Abstract
MicroRNAs are a class of small RNA regulators that are involved in numerous cellular processes, including development, proliferation, differentiation, and plasticity. The emerging concept is that microRNAs play a central role in controlling the balance between stem cell self-renewal and fate determination by regulating the expression of stem cell regulators. This review will highlight recent advances in the regulation of neural stem cell self-renewal and neurogenesis by microRNAs. It will cover microRNA functions during the entire process of neurogenesis, from neural stem cell self-renewal and fate determination to neuronal maturation, synaptic formation, and plasticity. The interplay between microRNAs and both cell-intrinsic and -extrinsic stem cell players, including transcription factors, epigenetic regulators, and extrinsic signaling molecules will be discussed. This is a summary of the topics covered in the mini-symposium on microRNA regulation of neural stem cells and neurogenesis in SFN 2010 and is not meant to be a comprehensive review of the subject.
Introduction
Neural stem cells are a subset of undifferentiated precursors that retain the ability to proliferate and self-renew, and have the capacity to give rise to neuronal and glial lineages in the nervous system (Gage, 2000). Neurogenesis, initiated from neural stem cells and resulting in functional new neurons, is a fundamental process for both embryonic neurodevelopment and adult brain plasticity. Consequently, the process of creating a properly functional neuron, including neural stem cell self-renewal and fate specification, neuronal migration, maturation, and integration, is regulated by the dynamic interplay between transcription factors, epigenetic control, microRNA (miRNA) regulators, and cell-extrinsic signals from the “niche” where stem cells reside (Shi et al., 2008). This review focuses on the role of miRNA regulators in neural stem cell self-renewal and neurogenesis.
miRNAs are short 20–22 nucleotide RNA molecules that are expressed in a tissue-specific and developmentally regulated manner and function as negative regulators of gene expression in a variety of eukaryotic organisms (Ambros, 2004; Bartel, 2004). These small RNA regulators are key post-transcriptional players in stem cell self-renewal and fate determination due to their important roles in the regulation of diverse genes through messenger RNA degradation or translational inhibition (Ambros, 2004; Bartel, 2004). Deletion of genes centrally involved in miRNA processing, such as Dicer or Argonaute family members, led to loss of stem cell populations (Carmell et al., 2002; Bernstein et al., 2003; Wienholds et al., 2003), strongly supporting a role for miRNAs in stem cell biology. Despite the vast interest by neuroscientists, studies of miRNA functions in the nervous system have been hindered by many road blocks. Research on how miRNAs regulate neural stem cell biology is still in its infancy. This mini-symposium will present the most recent advances in this emerging research area, covering miRNA functions in multiple steps of neurogenesis, from neural stem cell self-renewal and fate determination, to neuronal maturation and functional integration (Fig. 1).
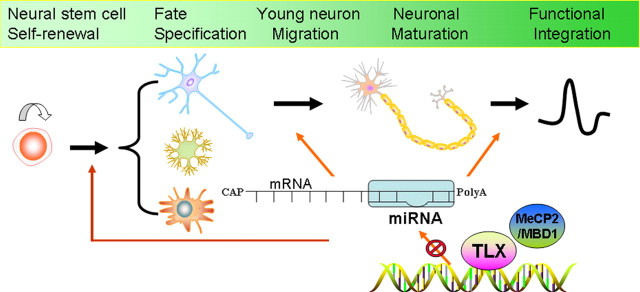
Figure 1.
miRNAs regulate multiple steps of neurogenesis. Neurogenesis includes self-renewal and fate specification of neural stem cells, migration and maturation of young neurons, and functional integration of new neurons into the neural circuitry. miRNAs have been shown to regulate all these steps of neurogenesis, by base-pairing with target mRNAs to regulate target gene expression. The expression of miRNAs themselves is also regulated by transcription factors, such as TLX, and epigenetic factors, such as MeCP2 and MBD1.
Regulatory loop between miRNAs and nuclear receptor TLX in neural stem cell self-renewal and fate determination (Y. Shi)
Nuclear receptors are a superfamily of phylogenetically related proteins that regulate the expression of genes critical for a variety of biological processes, including development, growth, differentiation, and homeostasis (Mangelsdorf et al., 1995; Evans, 2005). TLX (also called NR2E1) is an orphan nuclear receptor that was initially identified as a homolog of the Drosophila tailless and plays an important role in vertebrate brain functions (Yu et al., 1994; Monaghan et al., 1997; Shi et al., 2004; Liu et al., 2008; Zhang et al., 2008; Qu et al., 2010). Our recent studies revealed that the interplay between miRNAs and nuclear receptor TLX plays an important role in neural stem cell fate specification and determination (Zhao et al., 2009, 2010).
We have shown that TLX is an essential regulator of neural stem cell self-renewal, through regulating genes that are important in stem cell proliferation control (Shi et al., 2004; Qu and Shi, 2009; Qu et al., 2010; Sun et al., 2010; Zhao et al., 2010). miR-9 is a brain-specific miRNA that is expressed in the neurogenic areas of both embryonic and adult brains (Lagos-Quintana et al., 2002; Krichevsky et al., 2003; Deo et al., 2006; Kapsimali et al., 2007). Interestingly, miR-9 and TLX form a feedback regulatory loop, which in turn regulates neural stem cell proliferation and differentiation (Zhao et al., 2009). TLX is highly expressed in neural stem cells, the expression of which is reduced upon differentiation; in contrast, miR-9 expression is increased during differentiation. The temporal relationship between miR-9 and TLX expression supports the hypothesis that miRNAs induced during neural stem cell differentiation ensure cell fate transition by suppressing essential stem cell maintenance factors. miR-9 functions in neural stem cells through targeting TLX expression. Interestingly, TLX also acts as a transcriptional repressor of miR-9 genes (Zhao et al., 2009). This negative feedback loop between TLX and miR-9 would allow rapid transition from neural stem cells to differentiated cells.
In addition to TLX, several other target genes have been predicted and tested for miR-9, including those encoding transcription factors REST, FoxG1, Senseless, the Hairy/E (spl) transcription factors Her 5, Her 9, and components of the FGF signaling pathway, the Drosophila LIM only transcription factor dLMO, and stathmin, a protein that increases microtubule instability (Conaco et al., 2006; Li et al., 2006; Wu and Xie, 2006; Bredenkamp et al., 2007; Leucht et al., 2008; Biryukova et al., 2009; Delaloy et al., 2010). These studies raise an interesting possibility that miR-9 may act as a regulatory switch at different developmental stages and in different organisms by interacting with target mRNAs differentially expressed in a temporal and cellular context-specific manner (Uchida, 2010). The regulatory loop between miRNAs and transcription factors may serve as a general mechanism to control the balance between stem cell proliferation and differentiation and to allow cell fate determination in a timely manner.
Recently, we showed that miRNA lethal-7b (let-7b) also regulates neural stem cell fate decision (Zhao et al., 2010). Overexpression of let-7b led to inhibition of neural stem cell proliferation and accelerated neural differentiation, whereas antisense knockdown of let-7b resulted in increased proliferation of neural stem cells. Moreover, in utero electroporation of let-7b to embryonic mouse brains led to reduced cell cycle progression in neural stem cells. Interestingly, this study provided a direct link between let-7b and nuclear receptor TLX. let-7b regulates neural stem cell proliferation and differentiation by targeting TLX and its downstream effector, cyclin D1. Characterizing the let-7b-nuclear receptor signaling cascade provides new insight into the role of let-7b in neural stem cell fate determination. Let-7b has also been shown to target Hmga2 to reduce the self-renewal of neural stem cells in the aging brain (Nishino et al., 2008). Together with our observation on the role of let-7b in embryonic and adult neural stem cells through targeting TLX and cyclin D1, these studies suggest that let-7b functions in neural stem cells across a spectrum of developmental stages through targeting distinct key molecules.
Crosstalk between miRNA and epigenetic regulation in neural stem cells and neurogenesis (X. Zhao)
Epigenetic mechanisms, including DNA methylation and histone modification, are known to play significant roles in the modulation of stem cell proliferation and differentiation (Li and Zhao, 2008). DNA methylation represses gene transcription either through directly blocking the access of transcription factors to their binding sites or through indirectly recruiting proteins that bind methyl-CpGs (MBDs or Kaiso family proteins) (Klose and Bird, 2006), including MBD1, MBD2, MBD3, MBD4, and MECP2. The importance of epigenetic regulation in brain development and neurological disorders has been well documented (Chahrour et al., 2008). The de novo mutations in MeCP2 lead to Rett syndrome, an X-linked dominant pervasive neurodevelopmental disorder (Amir et al., 1999). MBD1 mutations have been found in a subset of autistic patients (Li et al., 2005; Cukier et al., 2010). MBD1 deficiency in mice results in impaired adult neurogenesis and hippocampal-dependent learning (Zhao et al., 2003; Allan et al., 2008; Li et al., 2008). Both MBD1 and MeCP2 have been shown to regulate the proliferation and differentiation of neural stem cells or the maturation of young neurons (Zhao et al., 2003; Kishi and Macklis, 2004; Smrt et al., 2007; Tsujimura et al., 2009). However, identification of their downstream targets by classical gene expression analyses have yielded limited results. We investigated whether these two proteins regulate neural stem cells by modulating noncoding RNAs, particularly miRNA expression.
We found that MeCP2 regulates specific miRNAs in mouse neural stem cells (Szulwach et al., 2010). We demonstrated that one of these miRNAs, miR-137, promotes proliferation and represses differentiation of neural stem cells via translational repression of Ezh2, a histone H3 lysine 27 methyltransferase and a member of the polycomb group (PcG) protein family (Boyer et al., 2006). On the other hand, miR-137 expression is also upregulated in MeCP2-deficient immature neurons and miR-137 has a significant impact on the dendritic morphogenesis of young hippocampal neurons (Smrt et al., 2010). Interestingly, the neuronal maturation function of miR-137 is achieved by translational repression of Mind bomb-1, a ubiquitin ligase known to be important for neurogenesis and neurodevelopment (Choe et al., 2007; Ossipova et al., 2009), rather than Ezh2. Therefore, miR-137 can have multiple functions during different stages of neurogenesis.
In a parallel experiment, we have discovered that MBD1 regulates the expression of a subset of miRNAs in adult neural stem cells and one of them is miR-184, an imprinted miRNA whose genomic region has been shown to be bound by MeCP2 in neurons (Nomura et al., 2008). We showed that in adult neural stem cells, miR-184 is regulated by MBD1, but not by MeCP2. miR-184 promotes neural stem cell proliferation and inhibits differentiation by targeting Numblike (Liu et al., 2010). Therefore, MBD1, miR-184, and Numblike may form a regulatory network that controls the balance between the proliferation and differentiation of neural stem cells.
Although the crosstalk between miRNA and DNA methylation is likely a common mechanism regulating critical cellular processes, evidence for this concept in mammalian neural stem cells is limited. Our results demonstrated that crosstalk between epigenetic regulation and miRNA pathway could play important roles in the modulation of adult neurogenesis.
The interplay between miRNAs and REST in neurogenesis (J. Hsieh)
Recent progress has been made in investigating the links between miRNAs and neurogenesis. Noncoding RNAs play key roles in the modulation of transcriptional networks and appear to have important functions in brain development and neurogenesis (Kosik and Krichevsky, 2005; Cao et al., 2006; Mehler and Mattick, 2007). Although studies have pointed to a global role of miRNAs in brain development, there are ∼500 miRNA genes identified in human and mouse to date and each miRNA may have multiple mRNA targets (Saini et al., 2008). Therefore, detailed studies of individual miRNAs are warranted to further establish the link between small noncoding RNAs and their contribution to neurogenesis.
One of the most fascinating stories regarding an individual miRNA and neuronal differentiation is that of miR-124, the most abundant miRNA in the adult brain (Lagos-Quintana et al., 2002). During neurogenesis, miR-124 expression is undetectable or expressed at low levels in progenitor cells and is upregulated in differentiating and mature neurons (Deo et al., 2006). Previous studies demonstrated that overexpression of miR-124 in HeLa cells led to decreased expression of many non-neuronal genes to reflect a gene expression program more similar to that of neuronal cells (Lim et al., 2005). Another set of studies showed that the repressor element-1 silencing transcription factor (REST, also known as NRSF) can silence the expression of miRNA-124 and the downregulation of REST and increased levels of miR-124, and neuronal gene expression is permissive for neuronal differentiation in mouse embryonal carcinoma cells (Conaco et al., 2006). However, inhibiting miR-124 did not affect neuronal differentiation, at least in chick neural tube (Cao et al., 2007). More recently, miR-124 was shown to promote neuronal differentiation and cell cycle exit of neural stem cells in the subventricular zone, one of the neurogenic niches of the adult mammalian brain (Cheng et al., 2009). MiR-124 may act in turn by silencing REST and switching on neuron-specific splicing by targeting PTBP1, a repressor of pre-mRNA splicing in non-neuronal cells (Makeyev et al., 2007).
The exact connection between miR-124 and REST remains largely unknown since REST plays an essential role in early development and REST-null mice are embryonic lethal (Chen et al., 1998). We therefore generated an inducible and conditional knock-out allele of REST to determine the role of REST during adult neurogenesis (Z. Gao and J. Hsieh, unpublished data). The deletion of REST in adult subventricular zone (SVZ) progenitors in vitro led to precocious neuronal differentiation, consistent with the observation upon overexpression of miR-124 (Cheng et al., 2009). These emerging studies suggest that the maintenance of neural stem cells and control of neurogenesis require the complex interplay between actions of both transcriptional regulators and miRNAs.
miRNAs in spatial patterning of the developing spinal cord (H. Wichterle)
Spatial patterning of neural tissue into discreet progenitor domains relies on cross repressive interactions among determinants of cell identity. To what extent miRNAs are involved in the segregation of neuronal subtypes in vertebrates remains unclear. The analysis of neural specific Dicer mutants in mammals established miRNA roles in the control of temporal transitions from early to late neural progenitors (Georgi and Reh, 2010) or from progenitors to postmitotic neurons (Fineberg et al., 2009). A role for miRNAs in spatial patterning has been demonstrated in the developing mesoderm where miR-196 is expressed in a spatially complementary manner with its target Hoxb8 (Mansfield et al., 2004; McGlinn et al., 2009). In contrast, there is currently no experimental evidence for miRNA involvement in the spatial patterning of neural progenitors (Fineberg et al., 2009). This might be in part due to the lack of a simple genetic system to probe miRNA function during the early stages of mammalian development when progenitor identity is established. A complete loss of Dicer function in mice leads to embryonic lethality before neural tissue formation (Bernstein et al., 2003) and selective disruption of Dicer function in early neural progenitors is complicated by the relative stability of existing miRNAs and the lack of suitable drivers expressed in prospective neural tissues (Davis et al., 2008).
To circumvent this limitation we used an embryonic stem (ES) cell model of spinal cord patterning (Wichterle et al., 2002). We developed an ES cell line in which Dicer function can be disrupted in a tamoxifen-regulated manner. Loss of ubiquitously expressed miRNAs was detectable 3–4 d after tamoxifen treatment. To determine whether miRNAs are essential for dorsoventral patterning of neural progenitors, tamoxifen-treated ES cells were induced to acquire distinct dorsoventral spinal progenitor identities by exposure of differentiating cells to retinoic acid and to increasing concentrations of sonic hedgehog agonist. Analysis of the expression pattern of progenitor markers revealed a selective loss of progenitors of V2 spinal interneurons in cultures exposed to a low concentration of sonic hedgehog agonists. The loss was compensated by an increase in the number of motor neuron progenitors, indicating that miRNAs play a role in the repression of motor neuron progenitors and promotion of V2 interneuron progenitor fate. These findings provide evidence that miRNAs are involved in the specification of spatial identity of neural progenitors and might be critical for the positioning of progenitor boundaries in the developing neuroepithelium. The stem cell-based approach used in this study can be easily adapted to examine miRNA-dependent patterning phenotypes at other neural territories and developmental time points.
miRNA in neuronal maturation and synaptogenesis (S. Impey)
miRNAs show striking developmental regulation in the nervous system. They are compartmentalized to distinct cellular domains, and have been implicated in neural differentiation, synapse development, and synaptic plasticity (Vo et al., 2010). The cAMP response element-binding (CREB) transcriptional pathway regulates dendritic growth, activity-regulated dendritic refinement, synaptogenesis, and use-dependent synaptic plasticity. We took a novel chromatin immunoprecipitation sequencing (ChIP-seq) methodology, termed SACO, to identify CREB target sites on a genome-wide basis (Impey et al., 2004). This led to the identification of a novel CREB- and activity-regulated noncoding RNA that encodes two activity-regulated miRNAs, miR-132 and miR-212 (Vo et al., 2005; Wayman et al., 2008). Subsequent work revealed that miR-132 regulates dendritic growth, activity-induced spine growth, spine morphology, hippocampal miniature EPSC (mEPSC) frequency, and circadian rhythms (Cheng et al., 2007; Wayman et al., 2008; Edbauer et al., 2010; Impey et al., 2010). Recently, high-throughput small RNA sequencing was used to clone and identify a novel “star” miRNA derived from the miR-132-212 cluster (Remenyi et al., 2010). This novel miRNA is highly regulated by synaptic activity and regulates spine formation in hippocampal neurons.
To further determine the role of miRNAs in neuronal maturation, we examined the expression of small RNAs in an embryonic stem cell model for neuronal differentiation and in brain-derived neural stem/progenitor cells. miR-132 and other miRNAs are dramatically upregulated at specific stages of neuro-ectodermal development. Furthermore, miR-132 plays a role in radial-glial stem cell self-renewal.
The variable kinetics of miRNA biogenesis and catabolism has made characterization of miRNA transcriptional circuits difficult. For example, in some tissues miRNAs have an estimated half-life of ∼24 h while, in other systems, miRNAs decay rapidly. To circumvent this limitation, we took advantage of high-throughput ChIP-Seq and RNA-Seq screens to characterize the regulation of miRNA-encoding genes by neuronal activity and CREB in hippocampal neurons. Integrated bioinformatic analysis of RNA polymerase II occupancy, transcription-associated histone methylation, and global RNA profiling facilitated the identification of novel CREB- and activity-regulated miRNA-encoding genes. These unbiased functional genomics studies also indentified novel classes of neuron-specific small RNAs, including mirtrons, miRNAs processed from loop sequences, and repeat-associated small RNAs. In particular, we will present data characterizing a genomic cluster of CREB- and activity-regulated RNAs that encode multiple novel and known miRNAs.
miRNA in synaptic plasticity (S. Banerjee)
Brain function is regulated by a vast neuronal network system made up of nerve junctions or synapses. Modification of these synapses or synaptic plasticity that occurs in response to neuronal activity underlies several neuronal circuit functions, including formation of long-term memory. Our current understanding of the fundamental basis of long-term memory formation is centered on how synapses can translate a neuronal signal into a long-lasting change in synaptic structure and function. Among the critical changes that underlie plasticity are those related to local translation.
Spatiotemporal control of protein synthesis at synaptodendritic compartments is regulated by multiple levels of control. An important modulator of localized protein synthesis is miRNAs (Kosik, 2006; Schratt, 2009). Mature miRNAs modulate post-transcriptional gene expression in association with a multiprotein complex, known as the RNA-induced silencing complex (RISC). They bind to the 3′-untranslated region (UTR) of mRNAs and result in translational inhibition (Kosik, 2006). Interestingly, it has been shown that impairment of key RISC factors, such as MOV10, a DEAD box helicase, can derepress translation (Chendrimada et al., 2007). However, a molecular mechanism of miRNA-mediated translation control of synaptic protein synthesis is largely unknown.
In a recent study we observed that MOV10, similar to its fly ortholog Armitage (Ashraf et al., 2006), is degraded rapidly through proteasome upon NMDA receptor activation or depolarization (Banerjee et al., 2009). Prompted by this observation and previous literatures, we hypothesized that the activity-regulated MOV10 degradation would lead to translational derepression of RISC-associated mRNAs at the synapse. We designed a novel method, known as a translation trap, to identify synaptic mRNAs that would be redistributed from the translationally suppressed pool to an actively translating polyribosomal fraction upon RNAi-mediated removal of MOV10. Of 54 transcripts tested, the translation trap identified a dozen synaptic mRNAs, including two known RISC-regulated mRNAs, LIMK1 and αCaMKII. Among these novel candidates, we studied lysophospholipase 1 (Lypla1) (also known as APT1), a depalmitoylating enzyme that regulates the cytoskeletal regulator, Gα (Duncan and Gilman, 1998). Palmitoylation is a reversible post-translational modification that has recently been implicated in synaptic plasticity (Kang et al., 2008). We identified that Lypla1 is targeted by miR-138, a brain-enriched miRNA. Furthermore, we found that Lypl1 and miR-138 colocalize in synapses, suggesting that the duplex exerts regulatory control over synthesis of Lypla1 protein at the synapse.
Using a photoconvertible translation reporter, Kaede, we observed that the proteasomal control of MOV10 degradation is a critical step toward Lypla1 3′-UTR-driven dendritic protein synthesis. Furthermore, our imaging analysis demonstrated coordination between degradative control of RISC and protein synthesis from RISC-associated mRNAs at individual synapses. In addition to our observations, another recent study has shown that Lypla1 plays a critical role in synaptic development (Siegel et al., 2009). Together, these observations suggest that miRNA-mediated control of dendritic protein synthesis is likely to regulate synaptic plasticity. Future experiments are directed toward testing the role of this degradative control over synaptic protein synthesis in long-term memory formation at the organismal level.
Conclusions
Stem cells have provided great hope for cell replacement therapies. Because neural stem cells have the potential to impact a wide spectrum of neurological diseases, there is a great deal of interest in understanding neural stem cell biology, from basic science to translational medicine. This mini-symposium, by addressing the emerging role of miRNAs in regulating neural stem cell fate, neuronal maturation, and synaptic development, will allow further understanding in the control of neural stem cell self-renewal and neurogenesis.
Footnotes
This work was supported by National Institutes of Health (NIH)–National Institute of Neurological Disorders and Stroke Grants R01 NS059546 and RC1 NS068370 (to Y.S.), an International Rett Syndrome Foundation grant, NIH Grants R01 MH080434 and R01 MH078972 (to X.Z.), NIH Grant R01AG032383 (to J.H.), NIH Grants R01 NS058502 and P01 NS055923 (to H.W.), and Keck Foundation Grant SB 060811 (to K.S.K). We apologize to colleagues whose work could not be cited due to space limitations.
References
- Allan AM, Liang X, Luo Y, Pak C, Li X, Szulwach KE, Chen D, Jin P, Zhao X. The loss of methyl-CpG binding protein 1 leads to autism-like behavioral deficits. Hum Mol Genet. 2008;17:2047–2057. doi: 10.1093/hmg/ddn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Biryukova I, Asmar J, Abdesselem H, Heitzler P. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev Biol. 2009;327:487–496. doi: 10.1016/j.ydbio.2008.12.036. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bredenkamp N, Seoighe C, Illing N. Comparative evolutionary analysis of the FoxG1 transcription factor from diverse vertebrates identifies conserved recognition sites for microRNA regulation. Dev Genes Evol. 2007;217:227–233. doi: 10.1007/s00427-006-0128-x. [DOI] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe EA, Liao L, Zhou JY, Cheng D, Duong DM, Jin P, Tsai LH, Peng J. Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. J Neurosci. 2007;27:9503–9512. doi: 10.1523/JNEUROSCI.1408-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukier HN, Rabionet R, Konidari I, Rayner-Evans MY, Baltos ML, Wright HH, Abramson RK, Martin ER, Cuccaro ML, Pericak-Vance MA, Gilbert JR. Novel variants identified in methyl-CpG-binding domain genes in autistic individuals. Neurogenetics. 2010;11:291–303. doi: 10.1007/s10048-009-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- Duncan JA, Gilman AG. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS) J Biol Chem. 1998;273:15830–15837. doi: 10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM. The nuclear receptor superfamily: a rosetta stone for physiology. Mol Endocrinol. 2005;19:1429–1438. doi: 10.1210/me.2005-0046. [DOI] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- Georgi SA, Reh TA. Dicer is required for the transition from early to late progenitor state in the developing mouse retina. J Neurosci. 2010;30:4048–4061. doi: 10.1523/JNEUROSCI.4982-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Impey S, Davare M, Lasiek A, Fortin D, Ando H, Varlamova O, Obrietan K, Soderling TR, Goodman RH, Wayman GA. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci. 2010;43:146–156. doi: 10.1016/j.mcn.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, Green WN, Yates JR, 3rd, Davis NG, El-Husseini A. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RH, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi N, Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol Cell Neurosci. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Kosik KS, Krichevsky AM. The elegance of the microRNAs: a neuronal perspective. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- Li H, Yamagata T, Mori M, Yasuhara A, Momoi MY. Mutation analysis of methyl-CpG binding protein family genes in autistic patients. Brain Dev. 2005;27:321–325. doi: 10.1016/j.braindev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Li X, Zhao X. Epigenetic regulation of mammalian stem cells. Stem Cells Dev. 2008;17:1043–1052. doi: 10.1089/scd.2008.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Barkho BZ, Luo Y, Smrt RD, Santistevan NJ, Liu C, Kuwabara T, Gage FH, Zhao X. Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells. J Biol Chem. 2008;283:27644–27652. doi: 10.1074/jbc.M804899200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu C, Teng ZQ, Santistevan NJ, Szulwach KE, Guo W, Jin P, Zhao X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6:433–444. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HK, Belz T, Bock D, Takacs A, Wu H, Lichter P, Chai M, Schütz G. The nuclear receptor tailless is required for neurogenesis in the adult subventricular zone. Genes Dev. 2008;22:2473–2478. doi: 10.1101/gad.479308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, Sharp PA, Tabin CJ, McManus MT. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- McGlinn E, Yekta S, Mansfield JH, Soutschek J, Bartel DP, Tabin CJ. In ovo application of antagomiRs indicates a role for miR-196 in patterning the chick axial skeleton through Hox gene regulation. Proc Natl Acad Sci U S A. 2009;106:18610–18615. doi: 10.1073/pnas.0910374106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- Monaghan AP, Bock D, Gass P, Schwager A, Wolfer DP, Lipp HP, Schütz G. Defective limbic system in mice lacking the tailless gene. Nature. 1997;390:515–517. doi: 10.1038/37364. [DOI] [PubMed] [Google Scholar]
- Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kimura M, Horii T, Morita S, Soejima H, Kudo S, Hatada I. MeCP2-dependent repression of an imprinted miR-184 released by depolarization. Hum Mol Genet. 2008;17:1192–1199. doi: 10.1093/hmg/ddn011. [DOI] [PubMed] [Google Scholar]
- Ossipova O, Ezan J, Sokol SY. PAR-1 phosphorylates Mind bomb to promote vertebrate neurogenesis. Dev Cell. 2009;17:222–233. doi: 10.1016/j.devcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q, Shi Y. Neural stem cells in the developing and adult brains. J Cell Physiol. 2009;221:5–9. doi: 10.1002/jcp.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q, Sun G, Li W, Yang S, Ye P, Zhao C, Yu RT, Gage FH, Evans RM, Shi Y. Orphan nuclear receptor TLX activates Wnt/β-catenin signaling to stimulate neural stem cell proliferation and self-renewal. Nat Cell Biol. 2010;12:31–40. doi: 10.1038/ncb2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi J, Hunter CJ, Cole C, Ando H, Impey S, Monk CE, Martin KJ, Barton GJ, Hutvagner G, Arthur JS. Regulation of the miR-212/132 locus by MSK1 and CREB in response to neurotrophins. Biochem J. 2010;428:281–291. doi: 10.1042/BJ20100024. [DOI] [PubMed] [Google Scholar]
- Saini HK, Enright AJ, Griffiths-Jones S. Annotation of mammalian primary microRNAs. BMC Genomics. 2008;9:564. doi: 10.1186/1471-2164-9-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- Shi Y, Chichung Lie D, Taupin P, Nakashima K, Ray J, Yu RT, Gage FH, Evans RM. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sun G, Zhao C, Stewart R. Neural stem cell self-renewal. Crit Rev Oncol Hematol. 2008;65:43–53. doi: 10.1016/j.critrevonc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hübel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt RD, Eaves-Egenes J, Barkho BZ, Santistevan NJ, Zhao C, Aimone JB, Gage FH, Zhao X. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol Dis. 2007;27:77–89. doi: 10.1016/j.nbd.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, Teng ZQ, Luo Y, Peng J, Bordey A, Jin P, Zhao X. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura K, Abematsu M, Kohyama J, Namihira M, Nakashima K. Neuronal differentiation of neural precursor cells is promoted by the methyl-CpG-binding protein MeCP2. Exp Neurol. 2009;219:104–111. doi: 10.1016/j.expneurol.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Uchida N. MicroRNA-9 controls a migratory mechanism in human neural progenitor cells. Cell Stem Cell. 2010;6:294–296. doi: 10.1016/j.stem.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo NK, Cambronne XA, Goodman RH. MicroRNA pathways in neural development and plasticity. Curr Opin Neurobiol. 2010;20:457–465. doi: 10.1016/j.conb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RT, McKeown M, Evans RM, Umesono K. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature. 1994;370:375–379. doi: 10.1038/370375a0. [DOI] [PubMed] [Google Scholar]
- Zhang CL, Zou Y, He W, Gage FH, Evans RM. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature. 2008;451:1004–1007. doi: 10.1038/nature06562. [DOI] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y. microRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A. 2010;107:1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, Nakashima K, Lein ES, Eadie BD, Willhoite AR, Muotri AR, Summers RG, Chun J, Lee KF, Gage FH. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci U S A. 2003;100:6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]