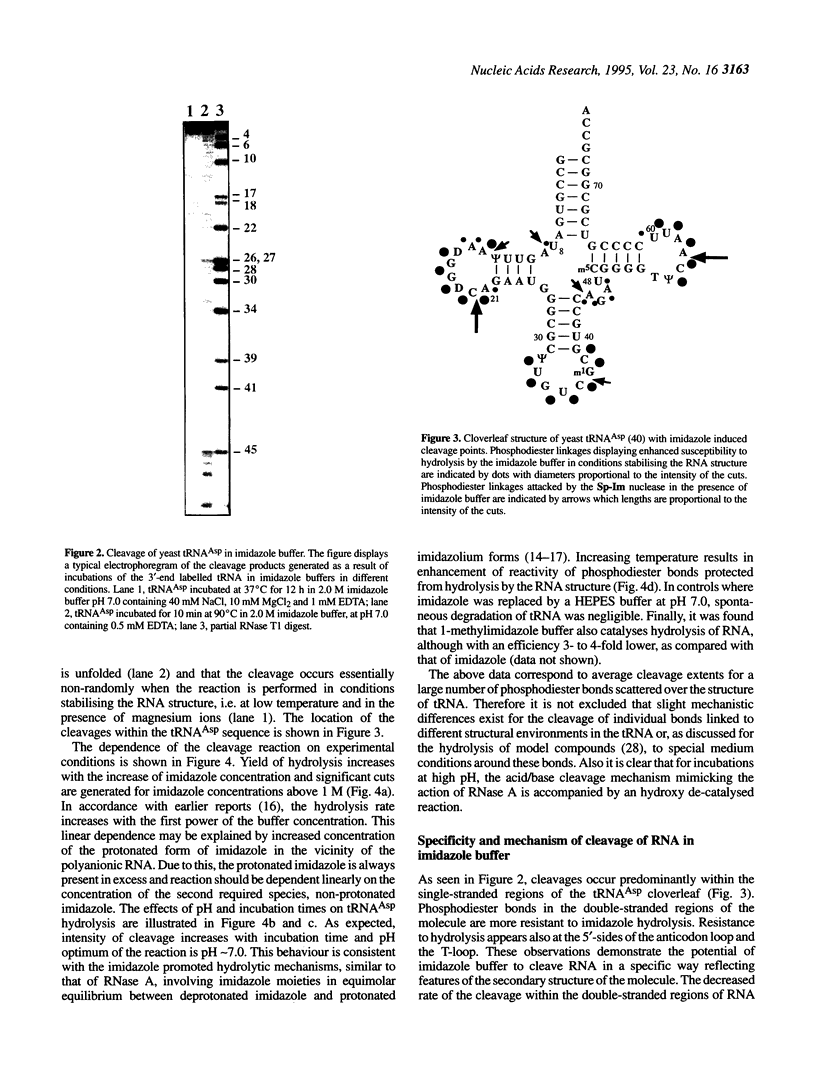
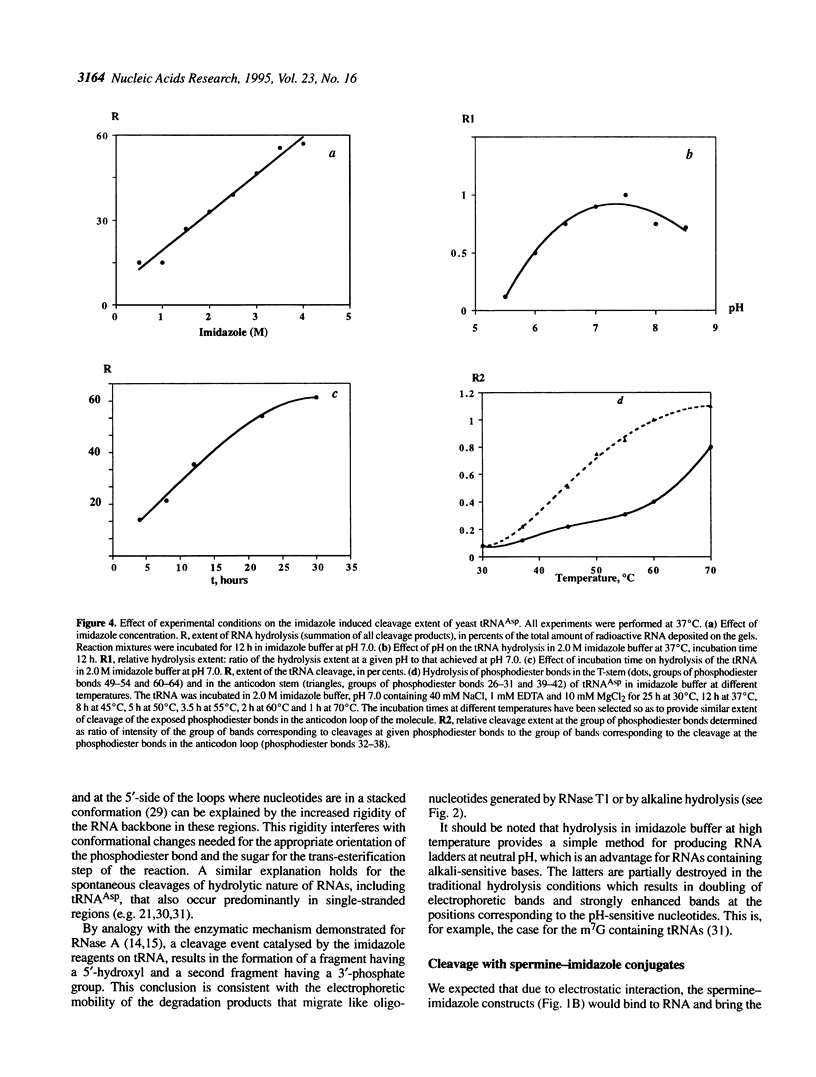
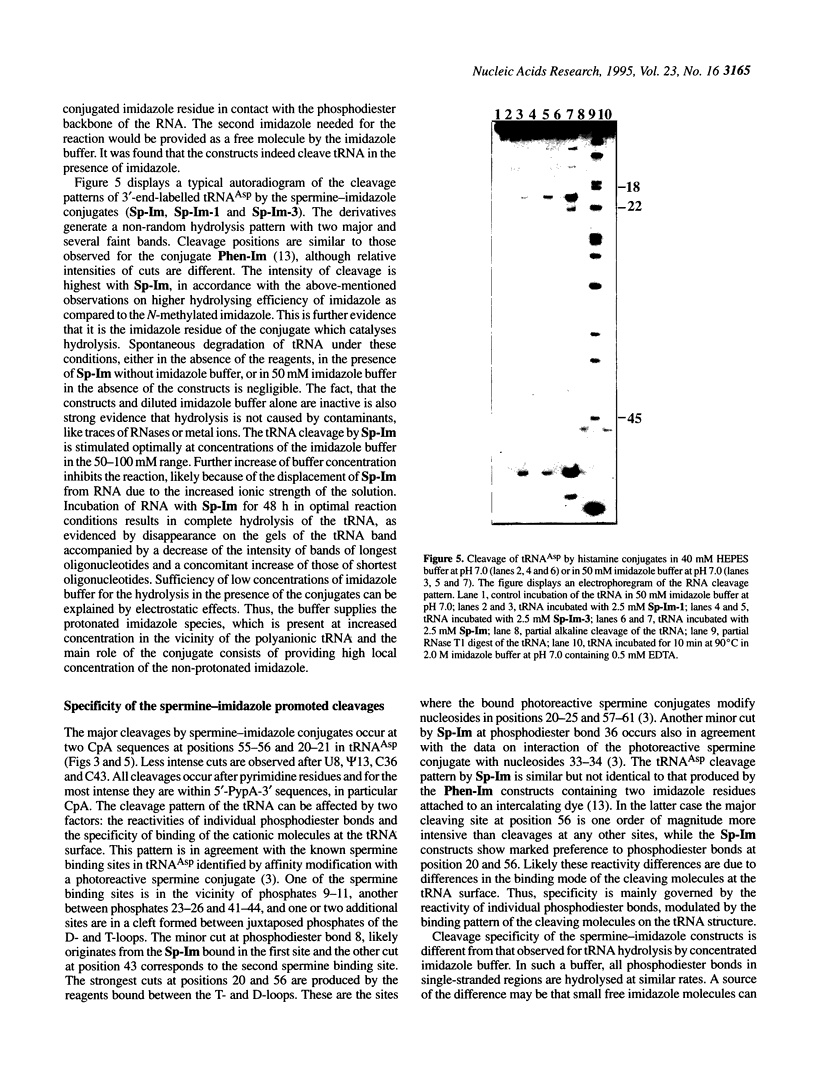
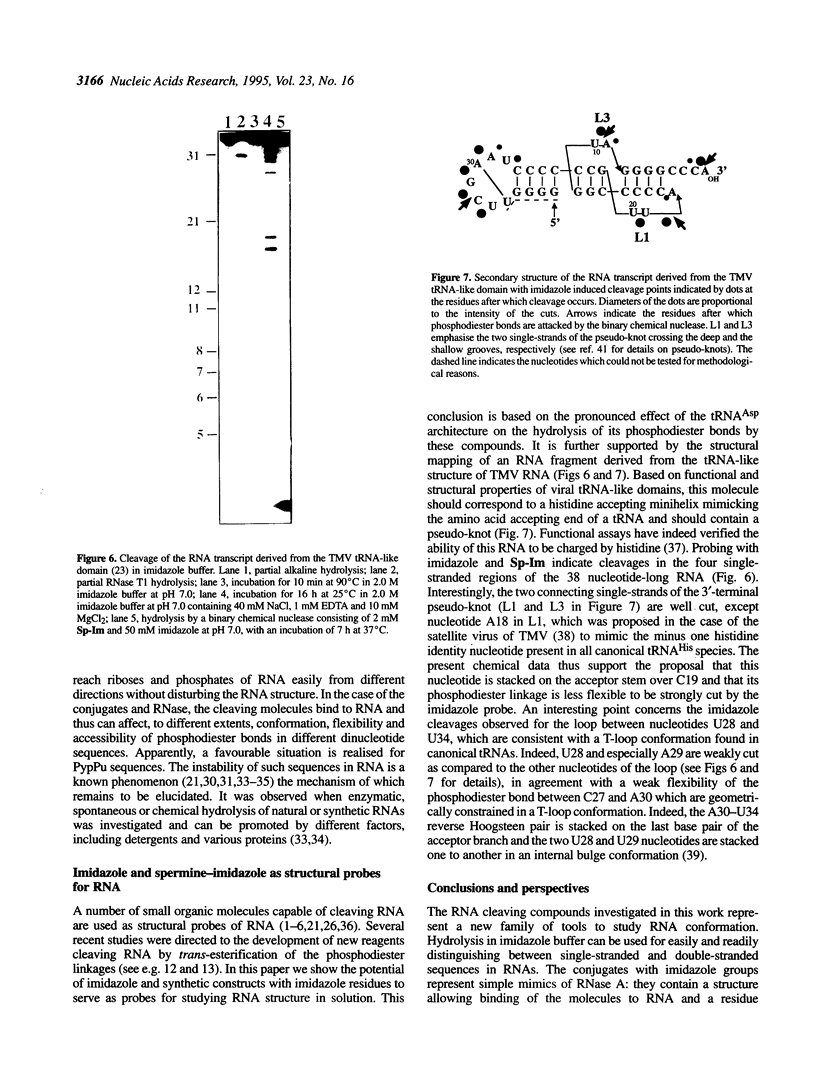
Abstract
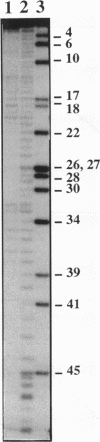
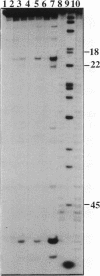
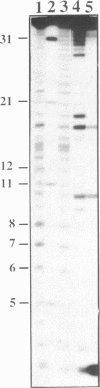
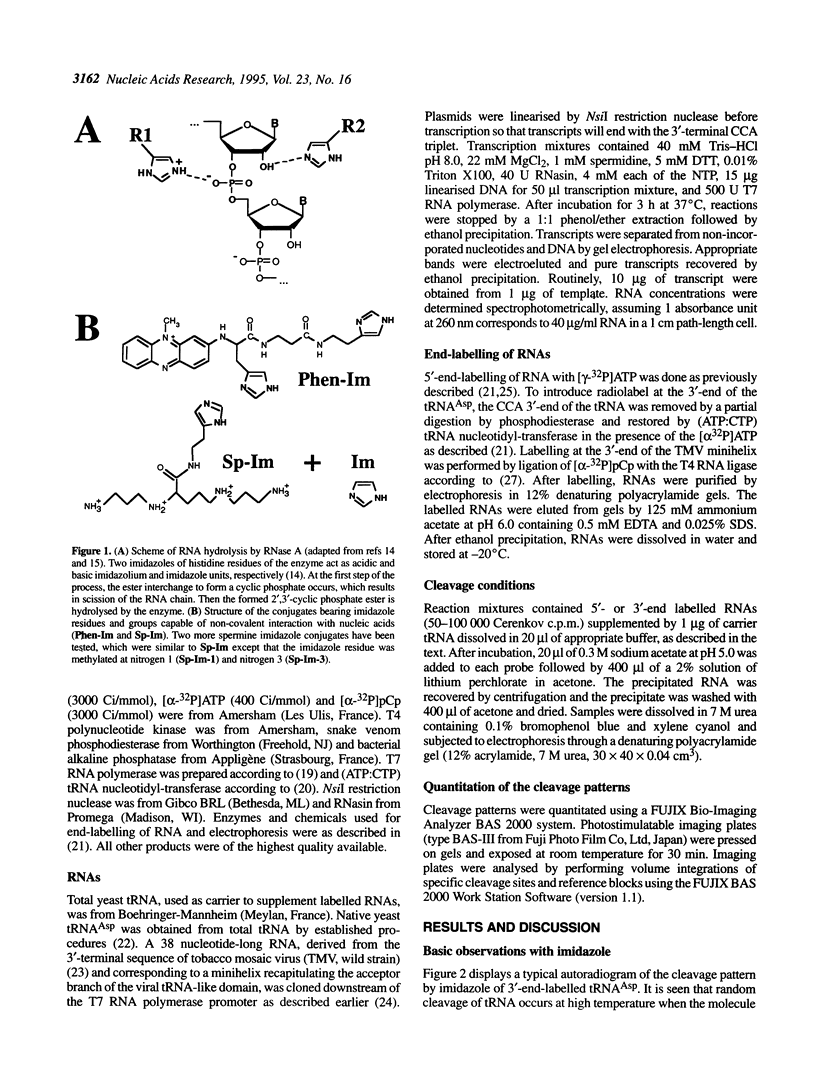
Hydrolysis of RNA in imidazole buffer and by spermine-imidazole conjugates has been investigated. The RNA models were yeast tRNA(Asp) and a transcript derived from the 3'-terminal sequence of tobacco mosaic virus RNA representing a minihelix capable of being enzymatically aminoacylated with histidine. Imidazole buffer and spermine-imidazole conjugates in the presence of free imidazole cleave phosphodiester bonds in the folded RNAs in a specific fashion. Imidazole buffer induces cleavages preferentially in single-stranded regions because nucleotides in these regions have more conformational freedom and can assume more easily the geometry needed for formation of the hydrolysis intermediate state. Spermine-imidazole constructs supplemented with free imidazole cleave tRNA(Asp) within single-stranded regions after pyrimidine residues with a marked preference for pyrimidine-A sequences. Hydrolysis patterns suggest a cleavage mechanism involving an attack by the imidazole residue of the electrostatically bound spermine-imidazole and by free imidazole at the most accessible single-stranded regions of the RNA. Cleavages in a viral RNA fragment recapitulating a tRNA-like domain were found in agreement with the model of this molecule that accounts for its functional properties, thus illustrating the potential of the imidazole-derived reagents as structural probes for solution mapping of RNAs. The cleavage reactions are simple to perform, provide information reflecting the state of the ribose-phosphate backbone of RNA and can be used for mapping single- and double-stranded regions in RNAs.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Breslow R., Huang D. L., Anslyn E. On the mechanism of action of ribonucleases: dinucleotide cleavage catalyzed by imidazole and Zn2+. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1746–1750. doi: 10.1073/pnas.86.6.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. S., Barton J. K. Transition metal complexes as probes of nucleic acids. Methods Enzymol. 1992;212:219–242. doi: 10.1016/0076-6879(92)12014-h. [DOI] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Moras D. Conformational changes and dynamics of tRNAs: evidence from hydrolysis patterns. Cold Spring Harb Symp Quant Biol. 1987;52:113–121. doi: 10.1101/sqb.1987.052.01.016. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry. 1978 May 30;17(11):2069–2076. doi: 10.1021/bi00604a008. [DOI] [PubMed] [Google Scholar]
- Felden B., Florentz C., McPherson A., Giegé R. A histidine accepting tRNA-like fold at the 3'-end of satellite tobacco mosaic virus RNA. Nucleic Acids Res. 1994 Aug 11;22(15):2882–2886. doi: 10.1093/nar/22.15.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentz C., Briand J. P., Romby P., Hirth L., Ebel J. P., Glegé R. The tRNA-like structure of turnip yellow mosaic virus RNA: structural organization of the last 159 nucleotides from the 3' OH terminus. EMBO J. 1982;1(2):269–276. doi: 10.1002/j.1460-2075.1982.tb01158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff J., Keith G., Ebel J. P., Dirheimer G. Structure of aspartate-tRNA from brewer's yeast. Nat New Biol. 1971 Mar 24;230(12):125–126. doi: 10.1038/newbio230125a0. [DOI] [PubMed] [Google Scholar]
- Garcia A., Giegé R., Behr J. P. New photoactivatable structural and affinity probes of RNAs: specific features and applications for mapping of spermine binding sites in yeast tRNA(Asp) and interaction of this tRNA with yeast aspartyl-tRNA synthetase. Nucleic Acids Res. 1990 Jan 11;18(1):89–95. doi: 10.1093/nar/18.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilley H., Jonard G., Kukla B., Richards K. E. Sequence of 1000 nucleotides at the 3' end of tobacco mosaic virus RNA. Nucleic Acids Res. 1979 Apr;6(4):1287–1308. doi: 10.1093/nar/6.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Hüsken D., Pieles U., Moser H. E., Häner R. Efficient sequence-specific cleavage of RNA using novel europium complexes conjugated to oligonucleotides. Chem Biol. 1994 Nov;1(3):185–190. doi: 10.1016/1074-5521(94)90008-6. [DOI] [PubMed] [Google Scholar]
- Hosaka H., Sakabe I., Sakamoto K., Yokoyama S., Takaku H. Sequence-specific cleavage of oligoribonucleotide capable of forming a stem and loop structure. J Biol Chem. 1994 Aug 5;269(31):20090–20094. [PubMed] [Google Scholar]
- Huber P. W. Chemical nucleases: their use in studying RNA structure and RNA-protein interactions. FASEB J. 1993 Nov;7(14):1367–1375. doi: 10.1096/fasebj.7.14.7693534. [DOI] [PubMed] [Google Scholar]
- Kierzek R. Hydrolysis of oligoribonucleotides: influence of sequence and length. Nucleic Acids Res. 1992 Oct 11;20(19):5073–5077. doi: 10.1093/nar/20.19.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek R. Nonenzymatic hydrolysis of oligoribonucleotides. Nucleic Acids Res. 1992 Oct 11;20(19):5079–5084. doi: 10.1093/nar/20.19.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama M., Inokawa T. Selective hydrolysis of tRNA by ethylenediamine bound to a DNA oligomer. J Biochem. 1994 Oct;116(4):719–720. doi: 10.1093/oxfordjournals.jbchem.a124586. [DOI] [PubMed] [Google Scholar]
- Krzyzosiak W. J., Marciniec T., Wiewiorowski M., Romby P., Ebel J. P., Giegé R. Characterization of the lead(II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNAPhe. Biochemistry. 1988 Jul 26;27(15):5771–5777. doi: 10.1021/bi00415a056. [DOI] [PubMed] [Google Scholar]
- Pan T., Dichtl B., Uhlenbeck O. C. Properties of an in vitro selected Pb2+ cleavage motif. Biochemistry. 1994 Aug 16;33(32):9561–9565. doi: 10.1021/bi00198a023. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia A., Puglisi J., Grosjean H., Ebel J. P., Florentz C., Giegé R. Conformation in solution of yeast tRNA(Asp) transcripts deprived of modified nucleotides. Biochimie. 1990 Oct;72(10):735–743. doi: 10.1016/0300-9084(90)90158-d. [DOI] [PubMed] [Google Scholar]
- Podyminogin M. A., Vlassov V. V., Giegé R. Synthetic RNA-cleaving molecules mimicking ribonuclease A active center. Design and cleavage of tRNA transcripts. Nucleic Acids Res. 1993 Dec 25;21(25):5950–5956. doi: 10.1093/nar/21.25.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rether B., Bonnet J., Ebel J. P. Studies on tRNA nucleotidyltransferase from baker's yeast. 1. Purification of the enzyme. Protection against thermal inactivation and inhibition by several substrates. Eur J Biochem. 1974 Dec 16;50(1):281–288. doi: 10.1111/j.1432-1033.1974.tb03896.x. [DOI] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Romby P., Moras D., Bergdoll M., Dumas P., Vlassov V. V., Westhof E., Ebel J. P., Giegé R. Yeast tRNAAsp tertiary structure in solution and areas of interaction of the tRNA with aspartyl-tRNA synthetase. A comparative study of the yeast phenylalanine system by phosphate alkylation experiments with ethylnitrosourea. J Mol Biol. 1985 Aug 5;184(3):455–471. doi: 10.1016/0022-2836(85)90294-3. [DOI] [PubMed] [Google Scholar]
- Romby P., Moras D., Dumas P., Ebel J. P., Giegé R. Comparison of the tertiary structure of yeast tRNA(Asp) and tRNA(Phe) in solution. Chemical modification study of the bases. J Mol Biol. 1987 May 5;195(1):193–204. doi: 10.1016/0022-2836(87)90336-6. [DOI] [PubMed] [Google Scholar]
- Tung C. H., Wei Z., Leibowitz M. J., Stein S. Design of peptide-acridine mimics of ribonuclease activity. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7114–7118. doi: 10.1073/pnas.89.15.7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassov V. V., Giegé R., Ebel J. P. Tertiary structure of tRNAs in solution monitored by phosphodiester modification with ethylnitrosourea. Eur J Biochem. 1981 Sep;119(1):51–59. doi: 10.1111/j.1432-1033.1981.tb05575.x. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Loop stereochemistry and dynamics in transfer RNA. J Biomol Struct Dyn. 1983 Oct;1(2):337–355. doi: 10.1080/07391102.1983.10507446. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W., Zachau H. G. A specific chemical chain scission of tRNA at 7-methylguanosine. FEBS Lett. 1970 Dec;11(3):160–164. doi: 10.1016/0014-5793(70)80518-x. [DOI] [PubMed] [Google Scholar]
- Wyatt J. R., Chastain M., Puglisi J. D. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques. 1991 Dec;11(6):764–769. [PubMed] [Google Scholar]