solve the problems of traditional models and answer the questions above, we developed a live imaging system to study and quantify the dynamic change of cell alignment and migration under mechanical stretch and the associated matrix remodeling.
2 Materials and methods
2.1 Mechanical force loading for live imaging systems - design and calibration
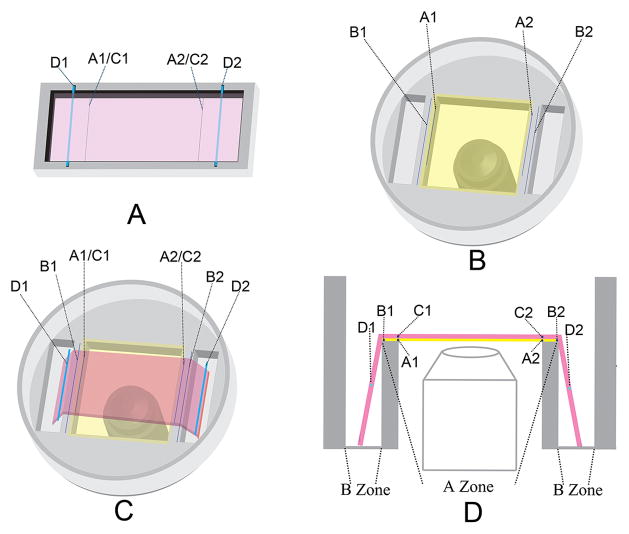
The system consists of two chambers custom machined from FDA approved polystyrene as shown in Figure 1. Chamber one has a rectangular shape well for preparing the cell-collagen construction. Chamber two has two rectangular wells for stretching the collagen hydrogel and a central square observation window for imaging. Line segments A1 and A2 were pre-marked on Chambers one and two respectively and vertical lengths between A1 and A2 were the same in the two chambers. Line segments B1 and B2 are also pre-marked on Chamber two to make the vertical length between B1 and B2 20% longer than that between A1 and A2. The cell-collagen mixture was added to Chamber one and two marker threads (C1, C2) and two stainless steel rods (D1, D2) were imbedded in the collagen solution before polymerization in Chamber one. The C1 and C2 threads exactly overlapped with line segments A1 and A2. After polymerization the cell-collagen construction was detached from the walls of Chamber one and transferred to Chamber two by lifting the stainless steel rods. The marker threads C1 and C2 were positioned to overlap line segments A1 and A2 in Chamber two. The degree of stretch is 20% by length, which is controlled by pushing the stainless steel rods downwards with a micropositioner until the two marker threads C1 and C2 overlap line B1 and B2 respectively.
Figure 1.
The schematic diagram of the alignment live imaging system. (A) The cell-collagen mixture was added to Chamber one and two marker threads (C1, C2) and two stainless steel rods (D1, D2) were imbedded in the collagen solution before polymerization. The C1 and C2 threads exactly overlapped with line segments A1 and A2. (B) Live imaging with two pair of line segments marked. B1B2 is 20% longer than A1A2. The vertical distances between line segments A1 to A2 are equal in chamber one and chamber two. (C) The collagen-SMCs construct is moved from chamber 1 to chamber 2 and a micromanipulator is used to stretch the hydrogel downwards to gain 20% stretch. (D) Side view of chamber 2 when collagen-SMCs are overlaid.
For any imaging assay, the long axis of an acquired image is set to be parallel to the direction of the mechanical force. Collagen-SMC constructs prepared in a traditional way[14] to generate cell alignment by cell contraction-generated force were used as controls.
2.2 Preparation of collagen-SMC constructs
SMCs were obtained from canine carotid arteries and identified and expanded by our previously published explant cell culture techniques[19, 20]. Bovine dermal type I collagen were prepared according to the instructions from the manufacture (PureCol™, Inamed BioMaterials, Fremont, CA). In brief, collagen stock was neutralized and mixed with 10X DMEM (Sigma-Aldrich, St. Louis, MO) to form a collagen solution. Trypsinized SMCs were resuspended in 1X DMEM (Invitrogen, Carlsbad, CA) and mixed with the collagen solution. The final concentration of the collagen was 2mg/ml and of SMCs was 20,000/ml.
2.3 Cell viability
SMC-collagen mixture was prepared as above in 2.2. A group prepared in the absence of stretch was used as the control. Cell viability was evaluated after 1h, 12h and 24h using calcein AM (Molecular probe, Invitrogen, Carlsbad, CA) according to the instruction of the manufacture and DAPI was used for nuclear counter staining.
2.4 Image processing software
The software used for image processing were Image J (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997–2010), and our custom designed software using JAVA(Sun Microsystems, Inc, Santa Clara, CA) and Matlab (The MathWorks, Inc., Natick, MA).
2.5 Cell alignment dynamics study under mechanical stretch using differential interference contrast (DIC) live microscopy
After the gel was stretched as described above, 15ml of CO2 independent media (Invitrogen was added and a temperature control unit was used to maintain the culture condition at 37°C Live imaging was then started immediately and images were acquired at 5 minute intervals using an Axiovert 200 inverted microscope (Carl Zeiss, Oberkochen, Germany) coupled to an Axiocam HRC camera using Axiovision software. High magnification objectives (40X, 63X) in DIC mode were used to track the morphology changes at a single cell level. Samples without stretch were used as controls.
2.6 Cell migration under mechanical force
2.6.1 Analyzing migration trajectories
The experiment was set up as above and a 10X objective in DIC mode was used to track a group of cells. Cells that moved out of the visual field were excluded. At least 50 cells were tracked for each assay. Cell migration was analyzed using Image J software and the coordinate of a cell at each time point was acquired. The trajectory of each cell was plotted with custom written software.
2.6.2 Cell migration directionality analysis
In order to analyze the directionality of a migrating cell, the direction of the mechanical fore in the groups under stretch or the horizontal line in the group without stretch was used as the X-axis. The coordinates of the geometric center of a cell was analyzed by our custom designed software and the migrating direction at each time point was calculated. The angles (α) between the migrating direction of the cell and the X-axis at each time point in the groups with or without mechanical stretch were calculated. The “directionality index” (DI) was calculated using the following equation:
where the N is the total number of time points. The directionality index equals 1 when a cell moves only horizontally no matter which direction it moves; equals 0 when a cell moves only perpendicularly no matter which direction it moves. The bigger the number of the DI is, the cell moves more directionally toward the mechanical force or the horizontal line.
2.6.3 Cell protrusion dynamics during migration
The directions of the cell protrusions, defined as the angle between the long axis of each protrusion and the X-axis (horizontal line) at each time point was analyzed by the software. The values of angles over time were plotted. SMC-collagen constructs prepared with the same protocol and concentration, cultured in the absence of mechanical stretch were used as the controls.
2.7 Cell morphology analysis by confocal microscopy
At the endpoint in each assay, collagen gels were rinsed with PBS and fixed with warm 4% paraformaldehyde for 30 minutes and rinsed again with PBS. After treatment with 0.1% Triton X-100 and blocking of non-specific binding with 2% BSA for 30 minutes, SMCs were stained using two methods for visualization: 1) Alexa Flour 488 staining: samples were stained with Alexa Flour 488 labeled Phalloidin (Molecular Probes, Invitrogen), which specifically binds to actin fibers in a cell, according to the manufacturer’s instructions and 2) α-SMA staining: as detailed previously, samples were incubated with a mouse monoclonal anti α-SMA antibody (Sigma, Carpinteria, CA), and then incubated with rhodamine-conjugated goat anti-mouse secondary antibody (MP Biomedicals, Solon, OH). Reflection confocal microscopy was used to visualize the collagen fibers and the fluorescent mode was used to visualize the actin fibers in SMCs using an Olympus Fluoview 300 confocal microscope with a 60X objective. Optical sections were performed and 3-D image reconstructions were made according to our previously published protocols[19–21].
2.8 Analysis and quantification of matrix remodeling and cell alignment
SMC-collagen constructs were prepared as above and after 16h of culture, collagen gels were fixed and imaged using confocal microscopy as above. The images were further processed by the software, including mosaic image stitching and image 3-D reconstructions in batch mode as we reported previously[19, 21].
Images of collagen fibers were stitched into a mosaic image and areas of interest were selected as follows: 1) immediately adjacent to the tips of cell protrusions: within 10 μm from the tip of a cell protrusion excluding any other cells and 2) immediately adjacent to the cell bodies: within 10 μm above or below the cell body in the acquired images excluding any other cells. Fast Fourier Transform (FFT) was used to quantify the orientation alignment of remodeled collagen fibers in response to the direction of the mechanical force as described before[19]. In brief, FFT transforms the image in the spatial domain into the frequency domain. After rotating the spectrum image 90° in the frequency domain, the relative intensity at different angles of the image in the frequency domain indicates the relative number of the fibers in the same angle in the image in the spatial domain. In order to quantify the degree of orientation, the spectrum images were transformed into histograms. In Cartesian coordinates (x, y), x represents the angle and y represents the average intensity at this direction. In polar coordinates R(θ-ϕ), θ represents the angle of the collagen fibers, ϕ represents the angle of the mechanical stretch and R represents the intensity in this direction. An “alignment index” (AI) was used, which was calculated using the following equation
Where,
where ϕ=0 in our assays, and the AI equals 0 when collagen fibers are in an ideal random distribution, +1 or −1 when collagen fiber are totally parallel or perpendicular to the direction of the mechanical stretch. Therefore, values closer to +1 mean greater alignment and values closer to 0 mean more random distribution
2.9 Statistics
Students t-tests were used to evaluate the differences between two groups with SPSS software (SPSS Inc, Chicago, IL) with the level of significance at P < 0.05. Data were expressed as mean ± SD.
3. Results
3.1 Characteristics of the new system
3.1.1 Capable of dynamic cell tracking
Because the mechanical force applied to the cells was from external instead of the internal force generated by cell contraction, no visible deformation of any in focus area in either X, Y or Z directions accrued during the image acquisition, which means there was no matrix contraction pulling cells out of focus or out of the visual field.
3.1.2 Compatible with high magnification low working distance objectives
As shown in Figure 1, because the collagen was stretched from two sides downwards, the cell-hydrogel construction in the A zone was tightly attached to the glass surface. The A zone is sealed with a class zero coverslip, which is about 80–130 μm thick, providing much greater depth of focus into the collagen hydrogel. For example, even when oil or water immersion 63X or 60X objectives were used, depending on different microscope brands, we were still be able to focus about 100–150 μm into the collagen gel. All this above makes it possible to use higher resolution objectives to visualize the detail of cell alignment.
3.1.3 Alignment generation at low cell concentrations enabling single cell tracking and eliminating cell-to-cell interference
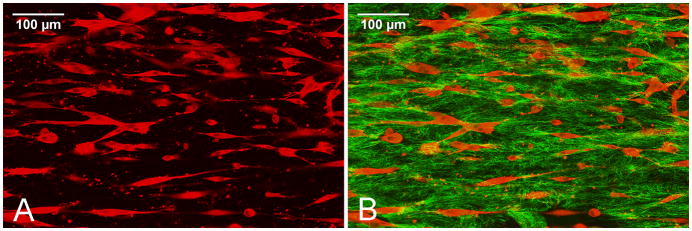
As seen in Figure 3 and Figure 6A&C, cells in our system are separated from each other eliminating cell contact and enabling investigation of the specific effect of the mechanical force on the matrix on a single cell. The cells in the traditional alignment assay contact each other because high cell concentrations were needed to generate tactile force (Figure 2). The morphology of the cells in our new alignment system are spindle shape compared with the polygonal shape of the contacted cells in the traditional alignment assay (see Figure 2).
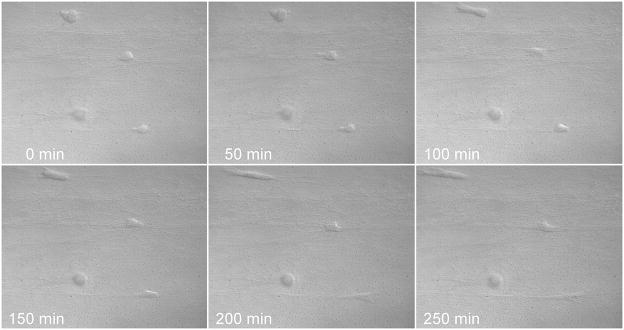
Figure 3.
Cell alignment dynamics were visualized by differential interface contrast microcopy over 250 minutes using a 40X objective in our cell alignment system.
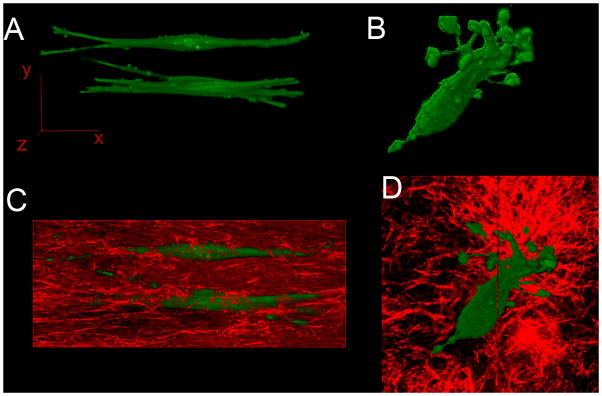
Figure 6.
A and B are the three dimensional reconstructed fluorescent confocal images of SMCs in the presence or absence of external stretch respectively. Figure 6A shows two aligned SMCs in elongated spindle shapes; both the cell bodies and protrusions are parallel to the direction of the mechanical stretch. Figure 6B shows an SMC in a polygonal shape with protrusions pointing in multiple directions. Figure 6C and 6D show the collagen matrix (red) by reflection confocal microscopy in which the SMCs (green) are embedded. A 60X oil immersion objective was used for all the image acquisitions.
Figure 2.
SMCs in traditional blocker-based alignment assays contact each other because of the high cell concentrations used and many SMCs are polygonally shaped. SMCs immunostained with rhodamine labeled anti α-SMA antibodies were visualized by fluorescent confocal microscopy (red in Fig 2A and 2B) and collagen fibers were visualized by reflection confocal microscopy (green in Fig. 2B). The scale bar represents 100 micrometers.
3.1.4 Rapid cell alignment, which can be accomplished in a few hours compared with a few days in traditional models
The new system is cell concentration independent. As shown in Figure 3, SMCs in the new chamber were highly aligned after 6h of incubation at a cell density of 20,000/ml. However, with the traditional chamber, SMCs at 20,000/ml did not show any alignment for up to a week and the SMCs at 500,000/ml took more than 24h to show alignment with many cells contacting with each other (Figure 2). In the new chamber, because mechanical force external to the hydrogel was applied immediately, the SMCs started to show alignment in less than two hours after collagen polymerization and more than 90% of the cells showed alignment within 8 hours. By contrast, in the traditional chamber, because the force applied to the cells is generated by hydrogel-compaction, with high cell density (≥500,000/ml), it takes more than 24hs for the SMCs to show visible alignment.
3.2 Cell viability in the stretched chamber
At every time point, the viability of SMCs cultured under stretch showed no difference compared with the ones cultured in the absence of stretch(P=0.24).
3.3 Alignment dynamics in the stretch chamber
Time lapse imaging started right after the collagen hydrogel was fully polymerized. The cells were in a spherical shape initially and there was no protrusion in any cell. Then the mechanical force was applied to the collagen hydrogel globally to achieve a 20% stretch. As shown in Figure3, the SMCs started to form protrusions as they began to spread and the protrusions were only formed in the direction of the mechanical force. The fact that SMCs reacted to the mechanical stretch before they were fully spread out indicated that SMCs sensed the mechanical signal while they are still in a spherical shape and alignment was the result of cell directional spreading.
3.4 Migration
3.4.1 Migration trajectory and directionality
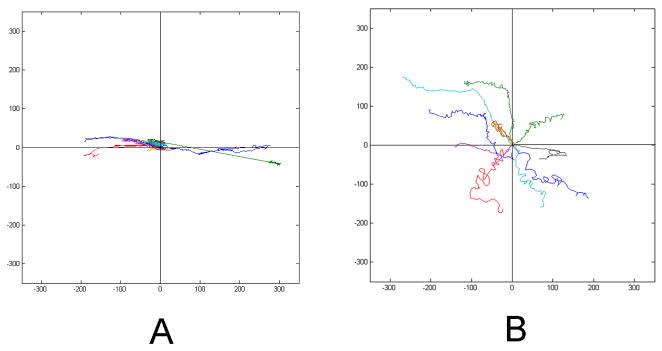
As seen from the cell migration paths in Figure 4, SMCs under stretch migrated parallel to the direction of the mechanical force compared with the non-directional migration of the cells in the absence of stretch. The SMCs in the presence of stretch had a significantly higher DI (0.803±0.074) than the ones in the absence of stretch (0.549±0.087) (P<0.001). As a cell migrated, it extended and withdrew its protrusions parallel to the direction of the mechanical force, which led to the directional migration of a cell.
Figure 4.
A and B are two representative plots of the SMCs migration paths in the presence or absence of mechanical stretch respectively. SMCs under stretch showed directional migration compared with random migration of the cells without external stretch. Each colored line represents an individual cell migration trajectory over time.
3.4.2 SMC protrusion dynamics during migration
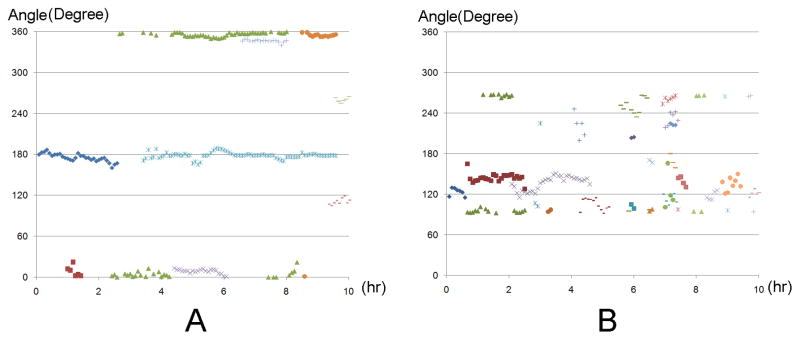
As shown in Video 1(Internet Explorer is the recommended software for better visualization of both Video 1 and Video 2, which are presented in the format of animated gif.), the extending and withdrawing of a SMC under mechanical force are mostly parallel to the direction of the mechanical force compared with the random direction of the SMCs without mechanical stretch (Video 2). As shown in Figure 5-A, the directions of protrusions of a cell under stretch varied slightly around that of the mechanical force, compared with almost randomly distributed protrusions from unstretched cells (Figure 5B).
Figure 5.
A and B are the representative plots of cell protrusion direction dynamics over the two hour period in the presence or absence of stretch respectively. Each colored dot represents one protrusion’s direction changes over time.
3.5 Morphology
SMCs in the new system were uniformly spindle shaped and had less protrusions(Figure 6A and C, Figure 7) compared with the SMCs in the traditional blocker assay which were more polygonal shaped and with more protrusions at both high (Figure 2) or low concentrations (Figure 6B and D). All the protrusions of the cells in the new system were parallel to the direction of the mechanical force and have elongated shapes (Figure 6A and C, Figure 7). In contrast, SMCs formed non-directional protrusions and each protrusion was curved when cultured at the same concentration in the traditional blocker assay (Figure 6B and D).
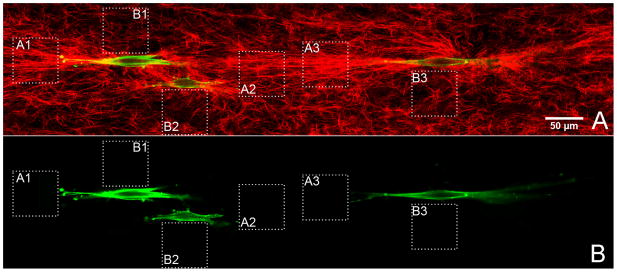
Figure 7.
A representative mosaic image of aligned SMCs (green), shown by fluorescent confocal microscopy, in collagen matrix (red), shown by reflection confocal microscopy. Square dashed line areas of A1, A2 and A3 are the representative areas used to analyze the AI immediately adjacent to cell protrusions and B1, B2 and B3 are the representative areas used to analyze the AI immediately adjacent to the cell bodies. Figure 7A is an overlay image of both collagen and cell channels and Figure 7B shows only the cell channel for better visualization of the cell margin in relation to the dashed area.
3.6 Cell alignment and matrix remodeling
Under the mechanical force, the SMCs also remodeled the collagen matrix to the direction of the mechanical stretch. As shown in Figure 7, the direction of the remodeled matrix in front of the cell protrusion is also highly aligned to the direction of the mechanical fore; however the direction of the collagen matrix below or above the cell body showed significantly low alignment. With the direction of the mechanical force as the reference, the AI calculated from in front of the cell protrusions (0.508±0.058) are about 3 fold greater than that calculated from adjacent to the cell bodies (0.178±0.049, P<0.001). Time lapse imaging showed that the protrusions of a cell mostly move in the direction of the mechanical force (Video 1), which may be one of the reasons for the directional matrix remodeling.
4. Discussion
If the mechanical characteristics of an engineered tissue do not resemble the equivalent natural tissue to a certain degree, it likely will not function optimally and may not be implantable. Mechanical force also plays a fundamental role in shaping an engineered tissue and in regulating its function [22, 23]. Understanding the mechanism of mechanical signal transduction and how cells are influenced by mechanical forces is a crucial step[24].
Seeing is believing- live imaging is a powerful tool to provide scientists with direct observation and a better understanding complex biological processes [25, 26]. Live imaging obviously can provide detailed information with which to characterize the dynamics of the cell under mechanical stimulation. However, because of the limitations of traditional blocker based stretch assays, to the best of our knowledge there is no high resolution dynamic imaging report about cell alignment under mechanical force in a 3-D environment. Because stretch, also referred to as displacement, is one of the most common types of biomechanical force, we applied it in our system. In this project, we developed a system for imaging cell dynamics and studied SMC alignment dynamics under mechanical stretch with quantification of stretch-induced SMC migration and SMC-mediated matrix remodeling.
To study the dynamics of cell characteristics under mechanical stretch, a critical function of a system is to enable specific individual cells to be observed in focus within the visual field during image acquisition, which is impossible in the traditional blocker assay due to a large degree to hydrogel compaction. However, the strategy of inhibiting hydrogel compaction to fix a cell in stable dimensions is not realistic because it is hydrogel compaction that generates the mechanical force. In order to solve this problem, in our new system mechanical force was applied externally enabling immediate mechanical stimulation without the hydrogel compaction as the force source. Low cell concentration eliminates the overall gel compaction and only maintains the microscale matrix deformation, which enables cells to remain in focus during image acquisition. Furthermore, the uniaxial stretch applied to the collagen hydrogel was generated by pulling the hydrogel downwards and the hydrogel is tightly attached to the bottom cover-glass after stretch.
Applying the force external not only eliminated the overall hydrogel displacement, but also has the following advantages. First of all, as in most cases in vivo, cells experience external forces rather than the force generated by themselves. Our new system mimics the in vivo environment better than the traditional assay in this regard. Secondly, our new system makes the degree of mechanical force applied tunable by tuning the degree of stretch. More importantly, applied external force is stable over a period of time and it generates rapid cell alignment. Our results demonstrated that most of the cells showed alignment within 8 hours and early alignment could be observed less than 2 hours after collagen gel polymerization. Externally applied force doesn’t require a high cell concentration. Theoretically, in terms of a successful cell alignment, the cell concentration could be as low as any number that the experiment required. We have successfully generated the same pattern of SMCs in the concentration of 2000/ml in the similar period of time (data not shown). The rapid alignment generation capability makes our chamber system an open platform for the dynamical study of any mechanical stretch-related biologic or pathologic phenomenon.
Using our new system, we observed five major phenomena of SMCs under mechanical stretch in 3-D: 1) sensing and responding to the external mechanical force before spreading, 2) directional overall cell migration, 3) directional cell protrusion extension and withdraw during cell spreading, 4) high polarization of cell bodies and reduced numbers of protrusions and 5) directional matrix remodeling. All five phenomena of the SMCs are highly integrated.
Mechanical signal sensing is the first step of alignment. In the traditional assay, it was not possible to determine when a cell began to sense and respond to the external mechanical force because all cells were in the system for many hours before they aligned; nor could dynamic tracking be performed. There might be two major possibilities: 1) after a cell spread out, because at this stage, there is an angle between the long axis of the cell body and the direction of the mechanical force, so that the cell may change its orientation to minimize the force or 2) a cell is able to sense and respond to the mechanical force before spreading.
We observed that a SMC sensed and responded to the mechanical force before it spread because spherical cells spread out directly to the direction of the mechanical force. This showed that mechanical sensing starts before spreading. As all the cells sensed and spread to the same direction of the mechanical force, they showed a high level of alignment. Therefore, the mechanism of cell alignment is directional cell protrusion extension or cell spreading. During migration, the cells continuously sense and respond to the mechanical stretch.
How SMCs migration is effected by stretch is also important because SMC migration is central to vascular remodeling under in vivo mechanical conditions[27]. Reports have suggested that mechanical pre-treatment affects the cell migration[28, 29] but the real time effects of mechanical force on cell migration, especially in 3-D, remains unclear. Using our imaging system, we found that SMCs showed highly directional migration under static mechanical stretch. We think that SMCs were able to sense and response to the mechanical force continuously.
We further analyzed the mechanism of the directional migration. By plotting the angle of the cell protrusions, we found that protrusion dynamic is also highly directional. The SMCs responded to the external mechanical signal with the directional protrusion extending and withdrawing, which leads to the directional movement of the whole cell body. The SMCs showed a highly polarized morphology under mechanical stretch in DIC time lapse images. 3-D reconstructed high resolution confocal images showed SMCs developed a greatly elongated spindle shape with long straightly extended protrusions, which were highly parallel to each other. These morphologies supported the dynamics phenomena of cell alignment.
It is well accepted that SMC migration is a major factor affecting the remodeling of a blood vessel wall[30, 31]. We predicted that the highly directional migration behavior of SMCs under mechanical stretch would lead to directional matrix remodeling. We explored this question using reflection confocal microscopy and Fast Fourier Transform image processing techniques. As we predicted, the orientation of remodeled matrix was also highly aligned to the direction of the mechanical force.
5. Conclusion
We developed a new system for dynamically studying cell alignment in three dimensional matrices under mechanical force. The system is capable of high resolution time lapse imaging, rapid cell alignment and concentration-independence of cell alignment. Using this system, we found that the mechanism of cell alignment is the result of directional cell protrusion extension prior to cell spreading. Mechano-sensing, directionalities in cell alignment, cell migration, protrusion dynamics and matrix remodeling are highly integrated. Our system provides an open system for studying the mechanical interplay between cell and extracellular matrix. We believe that it will be an effective tool for better biomaterials development and for enhancing the strategies in regenerative medicine.
Supplementary Material
Acknowledgments
This project was supported by grants from the NIH R01-HL41272 (HPG) and the Department of Veterans Affairs (HPG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 2.Ingber DE. Mechanical control of tissue morphogenesis during embryological development. Int J Dev Biol. 2006;50:255–66. doi: 10.1387/ijdb.052044di. [DOI] [PubMed] [Google Scholar]
- 3.le Noble F, Klein C, Tintu A, Pries A, Buschmann I. Neural guidance molecules, tip cells, and mechanical factors in vascular development. Cardiovasc Res. 2008;78:232–41. doi: 10.1093/cvr/cvn058. [DOI] [PubMed] [Google Scholar]
- 4.Lim CT, Zhou EH, Quek ST. Mechanical models for living cells--a review. J Biomech. 2006;39:195–216. doi: 10.1016/j.jbiomech.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Haga JH, Li YS, Chien S. Molecular basis of the effects of mechanical stretch on vascular smooth muscle cells. J Biomech. 2007;40:947–60. doi: 10.1016/j.jbiomech.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Yim EK, Reano RM, Pang SW, Yee AF, Chen CS, Leong KW. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials. 2005;26:5405–13. doi: 10.1016/j.biomaterials.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaunas R, Nguyen P, Usami S, Chien S. Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proc Natl Acad Sci U S A. 2005;102:15895–900. doi: 10.1073/pnas.0506041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reno F, Traina V, Cannas M. Mechanical stretching modulates growth direction and MMP-9 release in human keratinocyte monolayer. Cell Adh Migr. 2009;3:239–42. doi: 10.4161/cam.3.3.8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayakawa K, Sato N, Obinata T. Dynamic reorientation of cultured cells and stress fibers under mechanical stress from periodic stretching. Exp Cell Res. 2001;268:104–14. doi: 10.1006/excr.2001.5270. [DOI] [PubMed] [Google Scholar]
- 10.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–12. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 11.Keller PJ, Pampaloni F, Stelzer EH. Life sciences require the third dimension. Curr Opin Cell Biol. 2006;18:117–24. doi: 10.1016/j.ceb.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–45. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 13.Bell E, Ivarsson B, Merrill C. Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro. Proc Natl Acad Sci U S A. 1979;76:1274–8. doi: 10.1073/pnas.76.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellows CG, Melcher AH, Aubin JE. Association between tension and orientation of periodontal ligament fibroblasts and exogenous collagen fibres in collagen gels in vitro. J Cell Sci. 1982;58:125–38. doi: 10.1242/jcs.58.1.125. [DOI] [PubMed] [Google Scholar]
- 15.Huang D, Chang TR, Aggarwal A, Lee RC, Ehrlich HP. Mechanisms and dynamics of mechanical strengthening in ligament-equivalent fibroblast-populated collagen matrices. Ann Biomed Eng. 1993;21:289–305. doi: 10.1007/BF02368184. [DOI] [PubMed] [Google Scholar]
- 16.Roby T, Olsen S, Nagatomi J. Effect of sustained tension on bladder smooth muscle cells in three-dimensional culture. Ann Biomed Eng. 2008;36:1744–51. doi: 10.1007/s10439-008-9545-5. [DOI] [PubMed] [Google Scholar]
- 17.Berry CC, Shelton JC, Lee DA. Cell-generated forces influence the viability, metabolism and mechanical properties of fibroblast-seeded collagen gel constructs. J Tissue Eng Regen Med. 2009;3:43–53. doi: 10.1002/term.133. [DOI] [PubMed] [Google Scholar]
- 18.Lee EJ, Holmes JW, Costa KD. Remodeling of engineered tissue anisotropy in response to altered loading conditions. Ann Biomed Eng. 2008;36:1322–34. doi: 10.1007/s10439-008-9509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pang Y, Ucuzian AA, Matsumura A, Brey EM, Gassman AA, Husak VA, et al. The temporal and spatial dynamics of microscale collagen scaffold remodeling by smooth muscle cells. Biomaterials. 2009;30:2023–31. doi: 10.1016/j.biomaterials.2008.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pang Y, Wang X, Ucuzian AA, Brey EM, Burgess WH, Jones KJ, et al. Local delivery of a collagen-binding FGF-1 chimera to smooth muscle cells in collagen scaffolds for vascular tissue engineering. Biomaterials. 2010;31:878–85. doi: 10.1016/j.biomaterials.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pang Y, Greisler HP. Using a type 1 collagen-based system to understand cell-scaffold interactions and to deliver chimeric collagen-binding growth factors for vascular tissue engineering. J Investig Med. 2010;58:845–8. doi: 10.231/JIM.0b013e3181ee81f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziegler T, Nerem RM. Tissue engineering a blood vessel: regulation of vascular biology by mechanical stresses. J Cell Biochem. 1994;56:204–9. doi: 10.1002/jcb.240560215. [DOI] [PubMed] [Google Scholar]
- 23.Mammoto T, Ingber DE. Mechanical control of tissue and organ development. Development. 2010;137:1407–20. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dado D, Levenberg S. Cell-scaffold mechanical interplay within engineered tissue. Semin Cell Dev Biol. 2009;20:656–64. doi: 10.1016/j.semcdb.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Gerlich D, Ellenberg J. 4D imaging to assay complex dynamics in live specimens. Nat Cell Biol. 2003;(Suppl):S14–9. [PubMed] [Google Scholar]
- 26.Waters JC. Live-cell fluorescence imaging. Methods Cell Biol. 2007;81:115–40. doi: 10.1016/S0091-679X(06)81007-1. [DOI] [PubMed] [Google Scholar]
- 27.Gibbons GH, Dzau VJ. The emerging concept of vascular remodeling. N Engl J Med. 1994;330:1431–8. doi: 10.1056/NEJM199405193302008. [DOI] [PubMed] [Google Scholar]
- 28.Li L, Chaikof EL. Mechanical stress regulates syndecan-4 expression and redistribution in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2002;22:61–8. doi: 10.1161/hq0102.100314. [DOI] [PubMed] [Google Scholar]
- 29.Li C, Wernig F, Leitges M, Hu Y, Xu Q. Mechanical stress-activated PKCdelta regulates smooth muscle cell migration. FASEB J. 2003;17:2106–8. doi: 10.1096/fj.03-0150fje. [DOI] [PubMed] [Google Scholar]
- 30.Libby P, Tanaka H. The molecular bases of restenosis. Prog Cardiovasc Dis. 1997;40:97–106. doi: 10.1016/s0033-0620(97)80002-3. [DOI] [PubMed] [Google Scholar]
- 31.Newby AC, Zaltsman AB. Molecular mechanisms in intimal hyperplasia. J Pathol. 2000;190:300–9. doi: 10.1002/(SICI)1096-9896(200002)190:3<300::AID-PATH596>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.