Summary
Small RNA molecules play key regulatory roles in many bacterial species. However, little mechanistic data exists for the action of small regulatory RNAs (sRNAs) in the human pathogen group A Streptococcus (GAS). Here, we analyzed the relationship between a putative GAS sRNA and production of the secreted virulence factor streptokinase (SKA). SKA promotes GAS dissemination by activating conversion of host plasminogen into the fibrin-degrading protease plasmin. Homologues of the putative sRNA-encoding gene fibronectin/fibrinogen-binding/hemolytic-activity/streptokinase-regulator-X (fasX) were identified in four different pyogenic streptococcal species. However, despite 79% fasX nucleotide identity, a fasX allele from the animal pathogen Streptococcus zooepidemicus failed to complement a GAS fasX mutant. Using a series of precisely-constructed fasX alleles we discovered that FasX is a bona-fide sRNA that post-transcriptionally regulates SKA production in GAS. By base-pairing to the 5’ end of ska mRNA, FasX enhances ska transcript stability, resulting in a ~10-fold increase in SKA activity. Our data provide new insights into the mechanisms used by sRNAs to activate target mRNAs, and enhances our understanding of the regulation of a key GAS virulence factor.
Keywords: sRNA, post-transcriptional regulation, Streptococcus pyogenes, virulence factor, RNase
Introduction
Bacteria have evolved a complex array of transcriptional and post-transcriptional regulatory mechanisms that integrate internal and external signals into an optimized response. RNA-based mechanisms of regulation are a key component of a cells regulatory capacity (Waters & Storz, 2009, Gottesman, 2005). With respect to bacterial pathogens, small regulatory RNAs (sRNAs) have been described that regulate virulence factor production and fitness within the host (Toledo-Arana et al., 2007). Individual sRNAs can function through drastically different mechanisms, from sequestration of regulatory proteins (Babitzke & Romeo, 2007, Lenz et al., 2005), to base-pairing with target mRNA molecules to increase or decrease their translation and/or stability (Boisset et al., 2007, Grieshaber et al., 2006, Morita et al., 2006). For the subclass of sRNAs that activate gene expression through base-pairing (reviewed in Frohlich & Vogel, 2009), molecular mechanisms include (i) activation of mRNA translation by induction of structural rearrangements in the 5’ untranslated region (UTR) that unmasks the ribosome binding site (RBS) (Soper et al., 2010, Lybecker & Samuels, 2007), and (ii) enhancement of mRNA stability through undetermined mechanisms after binding to the mRNA 3’-UTR (Opdyke et al., 2004). Very recently, the Clostridium perfringens sRNA VR-RNA was shown to enhance target mRNA stability after binding to the 5’-UTR (Obana et al., 2010).
The human bacterial pathogen group A Streptococcus (GAS, S. pyogenes) causes a diverse array of infections ranging from self-limiting pharyngeal (strep throat) infections to severe invasive infections such as necrotizing fasciitis (the flesh-eating syndrome). Importantly, the disease potential of GAS is attributable to the coordinated expression of specific subsets of encoded virulence factors (Roberts & Scott, 2007, Gryllos et al., 2008, Trevino et al., 2009). A critical GAS virulence factor is the secreted protein streptokinase (SKA) (Sun et al., 2004). SKA subverts components of the host fibrinolytic system to promote GAS spread from a local fibrin-clot-encapsulated infection to a systemic infection (McArthur et al., 2008). SKA promotes bacterial spread through activation of host plasminogen into plasmin, a broad spectrum protease that dissolves the meshwork of fibrin fibers present in a blood clot (Lottenberg et al., 1994, Svensson et al., 2002).
Recently, we performed a microarray-based genome-wide search for GAS sRNAs that, together with a previous bioinformatic approach, yielded an estimate of 75 sRNAs in the genome of the serotype M1 GAS isolate MGAS2221 (Livny et al., 2006, Perez et al., 2009). To date, only three putative or proven GAS sRNAs have been investigated experimentally. The pleiotropic effect locus (PEL) sRNA enhances the abundance of several virulence factor-encoding mRNAs in a strain-specific manner (Li et al., 1999, Mangold et al., 2004, Perez et al., 2009). The RofA-like protein IV regulator X (RIVX) sRNA enhances the abundance of mRNAs encoding the virulence factors C5a peptidase, cysteine protease, and M protein (Roberts & Scott, 2007). Finally, the putative fibronectin/fibrinogen-binding/hemolytic-activity/streptokinase-regulator-X (FasX) sRNA enhances the abundance of ska mRNA, and reduces the abundance of fbp and mrp mRNAs (which encode fibronectin and fibrinogen-binding proteins, respectively) (Kreikemeyer et al., 2001). Concomitant with the mRNA level differences, a wild-type GAS strain has increased SKA activity and decreased binding to fibronectin and fibrinogen compared to an isogenic fasX mutant (Kreikemeyer et al., 2001). While not studied in detail, FasX is positively regulated by the upstream three-component regulatory system FasBCA (Figure 1A) (Kreikemeyer et al., 2001). The mechanisms by which PEL, RIVX, or FasX regulate virulence factor expression are unknown.
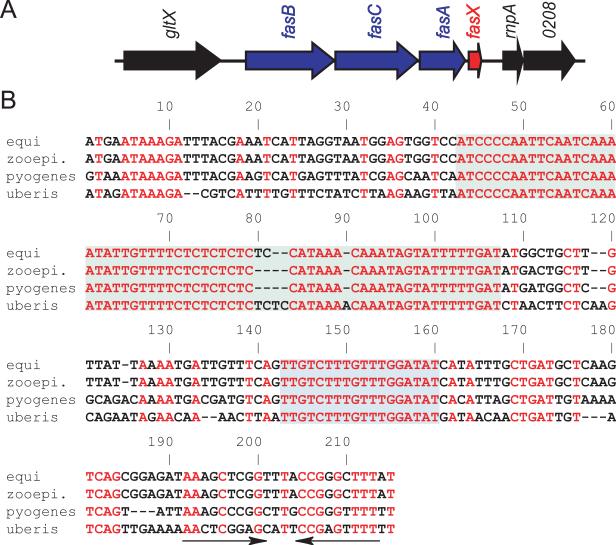
Figure 1. Distinct regions of FasX are conserved between different pyogenic streptococcal species.
(A) Arrangement of the fasBCAX locus in GAS. Genes are represented by block arrows facing the direction of transcription. Genes that lie within the fas locus are colored blue (fasBCA) or red (fasX). (B) Nucleotide sequence alignment of fasX alleles from representative GAS, S. zooepidemicus, S. equi, and S. uberis strains. The two conserved regions are shaded green and blue. The terminator hairpin is highlighted by inverted arrows.
Here, we discovered that FasX post-transcriptionally regulates SKA production through a process that requires FasX hybridization to the 5’-UTR of ska mRNA to enhance transcript stability. This is only the second description of a sRNA that stabilizes an mRNA target after binding to the 5’-UTR, and is the first for a sRNA that binds less than 30 nucleotides from the start codon, a binding location previously associated with negatively regulating sRNAs. Thus, we provide new insights into the mechanisms of sRNA-mediated positive regulation. Our data also elucidates a new layer of regulation in the expression of the critical GAS virulence factor SKA.
Results
Two regions of FasX are identical across pyogenic streptococcal species
Many sRNAs function by complementary base-pairing to target mRNAs. Therefore, fasX sequence comparisons may be informative by identifying regions of conserved and/or variable nucleotides. To assess conservation of the 205-bp fasX gene in serotype M1 GAS, we sequenced fasX from 48 serotype M1 isolates that were recovered from diverse geographical locations. The fasX gene was identical in all 48 strains (Table S1). To increase the potential for sequence variation we compared the fasX genes from the 13 available GAS genome sequences, which represent ten different serotypes. The resultant data was relatively uninformative as no more than two single nucleotide polymorphisms (SNPs) distinguished the contemporary M1 fasX allele from the fasX alleles of any other serotype (data not shown).
To further utilize available sequence information we expanded our fasX comparison to other streptococcal species. We compared the fasX allele of GAS isolate MGAS2221 with that of the Streptococcus equi isolate 4047 (Holden et al., 2009), the Streptococcus zooepidemicus isolate MGCS10565 (Beres et al., 2008), and the Streptococcus uberis isolate 0140J (Ward et al., 2009). S. equi and S. uberis are primarily opportunistic pathogens of horses and cows, respectively, although they can also cause bacteremia and meningitis in humans. S. zooepidemicus infects a wide-range of animals that include horses, cows, pigs, sheep, and dogs. The S. zooepidemicus (79% identity), S. equi (78% identity), and S. uberis (67% identity) fasX alleles all harbored multiple SNPs relative to the GAS allele (Figure 1B). Interestingly, two regions of the predicted FasX molecule were highly conserved between the four streptococcal species (green and blue shading in figure 1B). The locations of these conserved regions relative to the predicted FasX secondary structure are shown in figures S1A and S1B, which were generated using the bioinformatic programs RNAalifold and RNAfold, respectively (Hofacker et al., 2002, Hofacker, 2003). The paucity of variation in defined regions suggests that these nucleotides are critical to FasX function.
A S. zooepidemicus fasX allele does not complement the GAS fasX mutant strain 2221ΔFasX
To facilitate analysis of FasX-mediated regulation we created strain 2221ΔFasX, an isogenic fasX mutant of MGAS2221 (Figure S2). Strain 2221ΔFasX was complemented by introducing plasmid pFasXC, a fasX-containing derivative of the shuttle vector pDC123 (Chaffin & Rubens, 1998). To test whether a S. zooepidemicus fasX allele could complement strain 2221ΔFasX we also introduced plasmid pZOOFasX (Figure S3), which encodes FasX from the S. zooepidemicus strain MGCS10565. The mutation of fasX in strain 2221ΔFasX, and the restoration of FasX transcription in 2221ΔFasX containing plasmid pFasXC or pZOOFasX, was verified using Northern blot analysis (Figure 2A).
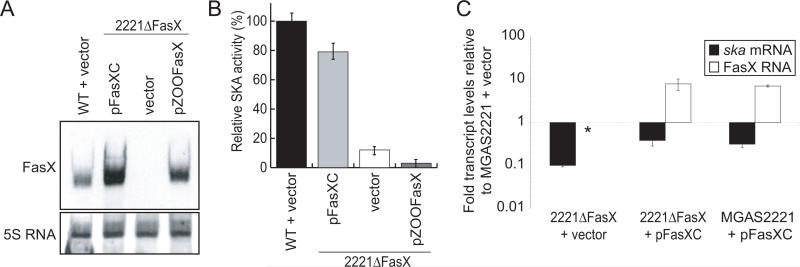
Figure 2. GAS fasX, but not fasX from S. zooepidemicus, can complement the GAS fasX mutant strain 2221ΔFasX.
(A) Northern blot showing restoration of fasX transcription in strain 2221ΔFasX containing either pFasXC (which contains GAS fasX) or pZOOFasX (which contains fasX from S. zooepidemicus). The Northern was probed with a FasX-specific probe, then striped and re-probed with a 5S RNA-specific probe for use as a loading control. (B) Indirect assay of streptokinase activity. Plasmid pFasXC, but not plasmid pZOOFasX, was able to complement the fasX mutant strain 2221ΔFasX. Data presented as percent SKA activity relative to that of parental strain MGAS2221 containing empty vector. The experiment was performed in triplicate with mean values (± standard deviation) shown. (C) Quantitative RT-PCR analysis. Complementation plasmid pFasXC, but not empty vector, enhanced the abundance of ska mRNA in mutant strain 2221ΔFasX. Data presented as fold-transcript levels relative to MGAS2221 containing the empty vector. The experiment was performed in quadruplicate with mean values (± standard deviation) shown. Asterisk highlights the fact that FasX RNA was not detected in the mutant strain containing empty vector.
SKA activity levels in the culture supernatants of strains MGAS2221 and 2221ΔFasX containing empty vector, and strain 2221ΔFasX containing pFasXC or pZOOFasX, were determined. As expected, plasmid pFasXC, but not empty-vector, restored high-level SKA activity to strain 2221ΔFasX (Figure 2B). The S. zooepidemics fasX allele did not complement strain 2221ΔFasX, indicating that the two regions of fasX conserved between GAS and S. zooepidemicus are not sufficient for regulation of GAS SKA activity (Figures 1B and 2B).
Using a quantitative RT-PCR approach we identified that fasX mutation leads to a drop in ska mRNA concentration to only 10% of that observed in the parent strain (Figure 2C). Surprisingly, the complementation plasmid pFasXC only restored ska mRNA levels to ~40% of wild-type levels, an unexpected finding given the high level of FasX RNA transcribed from the complementation plasmid relative to the parental strain (Figure 2A), and the restoration of 80% SKA activity in culture supernatants (Figure 2B). To address whether over-expression of FasX detrimentally effected the level of ska mRNA we introduced plasmid pFasXC into parental strain MGAS2221. The presence of plasmid pFasXC reduced ska mRNA levels in MGAS2221 to ~30% of that observed in MGAS2221 containing empty vector (Figure 2C). Thus, for uncharacterized reasons overproduction of FasX RNA has an inhibitory effect on this system.
The FasX RNA is the regulatory molecule
While postulated (Kreikemeyer et al., 2001), it has yet to be experimentally confirmed that the FasX transcript, and not a protein encoded within FasX, is the regulatory molecule. To facilitate testing of whether any of the five FasX open reading frames (ORFs; Figure S4A) were essential for regulatory activity, we performed site-directed mutagenesis to create plasmid-encoded fasX alleles mutated in one or more of the ORFs (Figure S5). As a consequence of overlap between ORFs only three 1-bp deletion-mutant alleles were required (plasmids pΔ1-3; Figures S4A and S4B). The complementation plasmid pFasXC and 1-bp deletion mutant derivatives pΔ1, pΔ2, and pΔ3, were transformed into strain 2221ΔFasX and their ability to restore SKA activity was tested. Each of the four plasmids restored SKA activity to wild-type levels (Figure S4C). Thus, the data are consistent with none of the FasX ORFs being required for SKA regulatory activity, and hence that FasX is a bonafide sRNA.
The deletion of single FasX nucleotides can result in the abrogation of SKA regulatory function
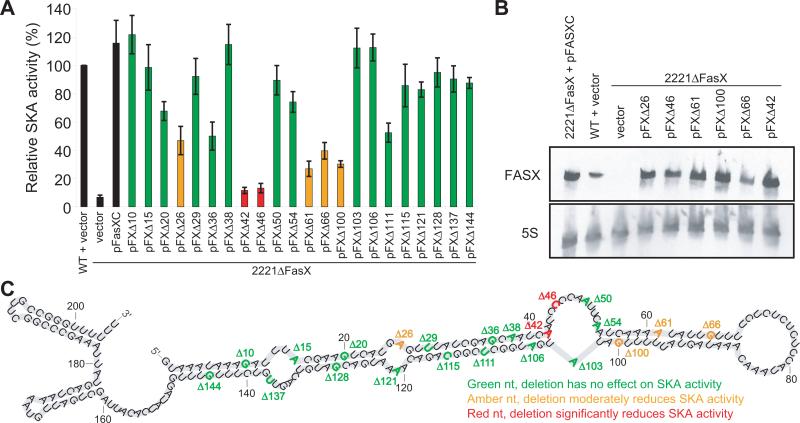
To conduct a systematic analysis of which FasX nucleotides were important for regulatory function we created 22 derivatives of the complementation plasmid pFasXC, each with a single nucleotide deletion. Each of the 22 pFasXC derivatives (pFXΔ plasmids) were transformed into strain 2221ΔFasX and SKA activity was tested. Significant variation was observed in the ability of the individual pFXΔ plasmids to restore SKA activity (Figure 3A). While the majority of pFXΔ-containing 2221ΔFasX derivatives had high SKA activity levels (green bars in figure 3A), four derivatives had only moderate activity (orange bars in figure 3A), and two derivatives had low activity (red bars in figure 3A). To ensure that 2221ΔFasX derivatives with moderate or low SKA activity levels produced FasX we performed Northern blot analysis. No appreciable difference in FasX abundance was observed between the tested 2221ΔFasX derivatives containing pFXΔ plasmids or the complementation plasmid pFasXC (Figure 3B). In addition, no appreciable difference in secondary structure was evident from bioinformatic predictions of the mutant FasX RNAs (data not shown). The locations of the nucleotides deleted in the 22 pFXΔ plasmids, and the effect of nucleotide deletion on SKA activity, are summarized in figure 3C.
Figure 3. Specific nucleotides of FasX are required for SKA regulatory activity.
The ability of plasmid-encoded wild-type or mutant fasX alleles to restore SKA activity to strain 2221ΔFasX was tested. (A) A series of twenty-two plasmid-encoded 1-bp deletion mutant fasX alleles (pFXΔ plasmids) were compared with the wild-type allele (pFasXC) via our indirect assay of SKA activity. Data presented as percent SKA activity relative to that of parental strain MGAS2221 containing empty vector. The experiment was performed in triplicate with mean values shown (± standard deviation). (B) Northern blot showing that the inability of particular mutant fasX alleles to restore SKA activity to strain 2221ΔFasX is not a result of an altered abundance of FasX transcripts. Northern blots were stripped and re-probed with a 5S RNA-specific probe to serve as a loading control. (C) Location and effect of nucleotide deletion within FasX. The single fasX nucleotide deleted in each pFXΔ plasmid is colored according to whether the mutation had no effect on SKA activity (green), had a negative effect (amber), or had a major negative effect (red). Colored nucleotides are numbered to highlight the pFXΔ plasmid that encodes each allele.
Nucleotides required for regulation of SKA activity are confined to a distinct region of the FasX molecule
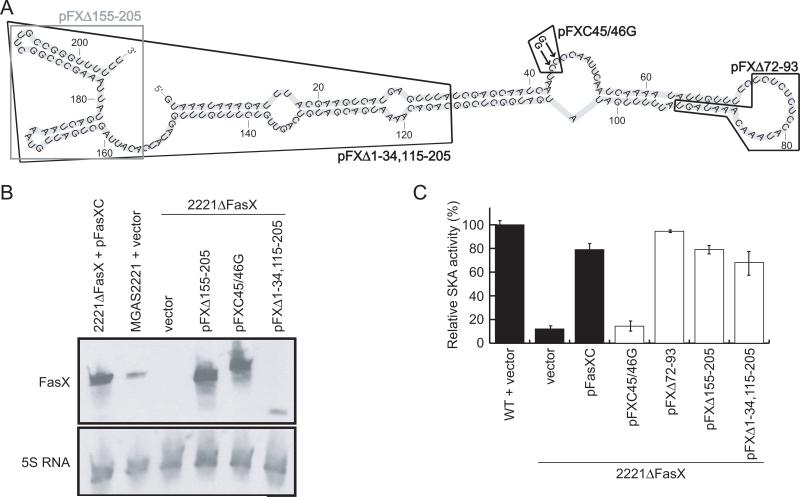
To further define FasX nucleotides required for regulatory activity we created four additional plasmid-encoded fasX mutant alleles. The first mutant harbored two SNPs, changing a C-repeat sequence CCCC to GGCC (plasmid pFXC45/46G; Figure 4A). The other three mutant alleles had large deletions of 22 nt (pFXΔ72-93), 51 nt (pFXΔ155-205), and 122 nt (pFXΔ1-34,115-205) (Figure 4A). After introduction into 2221ΔFasX, transcription of the mutant fasX genes was confirmed via Northern blot analysis (Figure 4B), and SKA assays were performed to assess the effect of the mutations on FasX activity (Figure 4C). Surprisingly, only plasmid pFXC45/46G failed to restore high level SKA activity to strain 2221ΔFasX. Thus, one or both of the C nucleotides that were substituted in plasmid pFXC45/46G appear critical for FasX function. In contrast, the extensive regions of FasX deleted in plasmids pFXΔ72-93, pFXΔ155-205, and pFXΔ1-34,115-205 are dispensable (Figure 4A).
Figure 4. A conserved region of FasX is necessary for SKA regulatory activity.
(A) Locations of the deletions present in the fasX alleles of plasmids pFXΔ155-205, pFXΔ1-34,115-205, pFXΔ72-93, and of the two nucleotide substitutions present in the fasX allele of plasmid pFXC45/46G. (B) Northern blot analysis of FasX abundance in 2221ΔFasX derivatives containing plasmid-encoded fasX alleles. RNA was isolated from exponential phase THY cultures of each GAS strain and used in Northern analysis with a FasX-specific probe. Note that due to the location of the probe, FasX levels could not be determined in strain 2221ΔFasX containing pFXΔ72-93. Blots were stripped and reprobed with a 5S RNA-specific probe to serve as a loading control. (C) Indirect assay of SKA activity showing that the fasX alleles of pFXΔ72-93, pFXΔ155-205, and pFXΔ1-34,115-205, but not pFXC45/46G, could restore high-level SKA activity to GAS strain 2221ΔFasX. The experiment was performed in triplicate with mean values (± standard deviation) shown.
FasX enhances ska mRNA stability
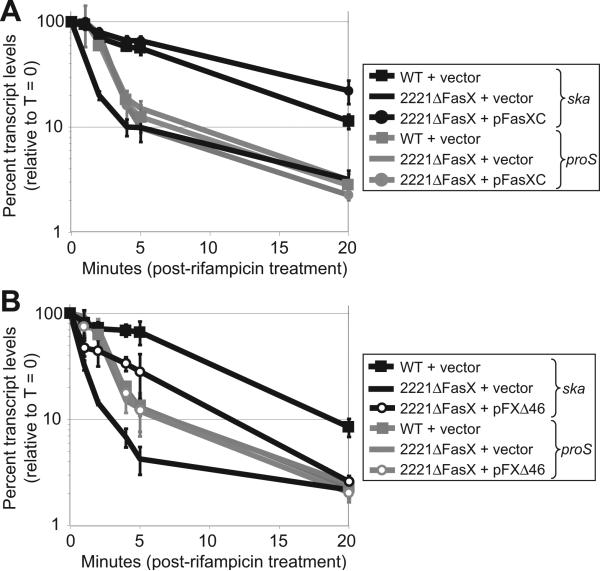
Deletion of fasX leads to a decrease in ska mRNA abundance (Figure 2C) (Kreikemeyer et al., 2001). Thus, FasX may act at the level of transcription, increasing the number of ska transcripts produced, or at the post-transcriptional level, by increasing the stability of ska transcripts. To test the hypothesis that FasX functions by increasing the stability of ska mRNA we used a quantitative RT-PCR approach. GAS cultures were treated with rifampicin to inhibit RNA synthesis and degradation of ska (test) and proS (control) mRNA transcripts were monitored over time. The proS mRNA transcripts were degraded at similar rates in each of the three strains tested (grey lines in figure 5A). In contrast, ska mRNA transcripts were highly unstable in the fasX mutant strain 2221ΔFasX, but more stable in the wild-type and complemented strains (black lines in figure 5A). Thus, the FasX-mediated regulation of SKA activity at least partially occurs at the post-transcriptional level by enhancing the stability of ska mRNA.
Figure 5. FasX enhances the stability of ska mRNA.
(A) Taqman quantitative RT-PCR analysis. RNA was isolated from rifampicin-treated GAS strains and the relative concentrations of ska and proS mRNAs determined. Data is presented as percent proS (control gene; grey lines) or ska (black lines) transcript levels relative to time-point zero. Experiment was performed in triplicate with mean values shown (±standard deviation). (B) Taqman quantitative RT-PCR analysis showing that the single nucleotide deletion present in the fasX allele of plasmid pFXΔ46 disrupts ability to enhance ska mRNA stability. Experiment was performed as in (A).
We next tested the hypothesis that the inability of the fasX allele present in plasmid pFXΔ46 to restore high level SKA activity to strain 2221ΔFasX (Figure 3A) was due to an inability to enhance ska mRNA stability. Using our quantitative RT-PCR approach we identified that while the stability of ska mRNA in strain 2221ΔFasX containing pFXΔ46 was increased relative to empty-vector (Figure 5B), the initial rate of ska mRNA degradation was similar, with 50% of the transcript being degraded after less than 1 min. In contrast, the ska transcript in strain MGAS2221 containing empty-vector had a half life of ~7.5 min. Thus, although the mutant FasX encoded within pFΔ46 afforded some protection to ska mRNA, the level of protection was significantly lower than that afforded by wild-type FasX.
Determination of the ska transcriptional start site
To facilitate analysis of the regulatory mechanism between FasX and ska mRNA we determined the transcriptional start site of the MGAS2221 ska gene. Our data identified the GAS ska transcriptional start site as being the first of four G residues located 32 nt upstream of the start codon, and is identical to that observed in S. equisimilis (Gase et al., 1995).
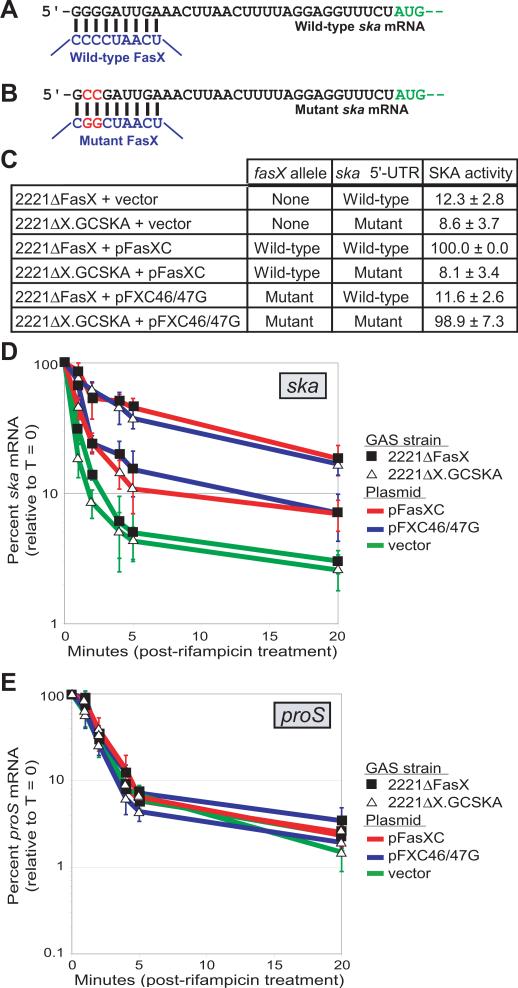
In vivo confirmation of FasX hybridization to the 5’-UTR of ska mRNA
Our data is consistent with the CCCC region of FasX being important for regulatory activity. To identify potential regions of complementarity between this region of FasX and ska mRNA we performed a bioinformatic analysis using the program TargetRNA (Tjaden et al., 2006). Nine nucleotides at the extreme 5’ end of ska mRNA were identified as being perfectly complementary to the CCCC region of FasX (Figure 6A). To facilitate testing whether this putative FasX:ska mRNA interaction occurs in vivo we created strain 2221ΔX.GCSKA, a 2221ΔFasX derivative in which the four G residues located upstream of ska were mutated to GCCG (Figure 6B). If the residues at the 5’ end of ska mRNA base-pair with FasX, and if this interaction is required to enhance ska mRNA stability, then the low level of SKA activity produced by strain 2221ΔX.GCSKA would not be complemented by introduction of pFasXC (which contains wild-type fasX), but would be complemented by introduction of pFXC46/47G (which contains a fasX-derivative in which the CCCC region has been mutated to CGGC; Figure 6B). To test this idea we used SKA activity assays (Figure 6C). The data are consistent with FasX interacting directly with the 5’ end of ska mRNA. That ska mRNA is stable only when FasX and the ska 5’-UTR are complementary to one another was confirmed by quantitative RT-PCR (Figures 6D and 6E). Thus, FasX post-transcriptionally regulates expression of the secreted virulence factor SKA by hybridizing to the 5’-UTR of ska mRNA and increasing transcript stability.
Figure 6. In vivo confirmation of interaction between FasX and the 5’-UTR of ska mRNA.
(A) Putative region of hybridization between wild-type FasX (blue) and the 5’-UTR of wild-type ska mRNA (black). The ska AUG start codon is colored green. (B) Putative region of hybridization between mutant FasX (blue; as is present in plasmid pFXC46/47G) and the 5’-UTR of mutant ska mRNA (black; as is present in GAS strain 2221ΔX.GCSKA). The two nucleotide substitutions present in the mutant fasX allele and in the mutant 5’-UTR of ska are colored red. (C) Table showing the percent SKA activity, relative to complemented strain 2221ΔFasX pFasXC, of six GAS strains tested via our indirect assay. Relative SKA activity is the mean value calculated from four independent experiments (± standard deviation). (D) Taqman quantitative RT-PCR analysis showing that FasX enhances the stability of ska mRNA only when complementary to the 5’-UTR of ska. Data is presented as percent ska transcript levels relative to time point zero. Strains analyzed were either 2221ΔFasX (filled squares) or 2221ΔX.GCSKA (open triangles) derivatives. Strains contained either empty vector (green lines), plasmid pFasXC (red lines), or plasmid pFXC46/47G (blue lines). Experiment was performed in triplicate with mean values shown (±standard deviation). (E) The stability of transcripts from the house-keeping gene proS are unaffected by the presence or absence of FasX. These control proS qRT-PCR reactions were ran the same time as the ska reactions in D, but are shown in a separate figure to enhance data visualization. Experiment was performed in triplicate with mean values shown. Error bars represent ±standard deviation.
Addition of complementary nucleotides to the 5’ end of ska mRNA enhances transcript stability in a FasX-independent manner
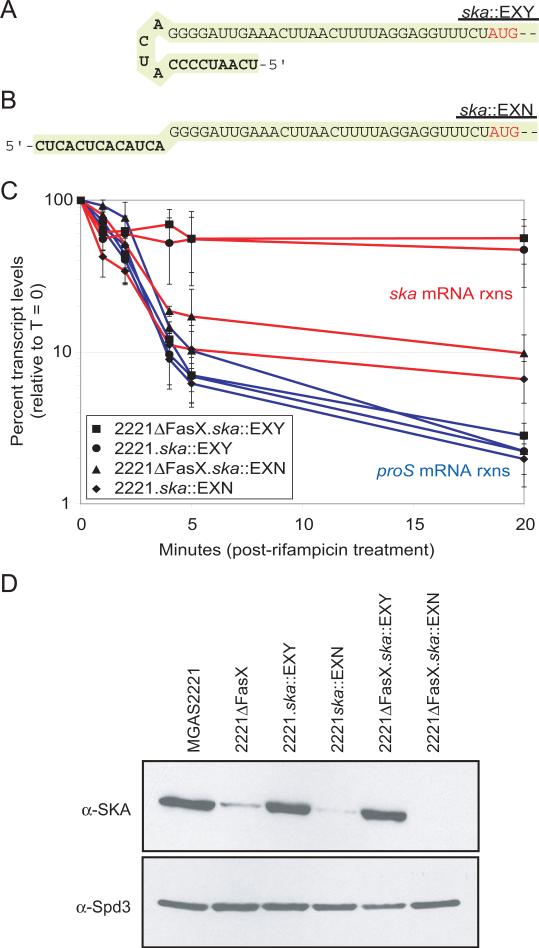
We hypothesized that addition of a complementary loop to the 5’-end of ska mRNA that mimics FasX binding would stabilize the transcript and uncouple ska mRNA stability from FasX-mediated regulation. In addition, we hypothesized that addition of a non-complementary sequence would result in unstable ska transcripts regardless of the FasX status of the cell. To test these hypotheses we used homologous recombination to insert complementary or non-complementary sequences at the ska transcriptional start site (Figures 7A & 7B). The insertions were made in both the MGAS2221 background and the 2221ΔFasX background. The stability of ska mRNA in the four strains was tested by quantitative RT-PCR following addition of rifampicin. As predicted, strains containing the complementary extension to the 5’-end of ska mRNA had highly stable ska transcripts, both in the presence (strain 2221.ska::EXY) and absence (strain 2221ΔFasX.ska::EXY) of FasX, while strains with the non-complementary extension had unstable ska transcripts in the presence (strain 2221.ska::EXN) and absence (strain 2221ΔFasX.ska::EXN) of FasX (Figure 7C). Western blot analysis of SKA expression supported the ska transcript stability data (Figure 7D).
Figure 7. Complementary, but not uncomplementary, nucleotides added to the 5’-end of ska mRNA enhances stability in a FasX-independent manner.
(A) Nucleotide sequence of the modified ska 5’-UTR from strains containing a complementary extension (strains ending ska::EXY). The 13 nucleotides added at the ska 5’ end are in bold. The ska start codon is colored red. (B) Nucleotide sequence of the modified ska 5’-UTR from strains containing an uncomplimentary extension (strains ending ska::EXN). The 13 nucleotides added at the ska 5’ end are in bold. The ska start codon is colored red. (C) Quantitative RT-PCR-based analysis of ska (red lines) and proS (blue lines) mRNA stability in MGAS2221 and 2221ΔFasX derivatives containing complementary and uncomplementary extensions to ska. Samples from duplicate experiments were ran in triplicate with mean values shown (± standard deviation). (D) Western blot analysis assaying for SKA protein levels in the exponential phase culture supernatants of the indicated strains. The non-FasX regulated secreted protein Spd3 was assayed for use as a loading control.
FasX does not enhance ska mRNA stability by inhibiting the ribonucleases CvfA and PNPase
In Bacillus subtilis the initial, and rate-limiting, step in the mRNA decay pathway is endonucleolytic cleavage of the 5’ end of a transcript (Commichau et al., 2009, Condon, 2003). In part because of this we hypothesized that hybridization of FasX with ska mRNA blocks the access of an endoribonuclease to the ska transcript, preventing cleavage and leading to the observed enhanced transcript stability. Conserved virulence factor A (CvfA) was recently characterized in GAS as being a membrane-spanning protein with putative endoribonuclease activity (Kang et al., 2010). Deletion of the cvfA gene has a major effect on transcript levels in a growth-phase and nutritional stress-dependent manner (Kang et al., 2010). To facilitate testing whether FasX enhances ska mRNA abundance by inhibiting CvfA-mediated cleavage of the transcript we created cvfA-mutant derivatives of MGAS2221 and 2221ΔFasX. SKA expression and ska mRNA stability were compared between the two cvfA mutant strains and two control strains. Neither cvfA mutant strain differed from their control strain with respect to the rate of ska mRNA degradation and the level of secreted SKA protein (Figures S6A and S6B). Thus, FasX does not enhance ska transcript stability by inhibiting CvfA-mediated cleavage. It is noteworthy that in contrast to ska mRNA, degradation of proS mRNA (which we monitored as a control transcript) was effected by cvfA mutation (Figure S6A).
The exoribonuclease polynucleotide phosphorylase (PNPase; encoded by the pnpA gene) contributes to the growth phase regulation of mRNA stability in GAS (Barnett et al., 2007). Given that PNPase regulates the stability of several GAS mRNAs we tested whether FasX regulates SKA production by inhibiting PNPase activity. Similar to the cvfA data, inactivation of pnpA had no effect on SKA expression, regardless of the presence or absence of FasX (Figure S6C and data not shown). Thus, FasX does not enhance ska transcript stability by inhibiting PNPase-mediated cleavage.
Discussion
The prevalence of sRNAs in bacterial genomes indicates that they represent a fundamental mechanism of regulation (Gottesman, 2005, Waters & Storz, 2009). Not confined to a single level of regulation, sRNAs have been described in E. coli that regulate transcriptionally (e.g. the 6S RNA inhibits transcription from σ70 promoters (Wassarman, 2007)), post-transcriptionally (e.g. the sRNA RyhB represses the translation and stability of sodB mRNA (Afonyushkin et al., 2005)), and post-translationally (e.g. the sRNA CsrB sequesters the global regulatory protein CsrA (Babitzke & Romeo, 2007)). In part due to the absence of mechanistic data regarding sRNA-mediated regulation in the human pathogen GAS, the current study examined the regulation of the key virulence factor SKA by the sRNA FasX. Research into SKA production by beta-hemolytic streptococci began more than seven and a half decades ago (Tillett & Garner, 1933), and continues today in part due to the critical role of this enzyme during infection (Svensson et al., 2002, Sun et al., 2004, Khil et al., 2003). SKA activity is negatively regulated at the transcriptional level by the CovR/S (also known as CsrR/S) two-component regulatory system (Federle et al., 1999, Gryllos et al., 2008, Levin & Wessels, 1998, Sumby et al., 2005). Here, we have shown that FasX positively regulates SKA activity post-transcriptionally by binding to the 5’-UTR of ska mRNA to increase transcript stability. The highly regulated nature of SKA expression is consistent with the level and timing of SKA expression being important during infection.
Transcription of fasX is regulated by the upstream genes fasBCA (Figure 1A) (Kreikemeyer et al., 2001). The fasBC genes encode proteins with homology to sensor kinases, while fasA encodes a protein with homology to response regulators. Thus, fasBCA may encode a three-component system through which the predicted membrane-spanning proteins FasB and FasC recognize as as-yet-unknown signals, resulting in transduction of the signal through FasA (possibly via phosphorylation), activating fasX transcription. Other than fasBCA all being required for fasX transcription (data not shown)(Kreikemeyer et al., 2001), nothing else is known regarding the regulation of fasX transcription by FasBCA.
The fasBCAX operon structure is conserved in GAS, S. zooepidemicus, and S. equi (Holden et al., 2009, Beres et al., 2008). In contrast, S. equisimilis has one, and S. uberis has four, putative sensor kinases upstream of the fasAX genes (Ward et al., 2009, Steiner & Malke, 2002). Despite conservation of operon structure, the fasX gene of S. zooepidemicus strain MGCS10565 did not restore high-level SKA activity to GAS mutant strain 2221ΔFasX (Figure 2B). This data identified that the two conserved regions of fasX, as identified by comparisons of fasX from four streptococcal species (Figure 1B), were insufficient for SKA-regulatory activity. Furthermore, while the S. zooepidemicus fasX allele contains a SNP within the 9 nt complementary to the ska 5’-UTR, this does not fully explain the inability of this allele to regulate SKA activity. We generated a derivative of the S. zooepidemicus fasX allele in which the SNP was substituted to that observed in GAS, and this allele still failed to restore high-level SKA activity to GAS strain 2221ΔFasX (data not shown). Thus, it appears that these 9 nt are not sufficient for regulatory activity. Interestingly, despite conservation of 8 of the 9 nt involved in hybridizing to the ska 5’-UTR in GAS, neither S. zooepidemicus, S. equi, or S. uberis have the corresponding complementary nucleotides upstream of the ska gene. Thus, if FasX regulates ska mRNA stability in these other streptococcal pathogens then it does so through a different mechanism, or at least through different nucleotides, than in GAS.
That deletion of FasX nucleotides 42 and 46 abrogated ska-regulatory function was not surprising given that they lie within the 9 nt complementary to the ska 5’-UTR (Figure 3C). In addition, nucleotide 42 is predicted to contribute to FasX secondary structure by base-pairing with FasX nucleotide 104, providing another possible explanation of why removal of this nucleotide is detrimental to regulatory activity. Disruption of FasX secondary structure may also explain why deletion of nucleotides 61, 66, or 100 impedes regulation. However, as nucleotide 26 is located in a predicted single-stranded region of FasX then disruption of secondary structure cannot be implicated in the reduced regulatory activity of the Δ26 fasX allele (Figure 3C). Rather than affecting secondary structure it is possible that nucleotides 61, 66, and 100 overlap with a binding site for an as-yet-unknown RNA-binding protein that is required for efficient regulation.
The importance of the RNA-binding protein Hfq in promoting sRNA-mediated regulation has been well established (Lenz et al., 2004, Christiansen et al., 2004, Ding et al., 2004, Fantappie et al., 2009, Kulesus et al., 2008, Meibom et al., 2009, Sharma et al., 2010). However, in a select number of species (e.g. Staphylococcus aureus) the hfq gene can be deleted without a detectable effect on sRNA-mediated regulation (Bohn et al., 2007). Furthermore, many pathogens naturally lack a Hfq homologue (e.g. pathogens of the genera Streptococcus, Enterococcus, Helicobacter, and Mycobacterium) (Chao & Vogel, 2010, Sun et al., 2002). It is possible that a functional homologue of Hfq is encoded within the genomes of species that lack Hfq. Such a finding may provide an explanation as to why over-expression of FasX RNA has a negative effect on FasX-mediated regulation of SKA activity (Figure 2C), as excess FasX may titrate this protein out, reducing the efficiency of the system.
In addition to FasX positively regulating the abundance of ska mRNA, it also negatively regulates the abundance of the adhesin-encoding mRNAs mrp and fbp (Kreikemeyer et al., 2001). The decreased expression of adhesins, and increased expression of factors that promote bacterial spread from a site of infection, is suggestive of FasX regulating the transition of GAS from the colonization to dissemination phases of infection (Kreikemeyer et al., 2001). While the mechanisms by which FasX regulates mrp and fbp transcript levels are unknown, the library of fasX mutant alleles constructed here may facilitate their identification. There are no obvious regions of complementarity between FasX and the mrp / fbp genes. Given that a distinct conserved region of FasX appears to be required for regulation of SKA activity, it is possible that the other conserved regions of FasX mediate regulation of these other targets.
The FasX sequence complementary to the ska 5’ UTR is 5’-UCAAUCCCC-3’. The C rich region is reminiscent of a conserved UCCC sequence motif recently identified in 11 previously uncharacterized S. aureus sRNAs (Geissmann et al., 2009). One of these sRNAs was investigated and the UCCC motif identified as directly base-pairing to target mRNAs. Similarly, the well-described virulence factor-regulating S. aureus sRNA RNAIII has UCCC sequences in three hairpin loops, and each are known to interact with target mRNA sequences (Boisset et al., 2007). It has been suggested that the UCCC motif represents a novel class of S. aureus sRNAs that target mRNAs through a common mechanism (Geissmann et al., 2009), and it is possible that the same mechanism is also present in other low GC% Gram-positive pathogens such as GAS.
RNA turnover is the primary function of the degradosome, a multi-enzyme complex which promotes RNA decay (Carpousis, 2007). While primarily studied in E. coli (Morita et al., 2004), the degradosome has also been studied in the model Gram-positive organism B. subtilis (Commichau et al., 2009). Proteins within the B. subtilis degradosome include the endoribonuclease RNase J1, the 3’ to 5’ exoribonuclease PNPase, the glycolytic enzyme enolase, and the membrane-bound endoribonuclease RNase Y (Commichau et al., 2009). Initiation of mRNA decay in B. subtilis is through binding of an endoribonuclease to the 5’ end of the transcript and tracking along to a cleavage site (Condon, 2003, Commichau et al., 2009). We hypothesize that control of mRNA degradation in GAS is also 5’-end dependent in at least some cases. Recently, CvfA was identified as the RNase Y homologue in GAS and was also shown to interact with enolase, suggesting the presence of a degradosome in this organism (Kang et al., 2010). Given this information we tested the hypothesis that CvfA was responsible for initiation of ska mRNA degradation, and that this was inhibited by FasX base-pairing to the 5’ end of ska mRNA. However, isogenic cvfA mutant strains were unchanged in their rate of ska mRNA degradation, regardless of the presence or absence of FasX (Figure S6A and S6B), indicating that CvfA is not the RNase inhibited by FasX. As PNPase regulates the abundance of several GAS mRNAs we also tested whether this was the RNase inhibited by FasX (Barnett et al., 2007). Disruption of the pnpA gene also failed to enhance ska mRNA stability (Figure S6C). Our current working hypothesis is that one or both of RNases J1 and J2, both of which are essential endoribonucleases in GAS (Bugrysheva & Scott, 2009), are responsible for initiating ska mRNA degradation via a process that is inhibited by FasX:ska mRNA hybridization.
Derivatives of MGAS2221 and 2221ΔFasX that contained a complementary 13 nucleotide extension to the 5’ end of ska mRNA had highly stable ska transcripts (Figure 7B). The high and similar level of ska mRNA stability in strains 2221.ska::EXY and 2221ΔFasX.ska::EXY indicates that the added extension protects the mRNA in a FasX-independent manner. We believe that this is due to the extension folding back and forming a stem:loop structure at the 5’ end of the molecule as depicted in figure 7A. In B. subtilis the presence of a stem:loop structure at the 5’ end of an mRNA enhances its stability (Sharp & Bechhofer, 2005), which is thought to be due to blockage of endoribonuclease binding.
Derivatives of MGAS2221 and 2221ΔFasX that contained a non-complementary 13 nucleotide extension to the 5’ end of ska mRNA had unstable ska transcripts (Figure 7B). The inability of FasX to enhance ska transcript stability in strain 2221.ska::EXN indicates that nucleotides at the extreme 5’ end of ska mRNA, rather than downstream nucleotides, must be base-paired to protect against RNase-mediated degradation. This is supported from work in B. subtilis which identified that stem:loop structures could only enhance mRNA stability when they were located less than 5 nucleotides from the 5’-end of the transcript (Sharp & Bechhofer, 2005). In strain 2221.ska::EXN the FasX:ska base-pairing would start 14 nucleotides from the 5’ end of the transcript.
We propose that FasX is the second of what may be a large class of sRNAs that enhance the stability of their mRNA targets by formation of base-paired structures at, or very close to, the mRNA 5’ end. Interestingly, while FasX and the C. perfringens sRNA VR-RNA both enhance target mRNA stability after binding to the 5’-UTR their mechanisms differ. FasX binds to the 5’ end of ska mRNA to create secondary structure predicted to inhibit endoribonuclease-mediated degradation. In contrast, base-pairing between VR-RNA and its mRNA target leads to a cleavage event downstream of the base-pairing site, resulting in a processed mRNA that lacks 62 nucleotides at the 5’ end relative to the full-length transcript (Obana et al., 2010). Unlike the full-length transcript, which has a predicted 33 nucleotide single stranded region at the 5’ end, the 5’ end of the processed transcript is present within a protective stem:loop structure. A potentially important distinction between FasX and VR-RNA is that FasX must remain bound to its mRNA target to enhance stability while VR-RNA does not. The reversible nature of the FasX:ska mRNA interaction implies that the positive regulation afforded by FasX could be removed by decreasing FasX transcription, or increasing FasX turnover.
FasX base-pairs with ska mRNA nucleotides –32 to –24 (relative to the first nucleotide of the start codon). That FasX binding to this region increases SKA expression was surprising given the current convention that sRNA hybridization to an mRNA between nucleotides -35 to +15 leads to inhibition of translation (Storz et al., 2004, Morita et al., 2006, Bouvier et al., 2008, Marzi et al., 2007, Sharma et al., 2007, Frohlich & Vogel, 2009). While we have not ruled out that FasX also alters translation of ska mRNA, we do not believe that this is the case as the drop in ska mRNA abundance following fasX mutation (10% of wild-type; Figure 2C) is similar to the drop in SKA activity (10% of wild-type; Figure 6C). Perhaps the FasX:ska mRNA interaction is sufficiently stabile to interfere with RNase cleavage but not ribosome binding. Thus, reevaluation of our knowledge with respect to the consequences of sRNA binding in close proximity to an mRNA RBS may be warranted. In summation, our data provides new insights into the mechanisms used by sRNAs to positively regulate mRNA targets, as well as uncovering a new layer of regulation of a key GAS virulence factor.
Experimental procedures
Bacterial strains and culture conditions
MGAS2221 is representative of the highly virulent M1T1 GAS clone responsible for significant morbidity and mortality since the mid-1980s in the U.S., Canada, and Western Europe (Sumby et al., 2005). GAS were grown in Todd-Hewitt broth with 0.2% yeast extract (THY broth) at 37°C (5% CO2). Chloramphenicol (4 μg/ml) and/or spectinomycin (150 μg/ml) were added when required.
Construction of isogenic fasX mutant strain 2221ΔFasX
An isogenic fasX mutant of parental strain MGAS2221 was created by replacement of the fasX gene with a non-polar spectinomycin resistance cassette via a previously described PCR overlap extension method. PCR primers used in the construction of mutant strains are listed in table S2. Confirmation of isogenic mutant strain construction was gained via PCR, sequencing, and Southern blot analyses (Figure S2). The Southern blot was generated using EcoRV-digested genomic DNA. The blot was probed with a labeled PCR product generated using primers FasXC and FasXD (Table S2).
Complementation of isogenic mutant strain 2221ΔFasX
To complement isogenic mutant strain 2221ΔFasX we introduced the wild-type fasX allele into the E. coli – GAS shuttle vector pDC123 (Chaffin & Rubens, 1998, Sumby et al., 2006). Forward and reverse primers used to amplify fasX contained BglII and NsiI restriction enzyme sites, respectively (Table S2). PCR products digested with BglII NsiI were ligated into similarly digested pDC123. All inserts were sequenced to ensure no spurious mutations had arisen during PCR and cloning.
Cloning of fasX from S. zooepidemicus
The fasX gene from S. zooepidemicus strain MGCS10565 was amplified using primers FasXZOOF/R (Table S2), and cloned into pDC123 to create pZOOFasX. The plasmid insert was sequenced to ensure no spurious mutations had arisen during PCR and cloning.
Total RNA isolation
For the described Northern blot and quantitative RT-PCR analyses GAS strains were grown to mid-exponential phase (O.D.600 = 0.5) and aliquots added to two volumes of RNAprotect (Qiagen Inc.). After incubating at room temperature for 5 mins samples were centrifuged for 10 mins at 4°C and 5,000g. Cell pellets were quick frozen in liquid nitrogen and stored at -80°C until ready for use. To isolate total RNA frozen GAS cell pellets were resuspended in 100 μl TE buffer and transferred to 2ml tubes containing fine glass shards (lysing matrix B tubes, MP Biomedicals). Tubes were placed into a glass bead beater (FastPrep machine, THERMO 101) and processed for 15 seconds at speed 4. Tubes were centrifuged for 5 seconds at 14,000 g to reduce foaming and an additional processing in the FastPrep machine performed following addition of 650 μl of buffer RLT (Qiagen Inc.). Samples were centrifuged for 30 seconds at 14,000 g to collect contents and 600 μl transferred to a 1.5 ml tube containing 900 μl 100% ethanol. RNA samples were subsequently bound to, washed on, and eluted from, RNeasy columns (Qiagen Inc.) as per the manufacturers’ miRNeasy protocol. Contaminating genomic DNA was removed from eluted RNA samples via four 30 minute incubations at 37°C with 2 μl TURBO DNase-free (Applied Biosystems), with DNA removal being verified by PCR.
Northern blot analysis
Total RNA was isolated as described and 6 μg from each GAS strain was loaded onto a 5% TBE-Urea gel before separating by electrophoresis. RNA was transferred to nylon membrane via electroblotting, UV cross-linked, and probed overnight with an in vitro transcribed probe complementary to fasX or to the 5S RNA (loading control). In vitro transcribed probes were generated using the Strip-EZ T7 kit (Applied Biosystems). DNA templates for in vitro transcription reactions were generated by PCR, with one primer containing the T7 promoter sequence (Table S2). RNA probes were labeled with biotin prior to hybridization (Brightstar psoralen-biotin labeling kit, Applied Biosystems). Following washes Northern blots were developed (Brightstar biodetect kit, Applied Biosystems) and exposed to film.
Indirect SKA activity assay
GAS strains were grown to the mid-exponential phase of growth (O.D. = 0.5), pelleted by centrifugation, and the supernatants filter-sterilized using 0.22μm syringe filters. Filtered supernatants were stored at -20°C until ready for use. SKA activity within supernatants was measured indirectly via the SKA-induced cleavage of human plasminogen into plasmin, cleavage of the chromogenic substrate S-2251 by plasmin, and measuring the light emitted using a spectrophotometer set at 405 nm. Specifically, 70 μl of filtered GAS culture supernatants were added to wells of a 96-well microtiter plate and S-2251 (Diapharma Group Inc.) added to 400 μM. Subsequently, 2 μg of purified human plasminogen (Molecular Innovations Inc.) was added to each well and the plate placed in a temperature controlled (37 °C) spectrophotometer. The absorbance at 405 nm was measured every two minutes for three hours. Control reactions were utilized that lacked plasminogen (negative control for SKA activity) or contained known amounts of purified streptokinase from GCS (Sigma-Aldrich; positive control for SKA activity).
Construction of plasmid-encoded mutant fasX alleles
Construction of plasmid-encoded 1 bp deletion mutants of fasX was through use of an overlap extension PCR method as outlined in figure S3. Briefly, using the complementation plasmid pFasXC, we used two vector embedded primers (DC123ECORV and DC123BGLII; Table S2) and two complementary mutagenesis primers (e.g. FDEL1F and FDEL1R; Table S2) to amplify the fasX gene from plasmid pFasXC while at the same time deleting 1 bp as dictated by the sequence of the mutagenesis primers. The resultant PCR product was digested with BglII NsiI and ligated into plasmid pDC123 that had been cut with the same enzymes. Ligations were transformed into E. coli (plated onto LB agar containing 20 μg/ml chloramphenicol) and minipreps performed on transformants. After PCR and sequencing to confirm the correct plasmid sequences they were transformed into strain 2221ΔFasX. Plasmid inserts were resequenced once transformed into GAS to ensure no spurious mutations had arisen.
Mutant fasX alleles containing deletions larger than 1 bp were constructed in a similar manner to the 1 bp deletion mutant plasmids. However, for pFXΔ155-205 an unrelated terminator sequence was added to the 3’ end of fasX to replace the terminator deleted in this mutant. This was required as in the absence of a terminator the mutant fasX allele failed to complement strain 2221ΔFasX, presumably due to transcript instability. For plasmid pFXΔ1-34,115-205, since the promoter and terminator region of the fasX allele were deleted we added a terminator to the 3’ end and placed the Pspac promoter at the 5’ end. The Pspac promoter is a low level constitutive promoter that has been previously described (Biswas et al., 2008). Primers containing the terminator / Pspac promoter are listed in table S2.
Western blot analysis
GAS strains were grown to mid-exponential phase (O.D.600 of 0.5) in THY broth at which point 10 ml was centrifuged to pellet the bacteria. The supernatant was filtered through 0.22μm syringe filters and the proteins precipitated via addition of 3.5 volumes of ice-cold ethanol. After four hours at -20°C the samples were centrifuged and the protein pellets resuspended in 500μl of SDS-PAGE loading buffer. Western blots were made using standard protocols and probed with rabbit polyclonal antibodies raised against SKA and Spd3. Goat anti-rabbit horseradish peroxidase-conjugated secondary antibodies were used to detect primary antibody binding and to generate signals.
Quantitative RT-PCR analysis of ska mRNA and FasX RNA concentration
cDNA was synthesized from total GAS RNA using the reverese transcriptase Superscript III (Invitrogen Corp.) as per the manufactures’ instructions. TaqMan quantitative RT-PCR was performed using an ABI 7500 Fast System (Applied Biosystems). Gene transcript levels present in different strains were compared to that of parental strain MGAS2221 using the ΔΔCT method as described (Shelburne et al., 2008). TaqMan primers and probes for the genes of interest, and the internal control gene proS, are listed in Table S2.
Quantitative RT-PCR analysis of ska mRNA stability
GAS strains were grown to mid-exponential phase (O.D.600 of 0.5) in THY broth before the addition of rifampicin to 1 mg/ml to inhibit RNA synthesis. After addition of rifampicin 3 ml samples were recovered after 0, 1, 2, 4, 5, and 20 mins. Recovered samples were added to two volumes of RNAprotect, incubated at room temperature for 5 mins, pelleted by centrifugation, and quick frozen in liquid nitrogen. RNA was isolated from each sample, converted into cDNA, and used in Taqman analysis. TaqMan quantitative RT-PCR was performed using an ABI 7500 Fast System (Applied Biosystems). TaqMan primers and probes for ska, and for the control gene proS, are listed in Table S2. Data is presented as percent proS (control gene; blue lines) or ska (red lines) transcript levels relative to the amount of these transcripts at time-point zero. Experiment was performed in triplicate with mean values shown.
ska transcriptional start site determination
An overview of the protocol used is shown in figure S7. Approximately 3μg of total RNA isolated from mid-exponential phase GAS (O.D.600 0.5) was used in a cDNA synthesis reaction with primer SKATMR (Table S2). RNA was removed via RNase digestion and the cDNA purified using the MinElute PCR purification kit (Qiagen Inc.). Purified cDNA was 5’ phosphorylated using T4 polynucleotide kinase (New England Biolabs), and circularized using T4 RNA ligase 1 (New England Biolabs). Circularized cDNA was purified using a second MinElute column and used in a PCR reaction with primers GSP1 and SKATMF (Table S2). A control reaction using uncircularized cDNA was also performed. PCRs were separated by electrophoresis and the band present in the test sample but absent from the control sample was extracted, TA cloned, transformed into E. coli, and sequenced from a dozen transformants. While this protocol does not distinguish primary transcript 5′- ends from internal 5′- processing sites the data was the same as that observed in S. equisimilus, which was determined using a discriminatory method (Gase et al., 1995).
Construction of double mutant strain 2221ΔX.GCSKA
Overlap extension PCR using primer pairs SKAMUT1/2 and SKAMUT3/4 (Table S2) was used to produce an ~2 kb product spanning the ska promoter region in which the two targeted nucleotides had been substituted. The PCR product was digested with BamHI and cloned into the BamHI site of the suicide vector pBBL740 (Zhu et al., 2009), and introduced into strain 2221ΔFasX. To select for loss of the integrated plasmid, and hence potential replacement of the chromosomally-derived GGGG sequence with the plasmid-derived GCCG sequence, chloramphenicol resistant transformants were grown for 5 passages in THY broth without antibiotics. Four of the 5 passages were for 4h each in fresh THY broth, while one passage was grown overnight in fresh THY broth. After passaging, cultures were serially diluted and plated onto blood agar plates. Individual colonies were patched onto THY agar plates with and without chloramphenicol. To test whether chloramphenicol sensitive colonies contained a wild-type or mutant 5’ ska region we performed PCR and sequencing.
Construction of MGAS2221 and 2221ΔFasX derivatives containing 5’ extensions to the ska mRNA transcript
MGAS2221 and 2221ΔFasX derivatives containing two different 13 bp extensions inserted at the ska transcriptional start site, and therefore incorporated at the 5’ end of ska mRNA, were constructed. Overlap extension PCR was used to create 2 kb regions spanning the ska transcriptional start site, with the required 13 bp sequences inserted via sequences constructed into the PCR primers (Table S2). These PCR products were cloned into the suicide vector pBBL740 to create plasmids pSKA::EXY (the transcribed 13 bp insert is complementary to the 5’ end of wild-type ska mRNA) and pSKA::EXN (the transcribed 13 bp insert is not complementary to the 5’ end of wild-type ska mRNA). Insertion of the 13 bp sequences into the GAS genome was performed by allelic exchange as described for creation of strain 2221ΔX.GCSKA. Strains were verified by PCR and sequencing.
Construction of cvfA mutant and non-mutant derivatives of MGAS2221 and 2221ΔFasX
To create cvfA mutants of strains MGAS2221 and 2221ΔFasX we disrupted the cvfA gene by insertional-inactivation. A central region of the cvfA gene was amplified by PCR, cloned into the suicide vector pBBL740 (creating plasmid pCVFAKO), and transformed into the two GAS strains. Disruption of the cvfA gene in chloramphenicol-resistant transformants was confirmed by PCR and sequencing. To ensure that mutant strain phenotypes were due to cvfA disruption and not due to a non-specific effect of pBBL740 integration into the chromosome we created a second pBBL740-based plasmid (pCVFAOK), this time containing the entire 3’ end of the cvfA gene. The design of pCVFAOK was such that integration of this construct into the chromosome did not disrupt the cvfA gene. Note that these plasmids were based upon those made in the published cvfA study (Kang et al., 2010).
Construction of pnpA mutant derivatives of MGAS2221 and 2221ΔFasX
To create pnpA mutants of strains MGAS2221 and 2221ΔFasX we disrupted the pnpA gene by insertional-inactivation. A central region of the pnpA gene was amplified by PCR, cloned into the suicide vector pBBL740 (creating plasmid pPNPAKO), and transformed into the two GAS strains. Disruption of the pnpA gene in chloramphenicol-resistant transformants was confirmed by PCR and sequencing.
Supplementary Material
Acknowledgments
This research was funded in part by grant 0865139F from the American Heart Association - South Central Affiliate (to P.S.), and by grants R21AI078159 and R01AI087747 from the National Institute of Allergy and Infectious Diseases (to P.S.). We thank B Lei for providing plasmid pBBL740. We also thank JM Musser, KJ Pflughoeft, and K Stockbauer for critical reading of the manuscript.
References
- Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Barnett TC, Bugrysheva JV, Scott JR. Role of mRNA stability in growth phase regulation of gene expression in the group A streptococcus. J Bacteriol. 2007;189:1866–1873. doi: 10.1128/JB.01658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Sesso R, Pinto SW, Hoe NP, Porcella SF, Deleo FR, Musser JM. Genome sequence of a Lancefield group C Streptococcus zooepidemicus strain causing epidemic nephritis: new information about an old disease. PLoS One. 2008;3:e3026. doi: 10.1371/journal.pone.0003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I, Jha JK, Fromm N. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology. 2008;154:2275–2282. doi: 10.1099/mic.0.2008/019265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn C, Rigoulay C, Bouloc P. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 2007;7:10. doi: 10.1186/1471-2180-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5' mRNA coding region inhibits translational initiation. Mol Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- Bugrysheva JV, Scott JR. The ribonucleases J1 and J2 are essential for growth and have independent roles in mRNA decay in Streptococcus pyogenes. Mol Microbiol. 2009;75:731–743. doi: 10.1111/j.1365-2958.2009.07012.x. [DOI] [PubMed] [Google Scholar]
- Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- Chaffin DO, Rubens CE. Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene. 1998;219:91–99. doi: 10.1016/s0378-1119(98)00396-5. [DOI] [PubMed] [Google Scholar]
- Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J Bacteriol. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, Hammer E, Volker U, Stulke J. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics. 2009;8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C. RNA processing and degradation in Bacillus subtilis. Microbiol Mol Biol Rev. 2003;67:157–174. doi: 10.1128/MMBR.67.2.157-174.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio choleraevirulence and downregulates sigma expression. Mol Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- Fantappie L, Metruccio MM, Seib KL, Oriente F, Cartocci E, Ferlicca F, Giuliani MM, Scarlato V, Delany I. The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect Immun. 2009;77:1842–1853. doi: 10.1128/IAI.01216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federle MJ, McIver KS, Scott JR. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Gase K, Ellinger T, Malke H. Complex transcriptional control of the streptokinase gene of Streptococcus equisimilis H46A. Mol Gen Genet. 1995;247:749–758. doi: 10.1007/BF00290407. [DOI] [PubMed] [Google Scholar]
- Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, Francois P, Vandenesch F, Gaspin C, Romby P. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009;37:7239–7257. doi: 10.1093/nar/gkp668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Grieshaber NA, Grieshaber SS, Fischer ER, Hackstadt T. A small RNA inhibits translation of the histone-like protein Hc1 in Chlamydia trachomatis. Mol Microbiol. 2006;59:541–550. doi: 10.1111/j.1365-2958.2005.04949.x. [DOI] [PubMed] [Google Scholar]
- Gryllos I, Tran-Winkler HJ, Cheng MF, Chung H, Bolcome R, 3rd, Lu W, Lehrer RI, Wessels MR. Induction of group A Streptococcus virulence by a human antimicrobial peptide. Proc Natl Acad Sci U S A. 2008;105:16755–16760. doi: 10.1073/pnas.0803815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J Mol Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- Holden MT, Heather Z, Paillot R, Steward KF, Webb K, Ainslie F, Jourdan T, Bason NC, Holroyd NE, Mungall K, Quail MA, Sanders M, Simmonds M, Willey D, Brooks K, Aanensen DM, Spratt BG, Jolley KA, Maiden MC, Kehoe M, Chanter N, Bentley SD, Robinson C, Maskell DJ, Parkhill J, Waller AS. Genomic evidence for the evolution of Streptococcus equi: host restriction, increased virulence, and genetic exchange with human pathogens. PLoS Pathog. 2009;5:e1000346. doi: 10.1371/journal.ppat.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SO, Caparon MG, Cho KH. Virulence Gene Regulation By CvfA, A Putative RNase: The CvfA-Enolase Complex In Streptococcus pyogenes Links Nutritional Stress, Growth Phase Control And Virulence Gene Expression. Infect Immun. 2010 doi: 10.1128/IAI.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil J, Im M, Heath A, Ringdahl U, Mundada L, Cary Engleberg N, Fay WP. Plasminogen enhances virulence of group A streptococci by streptokinase-dependent and streptokinase-independent mechanisms. J Infect Dis. 2003;188:497–505. doi: 10.1086/377100. [DOI] [PubMed] [Google Scholar]
- Kreikemeyer B, Boyle MD, Buttaro BA, Heinemann M, Podbielski A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol Microbiol. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- Kulesus RR, Diaz-Perez K, Slechta ES, Eto DS, Mulvey MA. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect Immun. 2008;76:3019–3026. doi: 10.1128/IAI.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol. 2005;58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Levin JC, Wessels MR. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Sledjeski DD, Kreikemeyer B, Podbielski A, Boyle MD. Identification of pel, a Streptococcus pyogenes locus that affects both surface and secreted proteins. J Bacteriol. 1999;181:6019–6027. doi: 10.1128/jb.181.19.6019-6027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Brencic A, Lory S, Waldor MK. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006;34:3484–3493. doi: 10.1093/nar/gkl453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottenberg R, Minning-Wenz D, Boyle MD. Capturing host plasmin(ogen): a common mechanism for invasive pathogens? Trends Microbiol. 1994;2:20–24. doi: 10.1016/0966-842x(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Lybecker MC, Samuels DS. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol. 2007;64:1075–1089. doi: 10.1111/j.1365-2958.2007.05716.x. [DOI] [PubMed] [Google Scholar]
- Mangold M, Siller M, Roppenser B, Vlaminckx BJ, Penfound TA, Klein R, Novak R, Novick RP, Charpentier E. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol Microbiol. 2004;53:1515–1527. doi: 10.1111/j.1365-2958.2004.04222.x. [DOI] [PubMed] [Google Scholar]
- Marzi S, Myasnikov AG, Serganov A, Ehresmann C, Romby P, Yusupov M, Klaholz BP. Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell. 2007;130:1019–1031. doi: 10.1016/j.cell.2007.07.008. [DOI] [PubMed] [Google Scholar]
- McArthur JD, McKay FC, Ramachandran V, Shyam P, Cork AJ, Sanderson-Smith ML, Cole JN, Ringdahl U, Sjobring U, Ranson M, Walker MJ. Allelic variants of streptokinase from Streptococcus pyogenes display functional differences in plasminogen activation. FASEB J. 2008;22:3146–3153. doi: 10.1096/fj.08-109348. [DOI] [PubMed] [Google Scholar]
- Meibom KL, Forslund AL, Kuoppa K, Alkhuder K, Dubail I, Dupuis M, Forsberg A, Charbit A. Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect Immun. 2009;77:1866–1880. doi: 10.1128/IAI.01496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Kawamoto H, Mizota T, Inada T, Aiba H. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol Microbiol. 2004;54:1063–1075. doi: 10.1111/j.1365-2958.2004.04329.x. [DOI] [PubMed] [Google Scholar]
- Morita T, Mochizuki Y, Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci U S A. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obana N, Shirahama Y, Abe K, Nakamura K. Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5' leader sequence. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07258.x. [DOI] [PubMed] [Google Scholar]
- Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez N, Trevino J, Liu Z, Ho SCM, Babitzke P, Sumby P. A Genome-Wide Analysis of Small Regulatory RNAs in the Human Pathogen Group A Streptococcus. PLoS ONE. 2009;4:e7668. doi: 10.1371/journal.pone.0007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Scott JR. RivR and the small RNA RivX: the missing links between the CovR regulatory cascade and the Mga regulon. Mol Microbiol. 2007;66:1506–1522. doi: 10.1111/j.1365-2958.2007.06015.x. [DOI] [PubMed] [Google Scholar]
- Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Sharp JS, Bechhofer DH. Effect of 5'-proximal elements on decay of a model mRNA in Bacillus subtilis. Mol Microbiol. 2005;57:484–495. doi: 10.1111/j.1365-2958.2005.04683.x. [DOI] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, Brennan RG, Musser JM. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci U S A. 2008;105:1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner K, Malke H. Dual control of streptokinase and streptolysin S production by the covRS and fasCAX two-component regulators in Streptococcus dysgalactiae subsp. equisimilis. Infect Immun. 2002;70:3627–3636. doi: 10.1128/IAI.70.7.3627-3636.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. Evolutionary origin and emergence of a highly successful clone of serotype M1 group a Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Ringdahl U, Homeister JW, Fay WP, Engleberg NC, Yang AY, Rozek LS, Wang X, Sjobring U, Ginsburg D. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science. 2004;305:1283–1286. doi: 10.1126/science.1101245. [DOI] [PubMed] [Google Scholar]
- Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson MD, Sjobring U, Luo F, Bessen DE. Roles of the plasminogen activator streptokinase and the plasminogen-associated M protein in an experimental model for streptococcal impetigo. Microbiology. 2002;148:3933–3945. doi: 10.1099/00221287-148-12-3933. [DOI] [PubMed] [Google Scholar]
- Tillett WS, Garner RL. The fibrinolytic activity of hemolytic streptococci. J Exp Med. 1933;58:485–502. doi: 10.1084/jem.58.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Trevino J, Perez N, Ramirez-Pena E, Liu Z, Shelburne SA, 3rd, Musser JM, Sumby P. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect Immun. 2009;77:3141–3149. doi: 10.1128/IAI.01560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PN, Holden MT, Leigh JA, Lennard N, Bignell A, Barron A, Clark L, Quail MA, Woodward J, Barrell BG, Egan SA, Field TR, Maskell D, Kehoe M, Dowson CG, Chanter N, Whatmore AM, Bentley SD, Parkhill J. Evidence for niche adaptation in the genome of the bovine pathogen Streptococcus uberis. BMC Genomics. 2009;10:54. doi: 10.1186/1471-2164-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM. 6S RNA: a small RNA regulator of transcription. Curr Opin Microbiol. 2007;10:164–168. doi: 10.1016/j.mib.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Liu M, Sumby P, Lei B. The secreted esterase of group a streptococcus is important for invasive skin infection and dissemination in mice. Infect Immun. 2009;77:5225–5232. doi: 10.1128/IAI.00636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.