Abstract
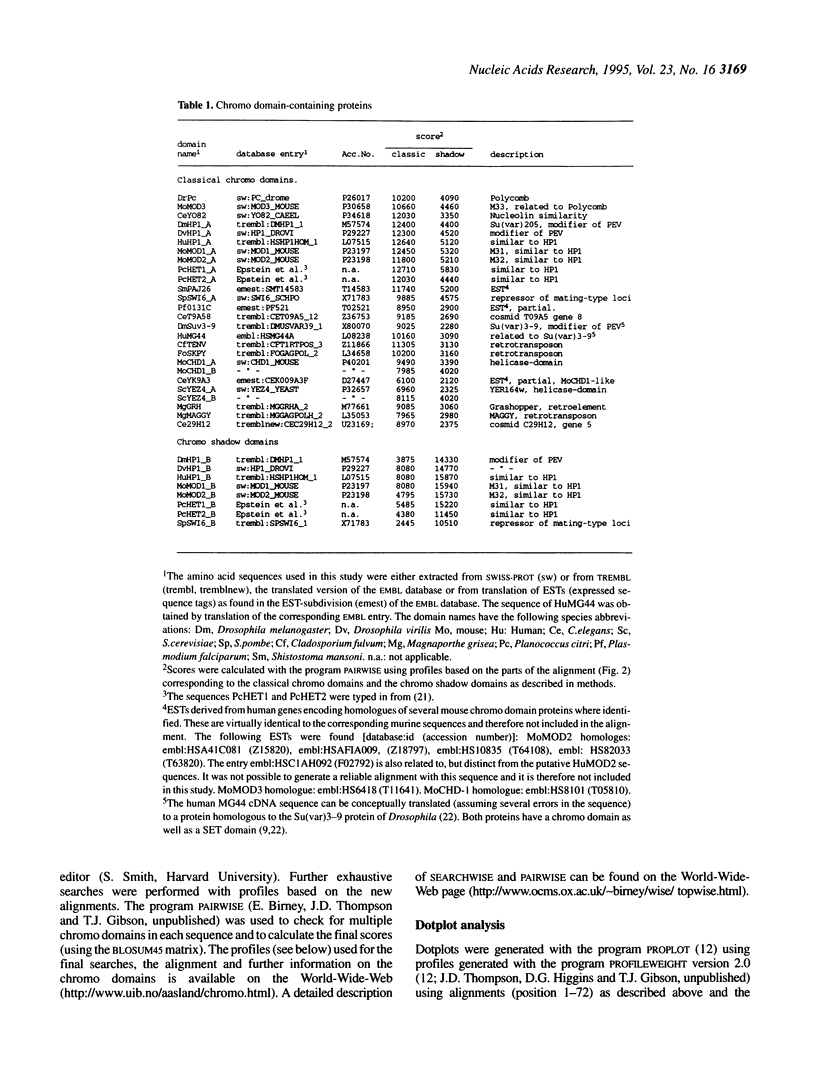
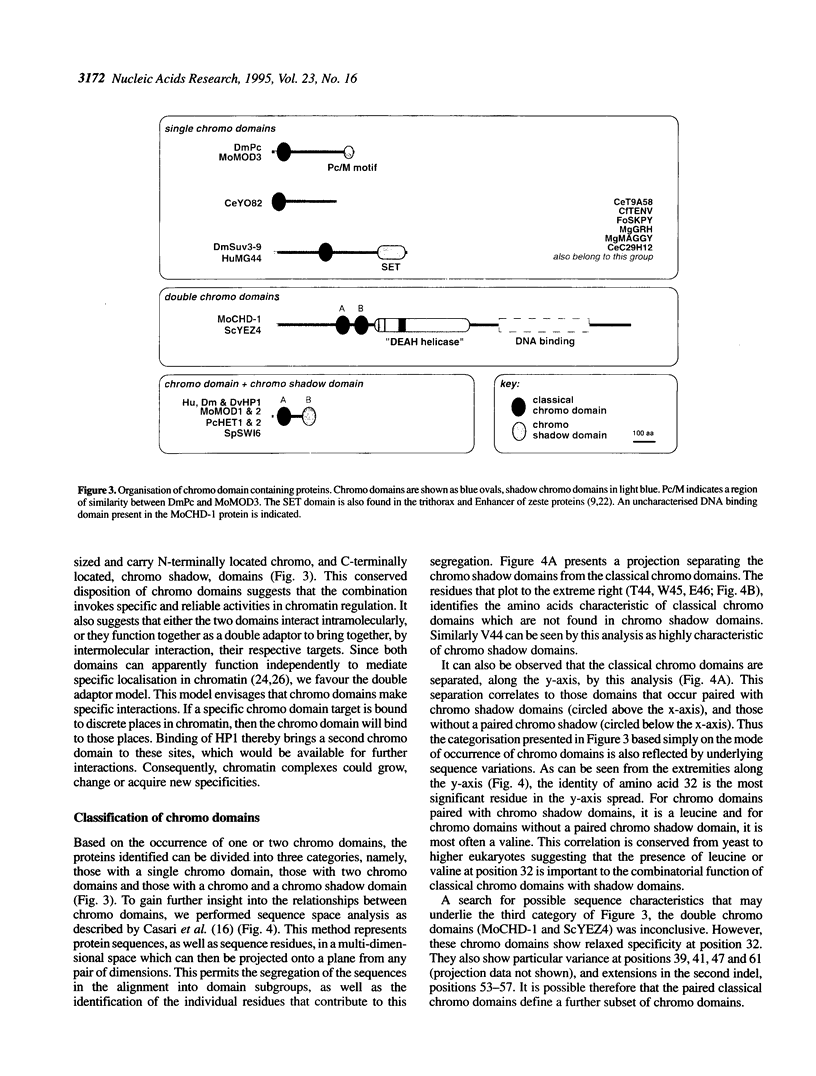
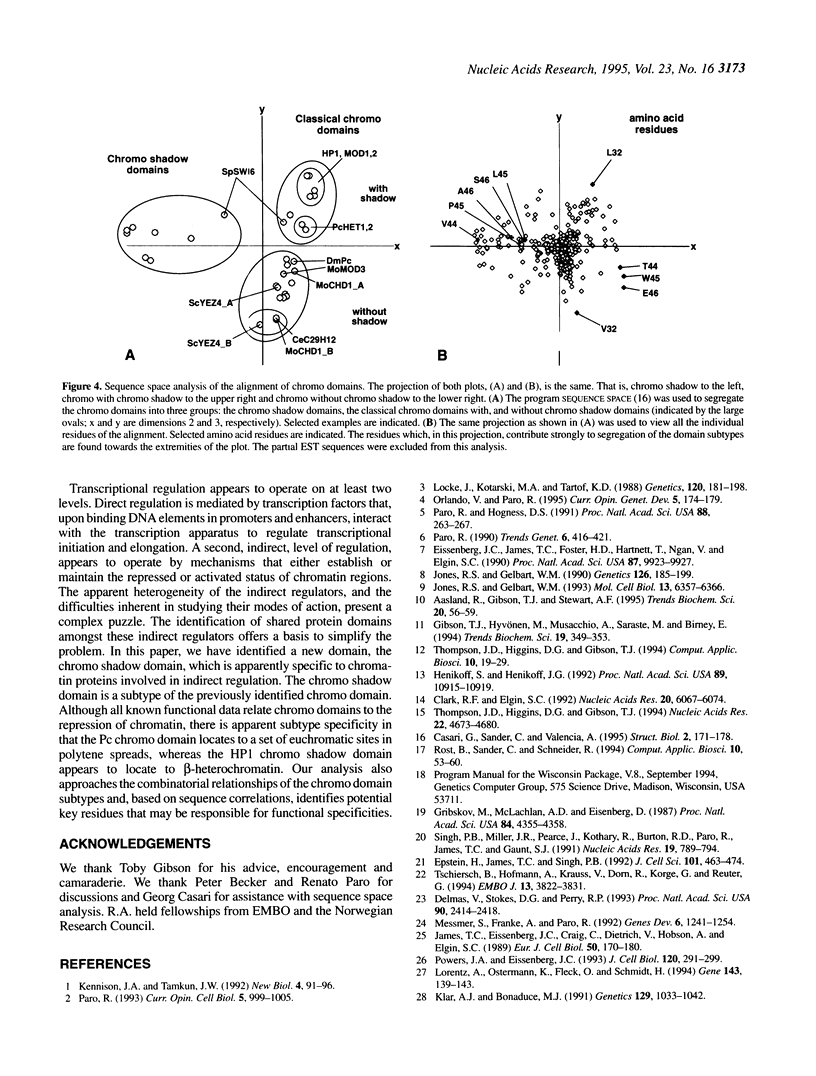
The chromo domain was originally identified as a protein sequence motif common to the Drosophila chromatin proteins, Polycomb (Pc) and heterochromatin protein 1 [HP1; Paro and Hogness (1991) Proc. Natl. Acad. Sci. USA, 88, 263-267; Paro (1990) Trends Genet., 6, 416-421]. Here we describe a second chromo domain-like motif in HP1. Subsequent refined searches identified further examples of this chromo domain variant which all occur in proteins that also have an N-terminally located chromo domain. Due to its relatedness to the chromo domain, and its occurrence in proteins that also have a classical chromo domain, we call the variant the 'chromo shadow domain'. Chromo domain-containing proteins can therefore be divided into two classes depending on the presence, for example in HP1, or absence, for example in Pc, of the chromo shadow domain. We have also found examples of proteins which have two classical chromo domains. The Schizosaccharomyces pombe SWI6 protein, involved in repression of the silent mating-type loci, is a member of the chromo shadow group. The similar modular architecture of SpSW16, HP1 and HP1-like proteins supports the model that the specificity of action of chromatin proteins is generated by combinations of protein modules.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aasland R., Gibson T. J., Stewart A. F. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995 Feb;20(2):56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Casari G., Sander C., Valencia A. A method to predict functional residues in proteins. Nat Struct Biol. 1995 Feb;2(2):171–178. doi: 10.1038/nsb0295-171. [DOI] [PubMed] [Google Scholar]
- Clark R. F., Elgin S. C. Heterochromatin protein 1, a known suppressor of position-effect variegation, is highly conserved in Drosophila. Nucleic Acids Res. 1992 Nov 25;20(22):6067–6074. doi: 10.1093/nar/20.22.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V., Stokes D. G., Perry R. P. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., James T. C., Foster-Hartnett D. M., Hartnett T., Ngan V., Elgin S. C. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein H., James T. C., Singh P. B. Cloning and expression of Drosophila HP1 homologs from a mealybug, Planococcus citri. J Cell Sci. 1992 Feb;101(Pt 2):463–474. doi: 10.1242/jcs.101.2.463. [DOI] [PubMed] [Google Scholar]
- Gibson T. J., Hyvönen M., Musacchio A., Saraste M., Birney E. PH domain: the first anniversary. Trends Biochem Sci. 1994 Sep;19(9):349–353. doi: 10.1016/0968-0004(94)90108-2. [DOI] [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T. C., Eissenberg J. C., Craig C., Dietrich V., Hobson A., Elgin S. C. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol. 1989 Oct;50(1):170–180. [PubMed] [Google Scholar]
- Jones R. S., Gelbart W. M. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics. 1990 Sep;126(1):185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. S., Gelbart W. M. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol Cell Biol. 1993 Oct;13(10):6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J. A., Tamkun J. W. Trans-regulation of homeotic genes in Drosophila. New Biol. 1992 Feb;4(2):91–96. [PubMed] [Google Scholar]
- Klar A. J., Bonaduce M. J. swi6, a gene required for mating-type switching, prohibits meiotic recombination in the mat2-mat3 "cold spot" of fission yeast. Genetics. 1991 Dec;129(4):1033–1042. doi: 10.1093/genetics/129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J., Kotarski M. A., Tartof K. D. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics. 1988 Sep;120(1):181–198. doi: 10.1093/genetics/120.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentz A., Ostermann K., Fleck O., Schmidt H. Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene. 1994 May 27;143(1):139–143. doi: 10.1016/0378-1119(94)90619-x. [DOI] [PubMed] [Google Scholar]
- Messmer S., Franke A., Paro R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992 Jul;6(7):1241–1254. doi: 10.1101/gad.6.7.1241. [DOI] [PubMed] [Google Scholar]
- Orlando V., Paro R. Chromatin multiprotein complexes involved in the maintenance of transcription patterns. Curr Opin Genet Dev. 1995 Apr;5(2):174–179. doi: 10.1016/0959-437x(95)80005-0. [DOI] [PubMed] [Google Scholar]
- Paro R., Hogness D. S. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990 Dec;6(12):416–421. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- Paro R. Mechanisms of heritable gene repression during development of Drosophila. Curr Opin Cell Biol. 1993 Dec;5(6):999–1005. doi: 10.1016/0955-0674(93)90084-4. [DOI] [PubMed] [Google Scholar]
- Powers J. A., Eissenberg J. C. Overlapping domains of the heterochromatin-associated protein HP1 mediate nuclear localization and heterochromatin binding. J Cell Biol. 1993 Jan;120(2):291–299. doi: 10.1083/jcb.120.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Sander C., Schneider R. PHD--an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994 Feb;10(1):53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- Singh P. B., Miller J. R., Pearce J., Kothary R., Burton R. D., Paro R., James T. C., Gaunt S. J. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 1991 Feb 25;19(4):789–794. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994 Nov 11;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. Improved sensitivity of profile searches through the use of sequence weights and gap excision. Comput Appl Biosci. 1994 Feb;10(1):19–29. doi: 10.1093/bioinformatics/10.1.19. [DOI] [PubMed] [Google Scholar]
- Tschiersch B., Hofmann A., Krauss V., Dorn R., Korge G., Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994 Aug 15;13(16):3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]




