Abstract
Parkin, an E3 ubiquitin ligase implicated in Parkinson's disease, promotes degradation of dysfunctional mitochondria by autophagy. Using proteomic and cellular approaches, we show that upon translocation to mitochondria, Parkin activates the ubiquitin–proteasome system (UPS) for widespread degradation of outer membrane proteins. This is evidenced by an increase in K48-linked polyubiquitin on mitochondria, recruitment of the 26S proteasome and rapid degradation of multiple outer membrane proteins. The degradation of proteins by the UPS occurs independently of the autophagy pathway, and inhibition of the 26S proteasome completely abrogates Parkin-mediated mitophagy in HeLa, SH-SY5Y and mouse cells. Although the mitofusins Mfn1 and Mfn2 are rapid degradation targets of Parkin, we find that degradation of additional targets is essential for mitophagy. These results indicate that remodeling of the mitochondrial outer membrane proteome is important for mitophagy, and reveal a causal link between the UPS and autophagy, the major pathways for degradation of intracellular substrates.
INTRODUCTION
Parkin and PINK1 are Parkinson's disease (PD)-related proteins that operate in a common pathway to ensure mitochondrial integrity (1–5). Recent studies indicate that Parkin monitors the quality of the mitochondrial population and translocates from the cytosol onto dysfunctional mitochondria (6–11). Once on mitochondria, it promotes their degradation via mitophagy, an autophagic pathway specific for mitochondria (8). Loss of this surveillance mechanism presumably contributes to the accumulation of degenerative mitochondria observed in Parkin mutant flies (1,2,4).
Molecular models of Parkin function have evolved over the last decade. Parkin is an E3 ubiquitin ligase (12), and some disease alleles have impaired enzymatic activity (6,12,13). Because PD is characterized pathologically by intracellular protein aggregates termed Lewy bodies, early models postulated that Parkin functioned to promote the ubiquitin–proteasome system (UPS), which is activated by K48-linked polyubiquitination of substrate proteins (14). Mutation of Parkin would impair the ubiquitin–proteasome pathway (UPS) of protein degradation, leading to the toxic accumulation of misfolded or aggregated proteins. Since the discovery that Parkin promotes mitophagy (8), however, recent models have instead emphasized the ability of Parkin to mediate K63-linked polyubiquitin chains, distinct from the classic K48-linked polyubiquitin chains associated with the UPS. The topology of the polyubiquitin chain linkage determines the cellular outcome of polyubiquitination (15). It has been shown that the K63-linked ubiquitination of mitochondrial proteins by Parkin activates the autophagic machinery through recruitment of ubiquitin binding adaptors, such as HDAC6 and p62/SQSTM1 (6,13,16). The importance of this mechanism requires clarification, however, because p62/SQSTM1 null cells have no defect in Parkin-mediated mitophagy (17,18). Thus, the key molecular events occurring between Parkin-mediated ubiquitination of mitochondrial proteins and the degradation of mitochondria by the autophagic pathway remain unresolved.
To elucidate the proximal function of Parkin, we used quantitative proteomics to define, in an unbiased and highly comprehensive manner, how the mitochondrial proteome changes in response to Parkin activity. Our results indicate that in addition to K63-linked polyubiquitination, the K48-mediated UPS pathway has a major role in Parkin-dependent mitophagy. We observe robust recruitment of the 26S proteasome onto mitochondria, leading to widespread degradation of mitochondrial outer membrane proteins via the UPS. Strikingly, activation of the UPS not only precedes mitophagy, but is required for mitophagy. Inhibition of the UPS causes complete abrogation of mitophagy.
RESULTS
Parkin activation results in changes to the mitochondrial proteome
We performed stable isotope labeling by amino acids in cell culture (SILAC) analysis (19) to monitor changes in the mitochondrial proteome in a clonal Parkin-expressing HeLa S3 cell line after a 2 h treatment with carbonyl cyanide m-chlorophenylhydrazone (CCCP). CCCP dissipates the mitochondrial membrane potential, resulting in recruitment of Parkin to mitochondria and Parkin-dependent mitophagy (8). With this mass spectrometry-based approach, we quantified 2979 unique protein groups. Of these, 766 were mapped to proteins in the human MitoCarta inventory (20), which contains 1013 mitochondrial proteins. This represents a highly comprehensive coverage of the mitochondrial proteome, especially given that cultured cell lines express fewer mitochondrial proteins than tissues.
To sort through the proteins with altered SILAC ratios, we set a stringent threshold by considering only those with a calculated significance of <0.01 (Table 1 and Supplementary Material, Tables S1–S4). As expected, Parkin was highly enriched (13-fold) in mitochondria after CCCP treatment. Consistent with studies indicating that Parkin translocation leads to mitophagy, we found enrichment of several autophagy-related proteins, including p62/SQSTM1, NBR1, LC3 and the LC3 family member GABARAPL2. In addition, we found an increase in several subunits of the V-type proton ATPase, which is a component of acidic organelles, such as endosomes and lysosomes. The mitochondrial fission factor Drp1 was also substantially enriched, and probably contributes to the fragmentation of mitochondria observed after CCCP treatment.
Table 1.
Proteins with altered abundance in the mitochondria of CCCP-treated cells
| Protein | Biological function | SILAC ratioa | Significanceb |
|---|---|---|---|
| Increased proteins | |||
| PARKIN | E3 ubiquitin ligase | 13 | 4.42E−20 |
| DRP1 | Mitochondrial fission | 6.3 | 4.78E−07 |
| Autophagy related | |||
| NBR1 | Autophagy adaptor | 8.3 | 2.90E−06 |
| p62/SQSTM1 | Autophagy adaptor | 5.8 | 1.75E−06 |
| MAP1LC3B2;MAP1LC3B | Autophagosome component | 5.4 | 3.70E−06 |
| GABARAPL2 | Autophagosome component | 3.4 | 7.87E−06 |
| ATP6V1B2 | V-type proton ATPase subunit | 3.3 | 1.64E−05 |
| ATP6V1E1 | V-type proton ATPase subunit | 3.1 | 4.77E−03 |
| ATP6V1C1 | V-type proton ATPase subunit | 2.9 | 7.23E−03 |
| ATP6V1A | V-type proton ATPase subunit | 2.9 | 2.45E−03 |
| UPS related | |||
| Ubiquitin | Protein modification | 8.9 | 9.58E−15 |
| PSMA2 | 20S proteasome subunit | 4.2 | 8.08E−05 |
| PSMB5 | 20S proteasome subunit | 4.2 | 4.21E−04 |
| PSMA1 | 20S proteasome subunit | 4.0 | 5.38E−04 |
| PSMB3 | 20S proteasome subunit | 4.0 | 6.20E−04 |
| PSMB6 | 20S proteasome subunit | 3.9 | 2.30E−04 |
| PSMA6 | 20S proteasome subunit | 3.7 | 1.18E−03 |
| PSMB4 | 20S proteasome subunit | 3.7 | 1.96E−03 |
| PSMA7 | 20S proteasome subunit | 3.7 | 1.29E−03 |
| PSMA4 | 20S proteasome subunit | 3.6 | 4.70E−04 |
| PSMA3 | 20S proteasome subunit | 3.6 | 4.85E−04 |
| PSMB1 | 20S proteasome subunit | 3.3 | 2.66E−03 |
| PSMA5 | 20S proteasome subunit | 3.2 | 9.68E−04 |
| PSMD6 | 19S proteasome subunit | 2.7 | 6.22E−03 |
| PSMD2 | 19S proteasome subunit | 2.5 | 6.38E−03 |
| Decreased proteinsc | |||
| MFN1 | Mitochondrial fusion | 0.09 | 6.94E−18 |
| MFN2 | Mitochondrial fusion | 0.10 | 2.51E−35 |
| TOM70 | Mitochondrial import | 0.13 | 3.78E−40 |
| MIRO1/RHOT1 | Mitochondrial transport | 0.16 | 4.29E−15 |
| CPT1A | Fatty acid metabolism | 0.23 | 1.62E−21 |
| MOSC2 | Oxidoreductase | 0.26 | 6.67E−10 |
| MITONEET/CISD1 | Regulation of respiration | 0.26 | 8.74E−18 |
| GPAM | Glycerolipid synthesis | 0.42 | 3.39E−04 |
| MIRO2/RHOT2 | Mitochondrial transport | 0.44 | 4.94E−06 |
| FIS1 | Mitochondrial fission | 0.55 | 1.33E−03 |
Summary of the most significantly altered proteins in mitochondria isolated from Parkin-expressing HeLa S3 cells after treatment with or without CCCP for 2 h.
aCombined SILAC ratio from three independent mass spectrometry experiments consisting of two independent biological samples, and one technical replicate. In one of the experiments, the SILAC cells were reverse labeled with SILAC. The ratio represents the protein level in mitochondria of CCCP-treated cells divided by the level in untreated cells.
bCorresponds to the significance calculated in MaxQuant.
cOnly MitoCarta proteins annotated as 'outer membrane proteins’ in Uniprot are shown.
In addition to the autophagy pathway, the SILAC data provided evidence for a major induction of the ubiquitin–proteasome system (UPS). Firstly, ubiquitin was 9-fold more abundant in the mitochondria of CCCP-treated cells (Table 1). Secondly, numerous subunits of the proteasome were identified as enriched. This included subunits of both the 20S core particle and the 19S regulatory particle, thereby suggesting that 26S proteasomes are recruited to CCCP-treated mitochondria. Finally, we found a number of mitochondrial proteins that were substantially under-represented after CCCP treatment. Among them, there are proteins from all four mitochondrial sub-compartments. Because Parkin translocates to the surface of mitochondria, we focused on outer membrane proteins, which are more likely to represent proximal effects of Parkin translocation.
Mitochondrial outer membrane proteins were over-represented in the list of mitochondrial proteins with reduced abundance (Table 1). Although mitochondrial outer membrane proteins make up only 5% of the quantified mitochondrial proteins in our SILAC experiment, they constitute 20% of the reduced abundance proteins (P = 0.00014). The most severely reduced outer membrane proteins include the mitofusins Mfn1 and Mfn2, which mediate mitochondrial fusion. In addition, Miro1 and Miro2, which are involved in mitochondrial transport along microtubules, are also highly reduced. These data, along with the increase in Drp1, support the recent suggestion that Parkin, as part of a quality control mechanism, may serve to restrict defective mitochondria from fusing with neighboring mitochondria (11,21). Although several proteins involved in mitochondrial dynamics are degraded by Parkin, the list of rapidly degraded mitochondrial proteins is broad and includes ones involved in protein import and various biosynthetic pathways. For example, the mitochondrial import receptor subunit Tom70 is one of the most severely reduced proteins.
Parkin promotes degradation of mitochondrial outer membrane proteins
To extend these findings, we used immunoblotting to monitor the abundance of specific mitochondrial proteins in response to CCCP treatment. In agreement with our SILAC data, we found rapid degradation of Mfn1, Mfn2 and Tom70 upon CCCP treatment of Parkin-expressing cells (Fig. 1A). Almost complete degradation of these proteins occurred within the first hour of CCCP treatment. Such degradation was dependent on Parkin, because the levels of Mfn1, Mfn2 and Tom70 remained unchanged with CCCP in the parental HeLa S3 cells (Fig. 1A), which do not express Parkin (Supplementary Material, Fig. S1A). Similarly, we found Parkin-dependent degradation of other outer membrane proteins, although with slower kinetics. These proteins support a range of mitochondrial activities, including solute transport (VDAC1), regulation of apoptosis (Bak), mitochondrial fission (Fis1) and protein import (Tom20). Because of the slower kinetics, some of these proteins were not identified as reduced in the SILAC experiment, where cells were analyzed after 2 h of CCCP treatment. For Bak and Fis1, higher molecular weight species appeared after CCCP treatment (asterisks, Fig. 1A). No significant changes were observed for several mitochondrial matrix proteins, as well as the intermembrane space protein cytochrome c. In addition, there was only a slight reduction in the short isoforms of Opa1, an inner membrane protein. As reported previously, the long isoform of Opa1 is degraded in response to CCCP treatment (22,23), but in a Parkin-independent manner.
Figure 1.
Parkin mediates extensive proteolysis of outer membrane proteins via the UPS. (A) Parkin- and CCCP-dependent proteolysis of mitochondrial outer membrane proteins. HeLa S3 cells or clonal Parkin-expressing HeLa S3 cells were treated with vehicle or 20 μm CCCP to dissipate the mitochondrial membrane potential. Total cell lysates at the indicated time points were analyzed by immunoblotting for the indicated proteins. Outer membrane proteins: Mfn1, Mfn2, Tom70, VDAC1, Bak, Fis1, Tom20. Intermembrane space protein: cytochrome c. Inner membrane protein: Opa1. Matrix proteins: Hsp60, Sod2, F1β. Loading control: actin. (B) Inhibition of outer membrane protein degradation by the proteasome inhibitor MG132. Immunoblot analysis of Mfn1, Mfn2, VDAC1, Tom20 and actin (loading control) levels after CCCP with or without treatment with the proteasome inhibitor MG132 (10 μm). (C) Inhibition of outer membrane protein degradation by the proteasome inhibitor epoxomicin. Same as (B), except epoxomicin (2 μm), a more specific proteasome inhibitor, was used.
To determine whether the observed Parkin-mediated degradation of outer membrane proteins is dependent on the UPS, we incubated cells with proteasome inhibitors before treatment with CCCP (Fig. 1B and C). Indeed, inhibition of the proteasome with either MG132 or epoxomicin substantially blocked the degradation of Mfn1, Mfn2, VDAC1 and Tom20. In the case of Mfn1, Mfn2 and VDAC, this treatment also resulted in higher molecular weight species (asterisks, Fig. 1B and C). Therefore, we conclude that a proximal function of Parkin recruitment to depolarized mitochondria is to mediate the proteasome-dependent degradation of multiple mitochondrial outer membrane proteins. The degradation of outer membrane proteins was less efficient in HeLa cells expressing the R275W mutant, which has a partial defect in ubiquitination (Supplementary Material, Fig. S1B) (6).
We obtained similar protein degradation profiles by treating cells with valinomycin, a potassium ionophore that dissipates the electric potential across the inner membrane of mitochondria (Supplementary Material, Fig. S1C). In contrast, treatment with the complex I inhibitor rotenone did not result in outer membrane protein degradation (Supplementary Material, Fig. S1C). These results support the view that Parkin is activated by severe mitochondrial depolarization, rather than by mitochondrial dysfunction per se (24). Indeed, Parkin does not localize to mitochondria upon treatment with rotenone (25).
Degradation of outer membrane proteins occurs independently of autophagy
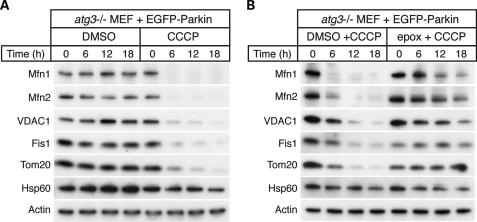
As in HeLa cells, expression of Parkin in mouse embryonic fibroblasts (MEFs) results in CCCP-induced mitophagy (8,17,18,26). We took advantage of the genetic tractability of this system to determine whether Parkin-dependent degradation of mitochondrial outer membrane proteins occurs independently of the autophagy pathway. Atg3 is an E2-like enzyme that is essential for conjugation of Atg8/LC3 to phosphatidylethanolamine, an early step in the induction of autophagosomes. Atg3-null MEFs do not show LC3 conjugation and are defective in autophagy (27). We found that Parkin-expressing Atg3-null MEFs underwent mitochondrial outer membrane protein degradation in response to CCCP (Fig. 2A). This degradation was blocked by epoxomicin (Fig. 2B). These results indicate that Parkin-mediated degradation of outer membrane proteins occurs via the proteasome and independently of the autophagy pathway.
Figure 2.
Proteolysis of the outer membrane occurs independently of the autophagy pathway. (A) CCCP-dependent degradation of mitochondrial outer membrane proteins in Atg3-null MEFs expressing Parkin. Atg3-null MEFs expressing EGFP-Parkin were treated with dimethyl sulfoxide (DMSO) (vehicle) or 20 μm CCCP for the indicated time. Total cell lysates were isolated and immunoblotted against the indicated proteins. (B) Inhibition of outer membrane protein degradation by the proteasome inhibitor epoxomicin. Atg3-null MEFs expressing EGFP-Parkin were treated with DMSO (vehicle) or the proteasome inhibitor epoxomicin (2 μm) for 2 h prior to treatment with 20 μm CCCP for the indicated times. Total cell lysates were isolated and immunoblotted against the indicated proteins.
Recruitment of the proteasome to depolarized mitochondria
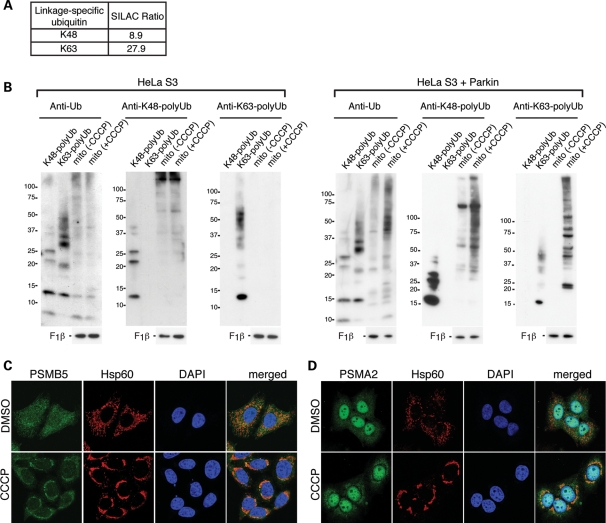
The UPS is classically associated with polyubiquitination via the K48 residue of ubiquitin (15). In our SILAC analysis, we found a substantial increase in both K48-linked (9-fold) and K63-linked (28-fold) polyubiquitination in mitochondria of cells treated with CCCP (Fig. 3A). We independently confirmed these mass spectrometric observations by immunoblot analysis using anti-polyubiquitin antibodies that are linkage specific (Fig. 3B). The increases in K48-linked and K63-linked polyubiquitin are both Parkin- and CCCP-dependent. Our SILAC data also indicated the accumulation of multiple 26S proteasome subunits to CCCP-treated mitochondria (Table 1). We confirmed this observation by immunostaining against α and β subunits of the proteasome. In Parkin-expressing HeLa cells, the anti-proteasome staining was diffusely cytosolic, but a clear enrichment on mitochondria was found upon CCCP treatment (Fig. 3C and D).
Figure 3.
Parkin activation results in mitochondrial K48-linked and K63-linked polyubiquitination and proteasome recruitment. (A) The SILAC ratio for K48-linked and K63-linked polyubiquitination obtained from mass spectrometric analysis of mitochondria isolated after 2 h of CCCP treatment. Both modifications yield unique diglycine signature peptides that can be quantified (41). Mitochondria were isolated under conditions where the activity of the 26S proteasome was not inhibited. (B) Accumulation of K48- and K63-linked polyubiquitinated proteins in mitochondria of CCCP-treated cells. Immunoblot analysis of mitochondria isolated from HeLa S3 or Parkin-expressing HeLa S3 cells, with or without CCCP treatment. Blots were probed with the following antibodies: anti-ubiquitin, anti-K48-linked polyubiquitin, anti-K63-linked polyubiquitin and anti-F1β (loading control). Purified polyubiquitin chains of the K48-linked or K63-linked type were used as controls to verify the specificity of the antibodies used. Cells were pretreated with the proteasome inhibitor MG132 (10 μm) together with CCCP, and mitochondria were isolated in the presence of N-ethylmaleimide (10 mm) to prevent deubiquitination. (C) Analysis of proteasome localization. HeLa cells expressing Parkin were treated with DMSO (vehicle) or 20 μm CCCP for 4 h. Formalin-fixed cells were stained for the β subunit PSMB5 of the proteasome (green), Hsp60 (red) and nuclei [4′,6-diamidino-2-phenylindole (DAPI), blue]. (D) Same as (C), except cells were immunostained for PSMA2 (green), an α subunit of the proteasome.
Degradation of mitochondrial outer membrane proteins precedes mitophagy
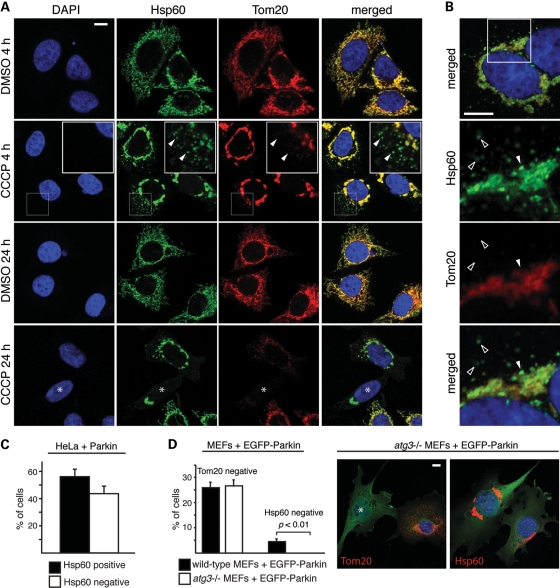
Given our finding that Parkin mediates proteolysis of outer membrane proteins, we investigated the temporal relationship of this process to mitophagy. We compared the behavior of the outer membrane marker Tom20 to the matrix marker Hsp60, which is not degraded by the UPS (Fig. 1A). When Parkin-expressing HeLa cells were treated with CCCP for 4 h, two populations of mitochondria could be distinguished (Fig. 4A). The bulk of the mitochondria aggregated around the perinuclear region, consistent with previous studies (6,10,17). These perinuclear aggregates were positive for both Tom20 and Hsp60, although discrete patches were negative for Tom20 (Fig. 4B, filled arrowhead). The second population consisted of individual mitochondria dispersed in the cell periphery. Remarkably, in all cells, this second mitochondrial population was strongly positive for Hsp60, but devoid of any Tom20 signal (Fig. 4A and B). These results indicate that the loss of Tom20 is distinct from mitophagy. Both the perinuclear clustering and loss of Tom20 signal were Parkin-dependent, as these events do not occur in HeLa cells lacking Parkin (Supplementary Material, Fig. S3A).
Figure 4.
Degradation of Tom20 occurs prior to mitophagy and does not require the autophagy pathway. (A) Degradation of Tom20 induced by CCCP treatment. After 4 and 24 h of CCCP (20 μm) or vehicle treatment, HeLa cells expressing Parkin were stained for Hsp60 (green), Tom20 (red) and nuclei (DAPI, blue). In the second row, the insets show enlarged views of the boxed area. Arrowheads mark examples of dispersed mitochondria that are positive for Hsp60 but negative for Tom20. In the fourth row, the asterisk indicates a cell with Tom20-negative/Hsp60-positive mitochondria. (B) Loss of Tom20 in both dispersed mitochondria and within patches of the mitochondrial aggregate. HeLa cells expressing Parkin were treated with CCCP for 4 h and stained for Hsp60 (green), Tom20 (red) and nuclei (DAPI, blue). The lower three panels correspond to the boxed area in the top panel. The filled arrowhead marks a patch in the perinuclear mitochondrial aggregate that is positive for Hsp60, but negative for Tom20. Unfilled arrowheads mark dispersed mitochondria that are Tom20 negative/Hsp60 positive. (C) Quantitation of the 24 h time point in (A). Cells that were Tom20 negative were scored for Hsp60 immunoreactivity. Error bars indicate standard deviations from three independent experiments; 100 cells were scored per experiment. (D) Tom20 degradation in Atg3-null MEFs. Wild-type and Atg3-null MEFs expressing EGFP-Parkin were treated with 20 μm CCCP for 24 h and immunostained for Tom20 or Hsp60. Cells were scored for complete loss of Tom20 or Hsp60. Representative images of Atg3-null cells are shown on the right. The left image shows an example of a Tom20-negative cell (asterisk). No Hsp60-negative cells were ever found (right image). Error bars represent standard deviations from three independent experiments; 100 cells were analyzed per experiment. The P-value was calculated using the t-test. All scale bars are 10 microns.
Later time points showed an even more dramatic discrepancy between Tom20 and Hsp60 staining. After 24 h of CCCP treatment, ∼20% of the cells showed complete loss of Tom20 immunostaining (Fig. 4A), consistent with previous studies. Among these cells, ∼40% have no detectable Hsp60 signal, whereas the other ∼60% retained strong Hsp60 signals despite an obvious reduction in overall mitochondrial biomass (Fig. 4A and C). The above observations are not specific to Hsp60, as immunostaining against two independent matrix proteins, TRAP-1 (Supplementary Material, Fig. S3B) and F1β (Supplementary Material, Fig. S3C), revealed a similar staining pattern. Previous studies have used the outer membrane protein Tom20 as a marker to monitor loss of mitochondria through Parkin-mediated mitophagy (6,8,9,13). Our observations indicate that, in some cases, the loss of Tom20 staining reflects Tom20 degradation by the UPS, rather than frank removal of the organelle. Our data therefore provide evidence that the Parkin-mediated proteolysis of outer membrane proteins precedes, and is distinct from, degradation of mitochondria via mitophagy.
We compared wild-type versus autophagy-deficient Atg3-null MEFs to further clarify the distinction between outer membrane protein degradation and mitophagy. After CCCP treatment, Parkin-expressing Atg3-null MEFs failed to undergo mitophagy (as indicated by preservation of Hsp60 staining) but still showed loss of Tom20 staining (Fig. 4D). These observations clearly demonstrate that degradation of mitochondrial outer membrane proteins, but not mitophagy, occurs independently of the autophagy machinery.
The UPS is essential for Parkin-mediated mitophagy
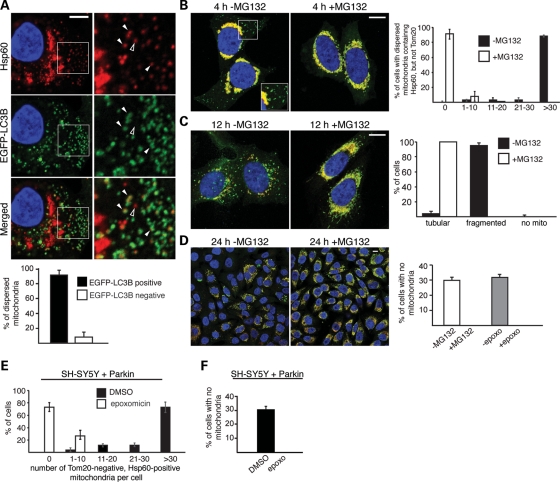
When Parkin-expressing cells were treated with CCCP for 4h, dispersed mitochondria at the cell periphery are Tom20 negative but Hsp60 positive (Figs 4A and B and 5B). Intriguingly, the autophagosome marker LC3B is selectively associated with these dispersed mitochondria (Fig. 5A). LC3B is present in ∼90% of the dispersed mitochondria, but generally absent from the aggregated mitochondria. Treatment with the proteasome inhibitor MG132 prevented the formation of dispersed, Tom20 negative, mitochondria at 4h after CCCP treatment (Fig. 5B), without affecting the recruitment of Parkin to mitochondria (Supplementary Material, Fig. S4A and B). Quantification showed that, in the absence of MG132, ∼90% of CCCP-treated cells show 30 or more Tom20-negative mitochondria that are dispersed in the cell periphery (Fig. 5B). In contrast, >90% of MG132-treated cells show no dispersed, Tom20-negative mitochondria (Fig. 5B). Similar results were obtained with epoxomicin (Supplementary Material, Fig. S4C and D). To understand the dynamics of Tom20 loss, we performed additional immunostaining experiments at early time points after CCCP treatment. In the absence of epoxomicin treatment, some Tom20-negative mitochondria arise prior to the overt perinuclear aggregation of mitochondria (Supplementary Material, Fig. S3D). After aggregation of the mitochondria, Tom20-negative mitochondria continue to accumulate. The latter observation suggests that the 26S proteasome may facilitate dispersion of mitochondria from the perinuclear aggregate, and thereby their uptake by the autophagy machinery. In addition, however, it is clear that Tom20-negative mitochondria can also arise without first being part of the perinuclear mitochondrial aggregate.
Figure 5.
Activation of the ubiquitin–proteasome pathway is essential for mitophagy. (A) Co-localization of dispersed mitochondria and LC3B in CCCP-treated cells. HeLa cells expressing Parkin and EGFP-LC3B were treated with 100 nm bafilomycin A1 and CCCP for 4 h, and stained for Hsp60 (red), EGFP-LC3B (green) and nuclei (DAPI, blue). Enlarged views of the boxed area are shown in the right column. Filled arrowheads mark examples of co-localization between dispersed mitochondria and EGFP-LC3B. The unfilled arrowhead marks an example of a dispersed mitochondrion that does not co-localize with EGFP-LC3B. Quantitation of this experiment is shown in the graph below. Error bars represent standard deviations from three independent experiments. Twenty cells were analyzed for each replicate, and ∼2400 dispersed mitochondria were manually assessed in total. In (B)–(D), Parkin-expressing HeLa cells were treated with CCCP in the presence or absence the proteasome inhibitor MG132 (10 μm). Cells were immunostained for Hsp60 (green), Tom20 (red) and nuclei (DAPI, blue). (B) MG132 inhibits Tom20 loss after 4 h of persistent CCCP treatment. Inset shows an enlarged view of the boxed area, highlighting the presence of dispersed, Tom20-negative mitochondria when MG132 is not present. The graph on the right shows quantification of this experiment. Each cell was scored into one of the five indicated bins, depending on the number of dispersed mitochondria that are Tom20 negative, but Hsp60 positive. Error bars indicate standard deviations from three independent experiments; 100 cells were scored per experiment. (C) MG132 preserves mitochondrial morphology at 12 h after a 100 min pulse treatment with CCCP. The graph on the right shows quantification of mitochondrial morphology. Cells were scored as having tubular mitochondria, fragmented mitochondria or no mitochondria. Error bars indicate standard deviations from three independent experiments; 500 cells were scored per experiment. (D) MG132 or epoxomicin abrogates CCCP-induced mitophagy. The images show cells 24 h after a 100 min pulse treatment with CCCP in the presence or absence of MG132 (10 μm). The graph on the right shows quantification of this experiment and a related one with epoxomicin (2 μm). Cells without mitochondria were defined by the complete lack of both Tom20 and Hsp60 signal. Error bars indicate standard deviations from three independent experiments. 1000 cells were scored for each MG132 experiment, and 200 cells were scored for each epoxomicin experiments. Scale bars equal 10 μm for (A)–(D). (E) Degradation of Tom20 in human neuroblastoma SH-SY5Y cells expressing exogenous Parkin. Cells were treated with DMSO (vehicle) or CCCP (20 μm) for 4 h. Cells were scored into one of the indicated five bins, depending on the number of dispersed mitochondria lacking Tom20, as described for (B). Error bars indicate standard deviations in three independent experiments; 100 cells were scored per experiment. (F) Epoxomicin abrogates CCCP-induced mitophagy in SH-SY5Y cells expressing Parkin. Cells were treated as in (D) in the presence or absence of epoxomicin (2 μm) and stained for Hsp60 and nuclei (DAPI). Cells without mitochondria were identified by complete loss of Hsp60 signal around the DAPI-stained nucleus. Error bars represent standard deviations from three independent experiments; 200 cells were analyzed per experiment.
Because Parkin-dependent proteolysis occurs prior to mitophagy, we asked whether this proteolysis is functionally coupled to mitophagy. When cells are treated with a 100 min pulse of CCCP and allowed to recover for 12 h, >90% of the cells show highly fragmented and dispersed mitochondria (Fig. 5C). By 24 h, ∼30% of the cells show complete loss of mitochondria, reflecting a high level of mitophagy (Fig. 5D). Pretreatment of cells with MG132 or epoxomicin dramatically changed the cellular outcome of the CCCP pulse. At 12 h, the cells showed an obvious preservation of mitochondria mass and tubular mitochondria, compared with cells without MG132 or epoxomicin (Fig. 5C and Supplementary Material, Fig. S4C and E). Most remarkably, at 24 h, all cells treated with MG132 or epoxomicin retained abundant mitochondria (Fig. 5D), indicating that the 26S proteasome is critical for Parkin-mediated mitophagy. Inhibition of the proteasome also blocked mitophagy in MEFs expressing Parkin (Supplementary Material, Fig. S4G). The degradation of mitochondria was reduced in HeLa cells overexpressing the K48R ubiquitin mutant (Supplementary Material, Fig. S2), consistent with the idea that K48-linked polyubiquitination is involved in Parkin-mediated mitophagy.
To determine whether these results apply to neuronal cells, we utilized the dopaminergic neuroblastoma cell line SH-SY5Y, which also undergoes CCCP-induced mitophagy upon expression of Parkin (6). As in HeLa cells, CCCP treatment of Parkin-expressing SH-SY5Y cells led to, at 4 h post-treatment, the appearance of peripheral mitochondria that were Tom20 negative but Hsp60 positive. Treatment with epoxomicin prevented the appearance of such Tom20-negative mitochondria (Fig. 5E and Supplementary Material, Fig. S4F). At 24 h post-CCCP treatment, epoxomicin also completely blocked mitophagy in the SH-SY5Y cells (Fig. 5F and Supplementary Material, Fig. S4F).
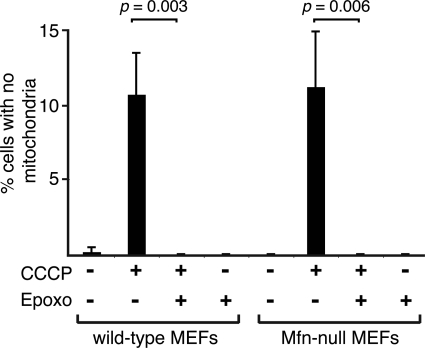
Because Mfn1 and Mfn2 are rapidly degraded downstream of Parkin in response to mitochondrial depolarization, we wondered whether inhibition of their degradation was responsible for the ability of proteasome inhibitors to block Parkin-mediated mitophagy. To address this issue, we tested whether epoxomicin could inhibit Parkin-mediated mitophagy in Mfn-null MEFs that lack both Mfn1 and Mfn2. We found that mitophagy in these cells, as in wild-type MEFs, was still completely blocked by epoxomicin (Fig. 6), indicating that degradation of proteins beyond Mfn1 and Mfn2 are essential for mitophagy caused by Parkin activation.
Figure 6.
Parkin-mediated mitophagy in Mfn-null cells is blocked by the proteasome inhibitor epoxomicin. EGFP-Parkin was expressed in wild-type (WT) and Mfn1/Mfn2-null MEFs containing matrix-targeted TagRFP-T. Cultures were treated with the indicated drugs, and EGFP-positive cells were scored for the presence of mitochondria. Error bars indicate standard deviations from three experiments; 200 cells were scored per experiment. The P-values were calculated using the t-test.
DISCUSSION
Previous studies showed that, upon recruitment to dysfunctional mitochondria, Parkin predominantly mediates K63-linked polyubiquitination, which facilitates recruitment of the autophagic adaptor p62 (6,13,16–18). However, p62 appears to be involved in controlling mitochondrial distribution and is not essential for degradation of mitochondria (17,18). In addition to the enrichment of K63-linked polyubiquitination, our proteomics and biochemical experiments revealed significant enrichment of K48-linked polyubiquitination, which is the linkage-type that classically targets proteins for degradation via the UPS. Given the rapid degradation of K48-linked proteins, effective detection of this type of polyubiquitination requires sensitive methods such as mass spectrometry, unless degradation of K48-linked proteins is prevented by inhibition of the UPS. Thus, it is not surprising that the role of K48-polyubiquitination in Parkin-mediated mitophagy was previously underappreciated. Taken together, these results indicate a dual molecular function for the ubiquitin ligase activity of Parkin. Polyubiquitination of mitochondrial proteins is used as an interaction module to recruit the autophagic machinery, as well as a signal for outer membrane protein degradation.
Several recent studies have focused on mitofusins as a degradation target of Parkin. In Drosophila, mitofusin (dmfn) is ubiquitinated and downregulated by Parkin (11,21). This relationship may underlie, in part, the strong genetic interactions between mitochondrial fusion/fission and the Pink1/Parkin pathway (28–30). In mammals, Mfn1 and Mfn2 are polyubiquitinated (31) and degraded (32) in response to Parkin activation. These studies in mammalian and Drosophila cells suggest that degradation of mitofusins may serve to segregate dysfunctional mitochondria, thereby enhancing the functional benefits of mitophagy (32–34).
During preparation of this manuscript, it was reported that Parkin-mediated degradation of mitofusins is prevented by over-expression of dominant-negative p97 or the proteasome inhibitor MG132 (32). Inhibition of either p97 or the proteasome also blocks mitophagy, but it is unclear whether this effect is due to the prevention of mitofusin degradation (32). Given that inhibition of the proteasome inhibits mitophagy, it is important to characterize the role of the UPS in this pathway and to evaluate whether mitofusins are the critical degradation targets of Parkin. Our proteomics and cell biological experiments demonstrate robust recruitment of the 26S proteasome to mitochondria and enrichment of K48-linked polyubiquitin, thus supporting a direct role of the UPS in Parkin-mediated mitophagy. In addition to mitofusins, we identified a broad range of mitochondrial outer membrane proteins that are degraded in a Parkin- and UPS-dependent manner. Importantly, we find that Parkin-mediated mitophagy in Mfn1/Mfn2-null cells is still blocked by inhibition of the proteasome. Therefore, although degradation of mitofusins may indeed serve to segregate dysfunctional mitochondria, our results provide strong evidence that additional remodeling of the mitochondrial outer membrane proteome, involving widespread proteolysis, is necessary to promote mitophagy. As a technical point, we note that accurate assessment of mitophagy in future studies will require the use of a marker protein located in the mitochondrial matrix, and not an outer membrane protein such as the commonly used Tom20.
Further work will be necessary to understand mechanistically how the UPS facilitates mitophagy. Two general models, not mutually exclusive, can be proposed. First, removal of outer membrane proteins may facilitate engulfment of mitochondria by autophagosomes. We speculate that aggregation of depolarized mitochondria depends on outer membrane proteins, and that degradation of such proteins by the UPS may help disperse mitochondria into small individual units so that they can be substrates for autophagy. This model is consistent with the broad degradation of outer membrane proteins by Parkin, and the observation that fragmentation of mitochondria is important for mitophagy (35). Alternatively, Parkin-mediated degradation may serve to remove one or more negative regulators of mitophagy on the mitochondrial surface, thereby triggering a signal for engulfment of a defective mitochondrion by an autophagosome.
By linking Parkin-mediated polyubiquitination to the UPS, our results support, in a general sense, earlier models postulating a role for Parkin in cellular protein quality control (14). In PD and other neurodegenerative disorders—including Alzheimer's, prion and polyglutamine diseases—defects in the UPS are thought to contribute to toxic accumulation of misfolded or aggregated proteins in the cytosol (14,36). Distinct from these earlier models, however, we show that for Parkin, the UPS operates within a separate cellular pathway involving mitochondrial quality control.
Our results provide evidence for the concept that the two major degradative pathways—the UPS and autophagy—are functionally linked (37). In future studies, it will be important to explore whether activation of the UPS is important for other forms of autophagy. In addition, although the UPS is clearly essential for Parkin-mediated mitophagy, it remains possible that activation of the UPS by Parkin has additional functions for mitochondria. Clarification of this issue will have important implications for the pathogenesis of PD.
MATERIALS AND METHODS
Cell culture and stable isotope labeling by amino acid in cell culture (SILAC)
Parkin-expressing cell lines were generated via lentivirus-mediated transduction of HeLa S3, HeLa cells and SH-SY5Y cells, followed by isolation of clones where indicated. Expression of Parkin is driven from the cytomegalovirus promoter. The wild-type and Atg3-null MEFs were kindly provided by Yu-Shin Sou and Masaaki Komatsu (27). Enhanced green fluorescent protein (EGFP)-tagged mouse Parkin was expressed in MEFs via retrovirus-mediated transduction, and EGFP-Parkin-expressing cells were selected by G418 (400 μg/ml). HeLa, HeLa S3 and MEF cell lines were cultured in Dulbeco's modified Eagle's medium (DMEM) containing 10% bovine serum (for HeLa and HeLa S3), or 10% fetal bovine serum supplemented with non-essential amino acids (for MEFs). For SILAC, DMEM lacking arginine and lysine was used, along with 10% dialyzed fetal bovine serum. For heavy labeling, Arg6 (U-13C6) and Lys8 (U-13C6, U-15N2) (Cambridge Isotopes) were supplemented at the same concentration as in the standard DMEM formulation. For light labeling, regular DMEM was used.
To dissipate the mitochondrial membrane potential, 20 µm carbonyl cyanide CCCP (Sigma) was used. For pulse treatments, cells were incubated in media with 20 µm CCCP for 100 min, followed by incubation in media without CCCP for the indicated time. For inhibition of the proteasome, cells were pretreated with 10 µm MG132 (Sigma), or 2 µm epoxomicin (Sigma) prior to treatment with CCCP.
Isolation of mitochondria
For immunoblot analysis, mitochondria were isolated as previously described (20). For SILAC, mitochondria were isolated from a 1:1 mixture of heavy and light SILAC-labeled, Parkin-expressing HeLa S3 cells. Cells were lysed using a nitrogen bomb (Parr) at 200 psi for 10 min, followed by mechanical homogenization with a glass–glass dounce homogenizer. Crude mitochondria were purified by differential centrifugation and further purified by a discontinuous Percoll gradient consisting of 80, 52 and 21% Percoll.
Isoelectric focusing of peptides
In-solution digested peptides (150 µg) were separated according to their isoelectric point with the Agilent 3100 OFFGEL fractionator (Agilent). The system was set up according to the manufacturers' guidelines (Agilent, 5969-1582), but strips were exchanged by 13 cm Immobiline™ DryStrip, pH 3–10 (GE Healthcare), and ampholytes were substituted by IPG buffer, pH 3–10 (GE Healthcare). Peptides were focused for 20 kilovolt hours at a maximum current of 50 µA, maximum voltage of 8000 V and maximum power of 200 mW into 12 fractions. Each peptide fraction was acidified by adding 1% trifluoroacetic acid, then desalted and concentrated on a reversed-phase C18 StageTips (Proxeon Biosystems) utilizing a reported adaptation (38). StageTips were first washed with 80% acetonitrile: 0.5% acetic acid, then methanol, followed by 2% acetonitrile: 1% triflouroacetic acid. Peptides were then loaded onto StageTips, washed with 0.5% acetic acid and eluted with 80% acetonitrile: 0.5% acetic acid.
Mass spectrometric analysis
All mass spectrometry experiments were performed on an EASY-nLC connected to a hybrid LTQ–Orbitrap classic (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Proxeon Biosystems) essentially as previously described (39) with some modifications. Peptides were separated on a 15 cm reversed-phase analytical column (75 µm internal diameter) in-house packed with 3 µm C18 beads (ReproSil-Pur C18-AQ medium; Dr Maisch GmbH) with a 160 min gradient from 5 to 35% acetonitrile in 0.2% formic acid at a flow rate of 350 nl per minute. The mass spectrometer was operated in data-dependent mode to automatically switch between full-scan MS and tandem MS acquisition. Survey full scan mass spectra were acquired in the Orbitrap (300–1700 m/z), after accumulation of 500 000 ions, with a resolution of 60 000 at 400 m/z. The top 10 most intense ions from the survey scan were isolated and, after the accumulation of 5000 ions, fragmented in the linear ion trap by collisionally induced dissociation (collisional energy 35% and isolation width 2 Da). Precursor ion charge state screening was enabled and all singly charged and unassigned charge states were rejected. The dynamic exclusion list was set with a maximum retention time of 90 s, a relative mass window of 10 ppm and early expiration was enabled.
Data analysis
Raw data files were analyzed by MaxQuant (v 1.0.13.13) (40) and searched against the IPI human database (v 3.54) with tryptic digestion, maximum of two missed cleavages, fixed carboxyamidomethyl modifications of cysteine, variable oxidation modifications of methionine and variable protein N-terminus acetylations with 1% false discovery rate thresholds for both peptides and proteins. At least two different peptide sequences were required for protein identification and two different ratio measurements were required for protein quantitation. Protein groups whose overall ratios were significantly different (P < 0.01) and whose ratios in at least two out of three replicates were also significantly different (P < 0.05) were considered to be significantly altered. Mitochondrial annotations were derived from MitoCarta (20) and mitochondrial outer membrane annotations were derived from Uniprot. Quantification of K48-linked and K63-linked polyubiquitin was performed by analysis of unique diglycine signature peptides associated with each modification (41).
Fluorescence microscopy
All images were acquired using a Plan-Apochromat 63X/1.4 oil objective on a LSM 710 confocal microscope with Zen 2009 software (Carl Zeiss) and processed (to crop out irrelevant areas, brightness and contrast adjustment) using ImageJ software.
Antibodies
The following commercially available antibodies were used: anti-actin (Mab1501R, Millipore), anti-Bak (Y164, Abcam), anti-cytochrome c (Mitosciences), anti-F1β (Mitosciences), anti-Fis1 (Alexis), anti-Hsp60 (SC-1052, Santa Cruz Biotech), anti-Mfn2 (Sigma), anti-Parkin (PRK8, Cell Signaling), anti-Parkin (Abcam), anti-Psma2 (BioMol), anti-Psmb5 (Abcam), anti-Sod2 (Abcam), anti-Tom20 (FL-145, Santa Cruz), anti-Tom70 (Novus Biologicals), anti-TRAP-1 (Abcam), anti-ubiquitin (Enzo), anti-K48-polyubiquitin (Cell Signaling), anti-K63-polyubiquitin (Enzo) and anti-VDAC (Molecular Probes). The following antibodies were raised in-house: anti-Opa1 monoclonal 1E8 and anti-Mfn1 (42).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the National Institute of Health (GM062967) and the Thomas Hartman Foundation for Parkinson's Research. Funding to pay the Open Access publication charges for this article was provided by the Howard Hughes Medical Institute.
ACKNOWLEDGEMENTS
We thank Ray Deshaies and the Chan lab for critical discussions and reading of the manuscript. We are grateful to Drs Yu-Shin Sou and Masaaki Komatsu for providing the Atg3-null MEFs, and to Willem den Besten and Ray Deshaies for providing ubiquitin plasmids.
Conflict of Interest statement: None declared.
REFERENCES
- 1.Clark I.E., Dodson M.W., Jiang C., Cao J.H., Huh J.R., Seol J.H., Yoo S.J., Hay B.A., Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. doi:10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 2.Greene J.C., Whitworth A.J., Kuo I., Andrews L.A., Feany M.B., Pallanck L.J. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl Acad. Sci. USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. doi:10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palacino J.J., Sagi D., Goldberg M.S., Krauss S., Motz C., Wacker M., Klose J., Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J. Biol. Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. doi:10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 4.Park J., Lee S.B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J.M., et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. doi:10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 5.Gautier C.A., Kitada T., Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl Acad. Sci. USA. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. doi:10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. doi:10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. doi:10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. doi:10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. doi:10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. doi:10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziviani E., Tao R.N., Whitworth A.J. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl Acad. Sci. USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. doi:10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 2000;25:302–305. doi: 10.1038/77060. doi:10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 13.Lee J.Y., Nagano Y., Taylor J.P., Lim K.L., Yao T.P. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J. Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. doi:10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciechanover A., Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. doi:10.1016/S0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 15.Deshaies R.J., Joazeiro C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. doi:10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 16.Ding W.X., Ni H.M., Li M., Liao Y., Chen X., Stolz D.B., Dorn Ii G.W., Yin X.M. Nix is critical to two distinct phases of mitophagy: reactive oxygen species (ROS)-mediated autophagy induction and Parkin-ubiqutin-p62-mediated mitochondria priming. J. Biol. Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. doi:10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okatsu K., Saisho K., Shimanuki M., Nakada K., Shitara H., Sou Y.S., Kimura M., Sato S., Hattori N., Komatsu M., et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narendra D.P., Kane L.A., Hauser D.N., Fearnley I.M., Youle R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. doi:10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.E., Walford G.A., Sugiana C., Boneh A., Chen W.K., et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. doi:10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poole A.C., Thomas R.E., Yu S., Vincow E.S., Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS ONE. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. doi:10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song Z., Chen H., Fiket M., Alexander C., Chan D.C. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. doi:10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griparic L., Kanazawa T., van der Bliek A.M. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 2007;178:757–764. doi: 10.1083/jcb.200704112. doi:10.1083/jcb.200704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin-induced mitophagy in the pathogenesis of Parkinson disease. Autophagy. 2009;5:706–708. doi: 10.4161/auto.5.5.8505. doi:10.4161/auto.5.5.8505. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda Y., Mitsui T., Kunishige M., Shono M., Akaike M., Azuma H., Matsumoto T. Parkin enhances mitochondrial biogenesis in proliferating cells. Hum. Mol. Genet. 2006;15:883–895. doi: 10.1093/hmg/ddl006. doi:10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- 26.Yao D., Gu Z., Nakamura T., Shi Z.Q., Ma Y., Gaston B., Palmer L.A., Rockenstein E.M., Zhang Z., Masliah E., et al. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc. Natl Acad. Sci. USA. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. doi:10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sou Y.S., Waguri S., Iwata J., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. doi:10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng H., Dodson M.W., Huang H., Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl Acad. Sci. USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. doi:10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole A.C., Thomas R.E., Andrews L.A., McBride H.M., Whitworth A.J., Pallanck L.J. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl Acad. Sci. USA. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. doi:10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y., Ouyang Y., Yang L., Beal M.F., McQuibban A., Vogel H., Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl Acad. Sci. USA. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. doi:10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gegg M.E., Cooper J.M., Chau K.Y., Rojo M., Schapira A.H., Taanman J.W. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. doi:10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M., Youle R.J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. doi:10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziviani E., Whitworth A.J. How could Parkin-mediated ubiquitination of mitofusin promote mitophagy? Autophagy. 2010;6:660–662. doi: 10.4161/auto.6.5.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pallanck L.J. Culling sick mitochondria from the herd. J. Cell Biol. 2010;191:1225–1227. doi: 10.1083/jcb.201011068. doi:10.1083/jcb.201011068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. doi:10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tai H.C., Schuman E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2008;9:826–838. doi: 10.1038/nrn2499. doi:10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 37.Korolchuk V.I., Mansilla A., Menzies F.M., Rubinsztein D.C. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol. Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. doi:10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rappsilber J., Ishihama Y., Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. doi:10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 39.de Godoy L.M.F., Olsen J.V., Cox J., Nielsen M.L., Hubner N.C., Frohlich F., Walther T.C., Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. doi:10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 40.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. doi:10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 41.Kirkpatrick D.S., Hathaway N.A., Hanna J., Elsasser S., Rush J., Finley D., King R.W., Gygi S.P. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. doi:10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 42.Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., Chan D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. doi:10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.