Summary
Programmed frameshifting in the RF2 gene (prfB) involves an intragenic Shine-Dalgarno (SD) sequence. To investigate the role of SD-ASD pairing in the mechanism of frameshifting, we have analyzed the effect of spacing between the SD sequence and P codon on P-site tRNA binding and RF2-dependent termination. When the spacing between an extended SD sequence and the P codon is decreased from 4 to 1 nucleotides (nt), the dissociation rate (koff) for P-site tRNA increases by >100-fold. Toeprinting analysis shows that pretranslocation complexes cannot be formed when the spacer sequence is ≤ 2 nt. Instead, the tRNA added secondarily to fill the A site and its corresponding codon move spontaneously into the P site, resulting in a complex with a 3-nt longer spacer between the SD-ASD helix and the P codon. While close proximity of the SD clearly destabilizes P-site tRNA, RF2-dependent termination and EF-Tu-dependent decoding are largely unaffected in analogous complexes. These data support a model in which formation of the SD-ASD helix in ribosomes stalled at the in-frame UGA codon of prfB generates tension on the mRNA that destabilizes codon-anticodon pairing in the P site and promotes slippage of the mRNA in the 5′ direction.
Keywords: protein synthesis, translation, ribosome, release factor, tRNA
Introduction
For most genes, the translational reading frame is faithfully maintained from the start codon to the stop codon. The average frequency with which ribosomes spontaneously shift frames is on the order of 10−5 to 10−4 (Kurland, 1979). For certain genes, though, sequence elements within the mRNA direct ribosomes to shift reading frames at specific sites with high frequency. These programmed frameshifting sites modulate gene expression by generating two alternative gene products and/or by regulating gene expression levels (reviewed in Farabaugh, 1996, Gesteland & Atkins, 1996). One critical feature of these sites is a “slippery” sequence, which allows the bound tRNA(s) to dissociate from the original (zero) frame codon(s) and re-pair with overlapping codon(s) in either the −1 or +1 frame. Generally, slippery sequences for −1 programmed frameshifting follow the consensus X_XXY_YYZ (where underscores denote the zero frame), indicating that two ribosome-bound tRNA molecules change register on the mRNA. Slippery sequences for programmed +1 frameshifting tend to be shorter and contain homopolymeric runs of 3 or 4 nucleotides (nt), consistent with repairing of one bound tRNA (i.e., peptidyl-tRNA). One or more additional mRNA elements typically contribute to programmed frameshifting. Examples of these stimulatory elements include pseudoknots or stem-loops that lie downstream from shift site and Shine-Dalgarno (SD) sequences that lie upstream.
One of the best-known examples of programmed frameshifting is that of the bacterial prfB gene, which encodes release factor 2 (RF2). Synthesis of full-length E. coli RF2 requires that the ribosome shift into the +1 reading frame when it encounters codon 26, an in-frame UGA (Craigen & Caskey, 1986). Frameshifting competes with RF2-dependent termination, generating an autoregulatory mechanism for RF2 synthesis. Genetic analysis of the region surrounding the prfB frameshift site has revealed that the efficiency of frameshifting depends largely on a combination of three elements: (1) a slippery sequence such as CUU_U, (2) an in-frame stop at codon 26 (presumably to induce ribosome pausing), and (3) an appropriately positioned Shine-Dalgarno (SD) sequence upstream of the frameshift site (Baranov et al., 2002, Bekaert et al., 2006, Curran, 1993, Curran & Yarus, 1988, Schwartz & Curran, 1997, Sipley & Goldman, 1993, Weiss et al., 1987, Weiss et al., 1988). The spacing between the SD sequence and frameshift site is critical—increasing it by only 1 nt reduces frameshifting by 17-fold (Weiss et al., 1987). It has been suggested that formation of a closely juxtaposed SD-ASD helix generates tension on the mRNA and thereby promotes frameshifting (Curran & Yarus, 1988). Consistent with this idea, inserting a second SD immediately upstream of the existing SD in the prfB gene, which would allow formation of an alternative SD-ASD helix, reduced frameshifting by 47-fold (Weiss et al., 1987).
While the genetic studies have identified the determinants of prfB programmed frameshifting and their relative importance, how these determinants act to promote frameshifting has remained unclear. Here, we compare ribosomal complexes with various spacer lengths between the SD sequence and P codon. We find that a close juxtaposition of the SD-ASD helix and P codon strongly destabilizes P-site tRNA but has little or no effect on RF2-dependent termination or EF-Tu-dependent decoding. These data suggest that the intragenic SD of prfB destabilizes pairing of peptidyl-tRNALeu to the zero-frame CUU and promotes directional movement of the mRNA template with respect to the bound tRNA.
Results
Short spacing between the SD sequence and P codon precludes the formation of a stable pretranslocation complex
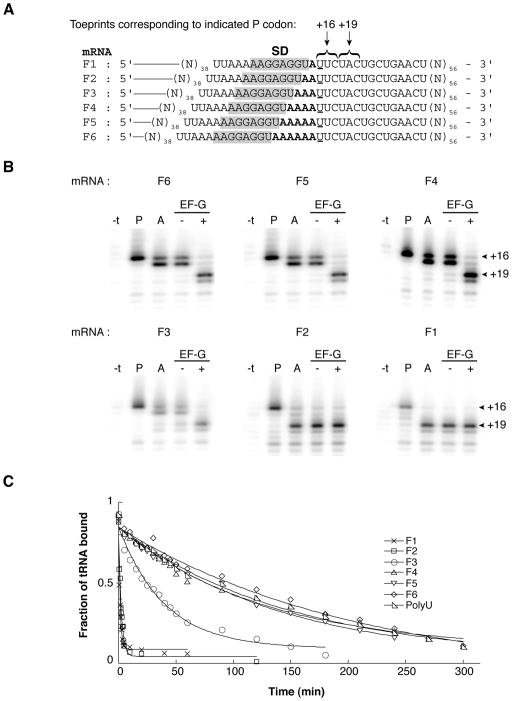
To investigate the influence of the SD-ASD pairing on tRNA binding, we made a series of model mRNAs in which the number of nucleotides between an extended SD element (AAGGAGGU) and P codon (UUC) was varied from 1 to 6 (Fig. 1A). Previous studies have shown that this extended SD sequence promotes very tight binding of mRNA to ribosomes (KD of the binary complex ≈ 15 pM) and pairs with the complementary nucleotides 1534–1541 (ACCUCCUU) of the 16S rRNA regardless of whether the P codon is relatively proximal (4 nt spacer) or distal (7 nt spacer) (Fahlman & Uhlenbeck, 2004, Jenner et al. 2010). We first used toeprinting to compare these mRNAs with respect to tRNA binding. Ribosomes were incubated with each of these mRNAs in the presence of tRNAPhe to bind the P site. In all cases, a strong toeprint was observed at position +16 (Fig. 1B, “P” lanes), indicating that the corresponding UUC codon was positioned in the 30S P site (Fig. 1A). In the case of mRNAs F2 and F1, the intensity of the +16 toeprint relative to the band at +22 (corresponding to the downstream codon UGC) was noticeably reduced. Subsequent addition of the peptidyl-tRNA analog N-acetyl-Tyr-tRNATyr (AcTyr-tRNATyr) to bind the A site resulted in a shift of the toeprint signal to +17, characteristic of A site occupancy (Jerinic & Joseph, 2000), for mRNAs F6, F5, F4, and F3 (Fig. 1B, “A” lanes). However, in the case of F2 and F1, there was little or no toeprint signal at +16/17, indicative of the pretranslocation (PRE) complex. Instead, a strong toeprint was observed at +19, corresponding to a complex containing the downstream UAC paired to AcTyr-tRNATyr in the P site. Presumably, complexes programmed with these mRNAs and containing tRNAPhe in the P site and AcTyr-tRNATyr in the A site are unstable, resulting in the spontaneous movement of tRNAPhe out of the P site and AcTyr-tRNATyr into it. Whether this movement occurs by spontaneous forward translocation or a mechanism involving complete dissociation of tRNAPhe and reassembly of the complex remains unclear. The observed rate of mRNA movement under these conditions is somewhat (~2-fold) slower than the dissociation rate (koff) of tRNAPhe from the P site (data not shown and see below), thus the latter mechanism clearly cannot be ruled out. As expected, in cases where the PRE complex was detected (i.e., F6, F5, F4, F3), addition of EF-G and GTP resulted in a strong toeprint at +19, consistent with efficient translocation of AcTyr-tRNATyr from the A site to the P site (Fig. 1B).
Figure 1. Effect of spacing between the SD sequence and P codon on tRNA binding.
(A) “F” series of mRNAs. Relevant codons and the Shine-Dalgarno sequence (SD) are indicated, spacer nucleotides are shown in bold type, and the position defined as +1 (the first nucleotide of the P codon) is underscored. (B) Toeprinting was used to map the position of mRNA within ribosome complexes after each of a number of additions. Ribosomes were incubated in polymix buffer with mRNA (as indicated) and tRNAPhe to fill the P site (P lanes), and AcTyr-tRNATyr2 was subsequently added to bind the A site (A lanes). Then, complexes were further incubated in the presence of GTP with or without EF-G (as indicated). The “−t” lane corresponds to primer extension of the mRNA in the presence of ribosomes but prior to addition of tRNA. (C) Examples of time courses to measure dissociation of [3′-32P]-tRNAPhe from the P site of ribosomes programmed with mRNA (as indicated).
Short spacing between the SD sequence and P codon increases the dissociation rate of tRNAPhe from the P site
To directly test how these various mRNAs affected the stability of P-site tRNA, we measured the dissociation rate (koff) of [3′-32P]-tRNAPhe from the P site using a double membrane filter binding method (Fahlman & Uhlenbeck, 2004). For ribosomes programmed with mRNAs with longer spacers (F5-F6), the koff was ≤ 0.005 min−1 (Fig. 1C, Table 1). When the spacer was shortened to 3, 2, and 1 nt, koff increased by 6-, 88-, and 160-fold, respectively. With polyU, which lacks a SD element, koff for tRNAPhe was identical to that seen for the mRNAs with longer (5–6 nt) spacers. These data show that the SD element can dramatically destabilize P-site tRNA when closely juxtaposed to the P codon.
Table 1.
Dissociation rate constants (koff) for P-site tRNA
| tRNA | mRNA | koff (min−1) |
|---|---|---|
| tRNAPhe | F1 | 0.78 ± 0.1 |
| F2 | 0.44 ± 0.004 | |
| F3 | 0.030 ± 0.004 | |
| F4 | 0.0081 ± 0.0006 | |
| F5 | 0.0042 ± 0.0006 | |
| F6 | 0.0051 ± 0.0007 | |
| Poly U | 0.0053 ± 0.0006 | |
| tRNAVal | V1 | 1.09 ± 0.1 |
| V2 | 0.71 ± 0.1 | |
| V3 | 0.45 ± 0.1 | |
| V4 | 0.23 ± 0.01 | |
| V5 | 0.062 ± 0.001 | |
| tRNAMet | M1 | 0.53 ± 0.01 |
| M2 | 0.28 ± 0.04 | |
| M3 | 0.014 ± 0.0007 |
Data represent the mean ± SEM from three independent experiments.
Short spacing between the SD sequence and P codon decreases the stability of P-tRNA in the prfB gene context
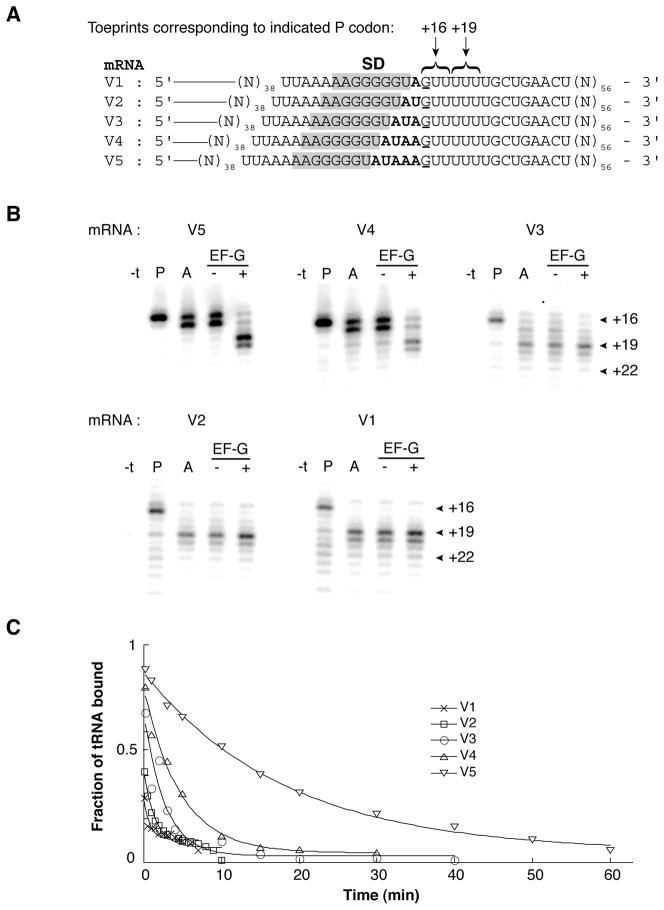
The above results show that the SD-ASD helix can strongly influence P-tRNA stability, consistent with the idea that the intragenic SD of prfB promotes +1 frameshifting by destabilizing codon-anticodon pairing in the P site. To more rigorously test this hypothesis, we repeated the analysis using a set of mRNAs based on the sequence of the E. coli prfB frameshift site. Message V2 of this series is identical to the relevant region of prfB except for a C to G substitution at position +1 (the first nucleotide of the zero frame P codon of the slippery site) (Fig. 2A). This substitution, which allows us to use the commercially-available native tRNAVal, confers only a modest (3- to 4-fold) decrease in the efficiency of programmed frameshifting in vivo (Curran, 1993, Weiss et al., 1987), suggesting that the mechanism is largely unperturbed. Consistent with the data of Fig. 1, PRE complexes could readily be formed on mRNAs V5 and V4, but were not observed for those mRNAs with shorter spacer regions (V1-V3) (Fig. 2). For this set of mRNAs, koff for P-site tRNA increased progressively over a 22-fold range as the distance between the SD and P codon decreased (Table 1).
Figure 2. Effect of spacing between the SD sequence and P codon on tRNA binding in the RF2 (prfB) context.
(A) “V” series of mRNAs, annotated as described in the legend to Fig. 1. (B) Toeprinting was used to map the position of mRNA within ribosome complexes after each of a number of additions. Ribosomes were incubated in polymix buffer with mRNA (as indicated) and tRNAVal to fill the P site (P lanes), and AcPhe-tRNAPhe was subsequently added to bind the A site (A lanes). Then, complexes were further incubated in the presence of GTP with or without EF-G (as indicated). The “−t” lane corresponds to primer extension of the mRNA in the presence of ribosomes but prior to addition of tRNA. (C) Examples of time courses to measure dissociation of [3′-32P]-tRNAVal from the P site of ribosomes programmed with mRNA (as indicated).
Effects of spacing between the SD sequence and P codon on RF2 function
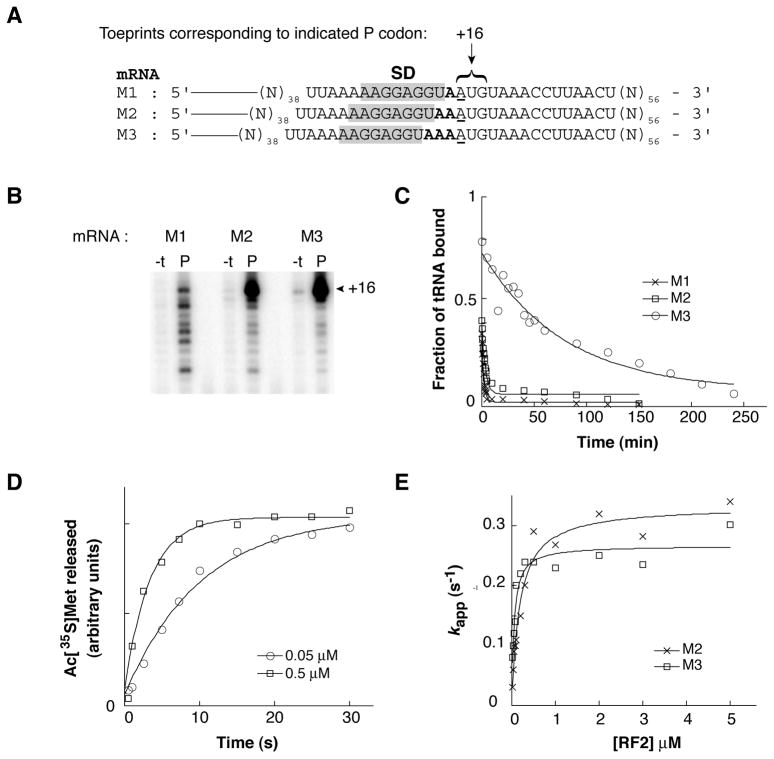
The amount of full-length RF2 produced in the cell depends on the rate of the frameshifting event versus the rate of RF2-dependent termination. The SD element of the programmed frameshifting site could increase the fraction of full-length product by stimulating peptidyl-tRNALeu slippage, inhibiting RF2-dependent termination, or both. In order to investigate whether the SD can influence RF2 activity, we designed another set of messages with sequential Met (AUG) and stop (UAA) codons positioned at various distances downstream from the SD sequence AAGGAGGU (Fig. 3A). Binding of AcMet-tRNAMet to the P site in the presence of mRNA M3 and M2 resulted in a strong toeprint at position +16 and several weaker toeprints corresponding to downstream positions (Fig. 3B). These data indicate that most of the ribosomes contain the AUG codon in the P site. In contrast, binding of AcMet-tRNAMet to the P site of ribosomes programmed with M1 gave toeprints of similar intensity at multiple positions, including +16, +18, +21, +22, and +25. This suggests that, with mRNA M1, complexes containing the cognate P codon (spaced near the SD) and those with a near- or non-cognate P codon (spaced further from the SD) exhibit similar stabilities. Analogous complexes were made in which Ac[35S]Met-tRNAMet was bound to the P site, and koff values were determined (Fig. 3C, Table 1). In ribosomes programmed with mRNA M3, dissociation of Ac[35S]Met-tRNAMet was relatively slow (koff = 0.014 ± 0.0007 min−1). This rate was 28-and 53-fold faster in the presence of mRNA M2 and M1, respectively. These data are in line with those presented in Figures 1 and 2 and underscore the generality of the results.
Figure 3. Effect of spacing between the SD sequence and P codon on RF2 function.
(A) “M” series of mRNAs, annotated as described in the legend to Fig. 1. (B) Ribosomes were incubated in polymix buffer with mRNA (as indicated) in the absence (−t) or presence (P) of AcMet-tRNAMet, and the complexes were analyzed by toeprinting. (C) Examples of time courses to measure dissociation of Ac[35S]Met-tRNAMet from the P site of ribosomes programmed with mRNA (as indicated). (D) Examples of time courses to measure RF2-catalyzed hydrolysis of Ac[35S]Met-tRNAMet. Ribosomes programmed with mRNA M3 and containing Ac[35S]Met-tRNAMet in the P site were rapidly mixed with RF2 for various times before quenching with 25% formic acid. The relative amount of Ac[35S]Met released at each time point was determined by ethyl acetate extraction and liquid scintillation counting. Data were fit to a single exponential function to obtain apparent rates (kapp). (E) Apparent rates were determined for several concentrations of RF2, which allowed kcat (M2 = 0.33 s−1; M3 = 0.27 s−1) and KM (M2 = 0.18 μM; M3 = 0.052 μM) for the single-turnover reaction to be deduced.
Next, RF2 was overexpressed and purified, and the effects of spacing on RF2-dependent termination were measured using a single-turnover quench-flow assay. Ribosomes programmed with either M2 or M3 and containing Ac[35S]Met-tRNAMet in the P site were mixed with RF2 for various periods of time before quenching with 25% formic acid and quantifying the amount of Ac[35S]Met released (Fig. 3D). The extent of the reaction in the presence of M3 was about twice that of M2, consistent with the predicted levels of bound Ac[35S]Met-tRNAMet (Fig. 3C). Apparent rates of Ac[35S]Met release were measured at several concentrations of RF2, which allowed us to estimate the maximal rate of peptide release (kcat) and the concentration of RF2 at which the half-maximal rate was observed (KM) (Fig. 3E). Shortening the spacer from 3 to 2 nt had virtually no effect on kcat and only modestly increased KM. It is known that RF2 is normally posttranslationally modified, and this modification (N5-methylation of Q252) increases kcat for the single-turnover reaction by about 10-fold (Dincbas-Renqvist et al., 2000). The kinetic parameters measured here are in line with previous experiments that employed overexpressed RF2, which lacks the modification (Dincbas-Renqvist et al., 2000, Zaher & Green, 2009).
Effects of spacing between the SD sequence and P codon on decoding
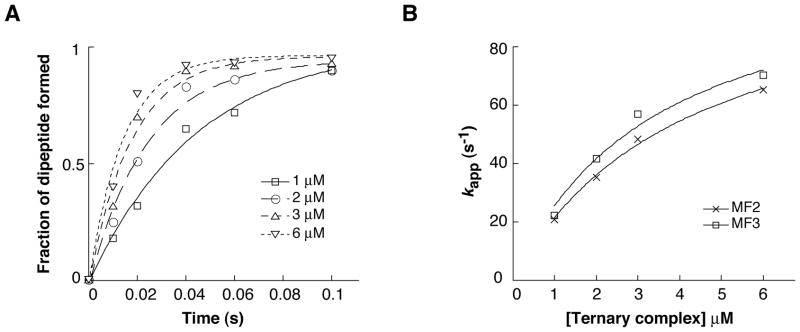
To further explore the potential effects of a proximal SD-ASD helix on ribosome function, decoding was analyzed. The UAA stop codon of mRNAs M2 and M3 was changed to UUC, and the resulting mRNAs (MF2 and MF3) were used to assemble 70S initiation complexes containing f[35S]Met-tRNAfMet. These complexes (0.2 μM) were rapidly mixed with EF-Tu•GTP•Phe-tRNAPhe (at various concentrations ≥ 1 μM), and the amount of f[35S]Met-Phe formed at various time points was quantified (Fig. 4A). The extent of the reaction was 50% lower with the shorter spacer, presumably due to a smaller fraction of f[35S]Met-tRNAfMet bound to the P site prior to addition of the ternary complex. Apparent rates, estimated by single-exponential curve fitting, were virtually identical for the two complexes at each concentration of ternary complex tested (Fig. 4B), indicating that the closely juxtaposed SD had no effect on decoding rate. Fitting the secondary plots to a hyperbolic function suggested a maximal rate (kcat) of 110 s−1 and a KM value in the neighborhood of 3 to 4 μM. These parameters are consistent with those obtained previously under similar experimental conditions (Johansson et al., 2008).
Figure 4. Effect of spacing between the SD sequence and P codon on decoding.
(A) Examples of experiments that measure the formation of f[35S]Met-Phe as a function of time. Ribosomes programmed with mRNA MF3 and containing f[35S]Met-tRNAfMet in the P site (0.2 μM) were rapidly mixed with EF-Tu•GTP•Phe-tRNAPhe (various concentrations as indicated) at 25° C for various times before quenching with 0.5 M KOH formic acid. The data were fit to a single exponential function to estimate apparent rates. (B) Apparent rates were plotted as a function of ternary complex concentration and the data were fit to a hyperbolic function, yielding kcat (MF2 = 110 s−1; MF3 = 110 s−1) and KM (MF2 = 4.2 μM; MF3 = 3.4 μM) parameters for the single-turnover reaction.
Discussion
In the case of prfB, ribosomal frameshifting is highly efficient, occurring at a frequency of about 50% during steady-state growth. This level of frameshifting depends on a closely juxtaposed SD element, but the mechanism by which the SD stimulates frameshifting has been unclear. In theory, the SD could increase the efficiency of frameshifting by either (1) accelerating dissociation/re-pairing of peptidyl-tRNALeu on the mRNA or (2) slowing RF2-dependent termination at the in-frame UGA (Baranov et al., 2004). Here, we systematically varied the distance between the SD element and P codon and analyzed the effects on P-tRNA binding and RF2-dependent termination. We find that proximal positioning of the SD strongly destabilizes the P-site tRNA but has little effect (< 3-fold) on kcat/KM for RF2. These data indicate that the SD acts primarily by accelerating the frameshift event itself rather than by slowing RF2-dependent termination.
It was proposed early on that tension on the mRNA generated by the proximal SD-ASD helix promotes dissociation of peptidyl-tRNA from the zero-frame CUU and facilitates re-pairing to the downstream overlapping UUU (Curran & Yarus, 1988). This model gains strong support from the biochemical data presented here and from recent x-ray crystal structures of ribosomal complexes with well-resolved mRNAs (Jenner et al., 2010). Two structures have been described, one containing two tRNAs (P and E) and 4 nt between the SD sequence and P codon and the other containing three tRNAs (A, P, and E) and 7 nt between the SD sequence and P codon. The SD sequence of the F and M series of mRNAs described here is identical to that employed in the structural studies, thus the ribosomal complex containing mRNA F4 and P-site tRNAPhe (Fig. 1) is analogous to the complex with the 4 nt spacer solved by crystallography. In this structure, the distance from the last nucleotide of the SD to the first nucleotide of the P codon is about 17 Å. The 4 nt spacer of the bound mRNA easily spans this distance—its ribose-phosphate backbone is not fully extended and instead exhibits a turn before and after the first nucleotide of the E codon. A shorter spacer of 3 nt with a fully-extended backbone should be able to span the distance, allowing both SD-ASD and P-site codon-anticodon pairing to occur without the need to reposition the SD-ASD helix. In contrast, shorter spacers of 2 and 1 nt will be unable to cover this distance, requiring that the SD-ASD helix move toward the P site by ~6 Å and ~13 Å, respectively, if both rRNA-mRNA and tRNA-mRNA interactions are to be retained. Movement of the SD-ASD helix is constrained by its connection to h45 and presumably by adjacent elements such as h28. Consequently, such large-scale movements of the SD-ASD helix toward the P site are predicted to be unfavorable and generate tension on the P codon, thereby destabilizing codon-anticodon pairing in the P site. Consistent with these structural predictions, we find with the analogous mRNAs that koff for P-site tRNA increases to some degree when the spacer is shortened to 3 nt (e.g., 4-fold for F3) and increases dramatically when the spacer is further shortened (e.g., 55-fold for F2 and 28-fold for M2). In the case of the “V” series of mRNAs, P-tRNA generally binds less tightly, and stepwise removal of spacer nucleotides increase koff in a more incremental fashion. The predicted SD-ASD helix in these complexes contains a central G-U wobble pair and the sequence of the spacer is heterogeneous, but whether one or both of these differences contribute to the somewhat altered trend remains unclear. The intragenic SD of prfB tends to be either GGGGGU or GGAGGU, depending on the bacterial species (Baranov et al., 2002, Bekaert et al., 2006), suggesting that the stretch of 5 guanines is not critical to the frameshifting mechanism.
A growing body of evidence suggests that codon-anticodon pairing in the E site contributes to reading frame maintenance, and it has been proposed that a key function of the SD of prfB is to catalyze release of E-site tRNA (Marquez et al., 2004). The last nucleotide of the predicted SD sequence corresponds to the first nucleotide of the E codon, hence annealing of the SD-ASD helix would clearly preclude codon-anticodon pairing in the E site. While attractive from the structural point of view, this hypothesis is difficult to address experimentally. Sanders and Curran (2007) systematically mutagenized the E codon of the prfB frameshift site (which is normally UAU in E. coli). They observed an inverse correlation between the rate of frameshifting and the predicted stability of codon-anticodon pairing, consistent with a role for E-tRNA in frame maintenance (Sanders & Curran, 2007). Additionally, they found that E codons beginning with U supported higher levels of frameshifting than those beginning with C, consistent with participation of this nucleotide in SD-ASD helix formation. In a subsequent study, we targeted the ribosome by truncating the β-hairpin of ribosomal protein S7, which normally interacts with tRNA and mRNA in the 30S E site (Devaraj et al., 2009). Ribosomes harboring this E-site mutation (S7ΔR77-Y84) exhibited increased levels of both +1 and −1 spontaneous frameshifting, lending further support to the idea that codon-anticodon pairing in the E site contributes to frame maintenance. The effects of mutation S7ΔR77-Y84 on programmed frameshifting were also measured, using the set of prfB’-lacZ constructs in which the E codon was varied (Sanders & Curran, 2007, Devaraj et al., 2009). In most cases, an increase in frameshifting was observed. However, in seven cases (UAU, UCA, UCU, UCC, UGG, UUG, and UGU), the E-site mutation clearly failed to confer an effect. Intriguingly, this subset of E codons includes all of those well represented in nature (i.e., UAU, UCU, and UGU) (Baranov et al., 2002, Bekaert et al., 2006). One possible explanation of these data is that in the natural context, E-tRNA release is very rapid (i.e., catalyzed) and hence this step does not contribute to limiting the overall rate of frameshifting. Whereas for many of the synthetic constructs, the rate of this step relative one or more other steps in the pathway is substantially reduced. In these cases, E-tRNA release does contribute to limiting the overall rate, and hence S7ΔR77-Y84 confers an observable effect (Devaraj et al., 2009).
The rate of the prfB frameshift event itself is unknown, but its rate relative to that of termination and decoding has been determined for cells in steady-state growth (Curran & Yarus, 1989, Poole et al., 1995). Because the closely juxtaposed SD does not substantially influence the rate of either termination or decoding (Fig. 3 and 4), the rate of the frameshift event can be roughly approximated based on available data. Curran and Yarus (1989) replaced the in-frame UGA codon of prfB with 29 sense codons, allowing the rate of decoding versus frameshifting to be deduced (Curran & Yarus, 1989). While the relative rates of decoding spanned a 20-fold range, the mean value suggests that the average decoding rate is ~ 8-fold faster than the frameshifting event. Tate and coworkers similarly assessed termination in 12 different contexts and found the average rate to be comparable to that of the frameshifting event (Poole et al., 1995). While the rate of termination in vivo has not been measured, it must exceed that of initiation, ~ 0.5 s−1 (Kennell & Riezman, 1977, Mitarai et al., 2008), since there is no evidence for a kinetic bottleneck at the termination stage of translation. A recent estimate of the average rate of decoding during rapid growth is 30 s−1 (Lovmar & Ehrenberg, 2006). Taken together, these observations suggest that, under similar growth conditions, the prfB frameshift occurs at a rate of 0.5 < × < 8 s−1.
Programmed -1 frameshifting can also involve SD-ASD pairing. In these cases, exemplified by dnaX, the spacer length between the SD and P codon exceeds that which is optimal for translation initiation (Larsen et al., 1997, Larsen et al., 1994, Vellanoweth & Rabinowitz, 1992). The binding of P-site tRNA to 30S subunits programmed with mRNAs containing the SD sequence AAGGAGGU and two different spacer lengths, corresponding to 5 and 8 nt in our numbering scheme, has been measured (Antoun et al., 2006). It was found that the longer spacer destabilized tRNA, increasing koff by 6- to 8-fold. The corresponding spacer length for dnaX is 9 nt, suggesting that the SD destabilizes pairing of peptidyl-tRNA in the zero frame to promote both +1 and −1 frameshifting. Many new examples of programmed ribosomal frameshifting have been uncovered in bacterial insertion sequences and phages (Baranov et al., 2006). Nearly all of these contain a putative SD element, implying that SD-ASD interactions provide an effective and versatile means to direct ribosomal frameshifting in bacteria.
Experimental Procedures
Ribosomes from E. coli strain CSH142 [F- ara Δ(gpt-lac)5], translation factors, and mRNAs were purified as described (Boon et al., 1992, Fredrick & Noller, 2002, Fredrick & Noller, 2003). Purified tRNAs (purchased from Sigma or Chemical Block) were charged, acetylated, and/or radiolabeled as described (Fredrick & Noller, 2002, McGarry et al., 2005, Walker & Fredrick, 2008). His6-tagged E. coli RF2 was overexpressed and purified using Ni2+-NTA resin (Qiagen).
Ribosome complexes formed in polymix buffer (Ehrenberg et al., 1990) were analyzed by toeprinting as described previously (Shoji et al., 2006). Briefly, 32P-labeled primer was annealed to mRNA (0.5 μM) in 5 mM potassium phosphate (pH 7.3) and 95 mM KCl by heating to 60 °C and placing on ice. Polymix salts [5 mM Mg(OAc)2, 0.5 mM CaCl2, 5 mM NH4Cl, 8 mM putrescine, and 1 mM spermidine], DTT (1 mM), ribosomes (0.7 μM), and tRNA (1 μM) were added (final concentrations indicated throughout) and incubated at 37 °C for 20 min to bind the P site. Next, N-acetyl-aminoacyl-tRNA (1 μM) was added and incubated at 37 °C for 10 min to bind the A site. Finally, EF-G (1 μM) and GTP (300 μM) were added and the reactions were further incubated at 37 °C for 10 min. At each stage of the experiment, a 2 μL aliquot was removed and added to 10 μL of extension mix [10 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 60 mM NH4Cl, 6 mM βME, 400 μM each dNTP and 0.2 U/μL AMV reverse transcriptase] to extend the primer. Extension products were analyzed using denaturing PAGE.
The rate of dissociation of tRNA from the P site was measured using a double-membrane filtration method as described (Fahlman & Uhlenbeck, 2004). Typically, < 50 nM [3′-32P]-tRNA (~20,000 cpm/pmol), 0.5 μM mRNA and 0.7 μM ribosomes were incubated in polymix buffer at 37 °C for 20 min to bind the P site. At t = 0, 2 μL of the reaction was diluted 100-fold in polymix buffer containing 0.2 μM unlabeled tRNA, and 20 μL aliquots were filtered at various time points. Membranes were immediately washed with 200 μL of polymix buffer, dried and exposed to a phosphor screen (Amersham). Data were quantified and corrected for background. Dissociation rate constants (koff) were determined by fitting the data to a single exponential function, using the program KaleidaGraph (Synergy Software).
The rate of single-turnover peptide release was determined as described (Brunelle et al., 2008). Ribosomes (1.5 μM) were first incubated with mRNA (M2 or M3; 3.0 μM) and Ac[35S]Met-tRNAMet (0.4 μM) in polymix buffer for 15 min at 37°C to fill the P site. Equal volumes of the ribosome complex (0.2 μM final) and RF2 (various concentrations) were then rapidly mixed at 37°C in a quench-flow apparatus (KinTek RQF-3) for indicated times before quenching with 25% formic acid. Released Ac[35S]Met was extracted into ethyl acetate and quantified in a scintillation counter. The data were fit to a single exponential function using KaleidaGraph (Synergy Software) to determine apparent rates (kapp) of peptide release. Apparent rates were plotted as a function of RF2 concentration and fit to the quadratic equation kapp = kcat((A+B+KM) – ((A+B+KM)2 – 4AB)1/2)/(2A), where A and B represent the total concentrations of ribosomal complex and RF2, respectively, in the reaction. This yielded the kcat and KM parameters for the single-turnover reaction.
The rate of decoding was measured as follows. Initiation complexes were formed by incubating ribosomes (1.5 μM) with mRNA (3 μM) and f[35S]Met-tRNAfMet (0.4 μM) in the presence of initiation factors IF1, IF2 and IF3 (1.5 μM each) in polymix buffer for 1 h at 37°C. The initiation complexes were then purified from unbound components by centrifugation through 1.3 mL 1.1 M sucrose cushions in polymix. The pelleted complexes were dissolved in polymix buffer with 80% recovery. Ternary complexes were formed by incubating tRNAPhe (1 μM), phenylalanine (100 μM), PheRS (2 μM), ATP (2.5 mM), EF-Tu (4 μM), EF-Ts (1 μM), GTP (1 mM), pyruvate kinase (100 μg/mL) and PEP (3 mM) in polymix buffer at 37°C for 30 min. Equal volumes of the initiation complex (0.2 μM final) and ternary complex (various concentrations) were rapidly mixed in a KinTek machine, and reactions were quenched at various time points with 0.5 M KOH. Electrophoretic thin-layer chromatography was used to separate f[35S]Met from f[35S]Met-Phe (Youngman et al., 2004), and the fraction of dipeptide formed [i.e., fMet-Phe/(fMet-Phe + fMet)] was determined by phosphorimager analysis. Apparent rates of dipeptide formation were measured at several concentrations of ternary complex, and the resulting data were fit to a hyperbolic function to estimate kcat and KM.
Acknowledgments
We thank R. Green for the strain to overexpress His6-tagged RF2, B. Kraal for the strain to overexpress EF-Tu-His6, H. Roy for ATP(CTP):tRNA nucleotidyltransferase, S. Walker for purified EF-G, S. McClory for purified EF-Ts, S. Shoji and S. McClory for technical assistance, and J. Lee, D. Qin, and S. McClory for comments on the manuscript. This work was supported by NIH grant GM072528.
References
- Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol Cell. 2006;23:183–193. doi: 10.1016/j.molcel.2006.05.030. [DOI] [PubMed] [Google Scholar]
- Baranov PV, Fayet O, Hendrix RW, Atkins JF. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Baranov PV, Gesteland RF, Atkins JF. Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 2002;3:373–377. doi: 10.1093/embo-reports/kvf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov PV, Gesteland RF, Atkins JF. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA. 2004;10:221–230. doi: 10.1261/rna.5122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert M, Atkins JF, Baranov PV. ARFA: A program for annotating bacterial release factor genes, including prediction of programmed ribosomal frameshifting. Bioinformatics. 2006;22:2463–2465. doi: 10.1093/bioinformatics/btl430. [DOI] [PubMed] [Google Scholar]
- Boon K, Vijgenboom E, Madsen LV, Talens A, Kraal B, Bosch L. Isolation and functional analysis of histidine-tagged elongation factor Tu. Eur J Biochem. 1992;210:177–183. doi: 10.1111/j.1432-1033.1992.tb17406.x. [DOI] [PubMed] [Google Scholar]
- Brunelle JL, Shaw JJ, Youngman EM, Green R. Peptide release on the ribosome depends critically on the 2′ OH of the peptidyl-tRNA substrate. RNA. 2008;14:1526–1531. doi: 10.1261/rna.1057908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Curran JF. Analysis of effects of tRNA:message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site. Nucleic Acids Res. 1993;21:1837–1843. doi: 10.1093/nar/21.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran JF, Yarus M. Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J Mol Biol. 1988;203:75–83. doi: 10.1016/0022-2836(88)90092-7. [DOI] [PubMed] [Google Scholar]
- Curran JF, Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J Mol Biol. 1989;209:65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- Devaraj A, Shoji S, Holbrook ED, Fredrick K. A role for the 30S subunit E site in maintenance of the translational reading frame. RNA. 2009;15:255–265. doi: 10.1261/rna.1320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dincbas-Renqvist V, Engstrom A, Mora L, Heurgue-Hamard V, Buckingham R, Ehrenberg M. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 2000;19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg M, Bilgin N, Kurland CG. Design and use of a fast and accurate in vitro translation system. In: Spedding G, editor. Ribosomes and protein synthesis-- a practical approach. Oxford: IRL Press; 1990. pp. 101–129. [Google Scholar]
- Fahlman RP, Uhlenbeck OC. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry. 2004;43:7575–7583. doi: 10.1021/bi0495836. [DOI] [PubMed] [Google Scholar]
- Farabaugh PJ. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- Gesteland RF, Atkins JF. Recoding: dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- Jerinic O, Joseph S. Conformational changes in the ribosome induced by translational miscoding agents. J Mol Biol. 2000;304:707–713. doi: 10.1006/jmbi.2000.4269. [DOI] [PubMed] [Google Scholar]
- Johansson M, Bouakaz E, Lovmar M, Ehrenberg M. The kinetics of ribosomal peptidyl transfer revisited. Mol Cell. 2008;30:589–598. doi: 10.1016/j.molcel.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Kennell D, Riezman H. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J Mol Biol. 1977;114:1–21. doi: 10.1016/0022-2836(77)90279-0. [DOI] [PubMed] [Google Scholar]
- Kurland CG. Reading frame errors on ribosomes. In: Celis JE, Smith JD, editors. Nonsense mutations and tRNA suppressors. London: Academic Press; 1979. pp. 97–108. [Google Scholar]
- Larsen B, Gesteland RF, Atkins JF. Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50% J Mol Biol. 1997;271:47–60. doi: 10.1006/jmbi.1997.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B, Wills NM, Gesteland RF, Atkins JF. rRNA-mRNA base pairing stimulates a programmed -1 ribosomal frameshift. J Bacteriol. 1994;176:6842–6851. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovmar M, Ehrenberg M. Rate, accuracy and cost of ribosomes in bacterial cells. Biochimie. 2006;88:951–961. doi: 10.1016/j.biochi.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Marquez V, Wilson DN, Tate WP, Triana-Alonso F, Nierhaus KH. Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- McGarry KG, Walker SE, Wang H, Fredrick K. Destabilization of the P site codon-anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Mol Cell. 2005;20:613–622. doi: 10.1016/j.molcel.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitarai N, Sneppen K, Pedersen S. Ribosome collisions and translation efficiency: optimization by codon usage and mRNA destabilization. J Mol Biol. 2008;382:236–245. doi: 10.1016/j.jmb.2008.06.068. [DOI] [PubMed] [Google Scholar]
- Poole ES, Brown CM, Tate WP. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14:151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders CL, Curran JF. Genetic analysis of the E site during RF2 programmed frameshifting. RNA. 2007;13:1483–1491. doi: 10.1261/rna.638707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R, Curran JF. Analyses of frameshifting at UUU-pyrimidine sites. Nucleic Acids Res. 1997;25:2005–2011. doi: 10.1093/nar/25.10.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipley J, Goldman E. Increased ribosomal accuracy increases a programmed translational frameshift in Escherichia coli. Proc Natl Acad Sci U S A. 1993;90:2315–2319. doi: 10.1073/pnas.90.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellanoweth RL, Rabinowitz JC. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- Walker SE, Fredrick K. Preparation and evaluation of acylated tRNAs. Methods. 2008;44:81–86. doi: 10.1016/j.ymeth.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RB, Dunn DM, Atkins JF, Gesteland RF. Slippery runs, shifty stops, backward steps, and forward hops: -2, −1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss RB, Dunn DM, Dahlberg AE, Atkins JF, Gesteland RF. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988;7:1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]