Abstract
A network of Hox factors specifies the identity of motor neurons in the spinal cord. Studies of an essential Fox-class accessory factor illuminate the developmental and evolutionary subtlety of the process.
The vertebrate central nervous system (CNS) contains many classes of neurons that have specialized properties. Imagine the problem of creating this cellular diversity, in which neurons that look alike have in fact acquired fine-grained distinctions to ensure that the proper synaptic connections with other neurons are established and maintained. There are an estimated 100 trillion synapses in the brains of the most complex vertebrates, and neuro-biologists are finding that nature has devised a multitude of clever strategies to guide neuron development and connectivity.
Remarkable inroads into understanding these strategies have been made by examining the development of a relatively simple part of the CNS — the spinal cord1. Most notably, it has been discovered that the evolutionarily conserved Hox family of gene transcription factors operates in a sophisticated cross-regulatory network to specify individual subtypes of motor neurons that are preordained to connect with specific muscles and ganglia in the periphery of an animal2. Work published in Cell (Dasen et al.)3 and Neuron (Rousso et al.)4 peels away a further layer of the developmental onion. The two papers report that another transcription factor, called FoxP1, helps Hox proteins to regulate the genes that control motor-neuron diversification.
Motor neurons represent the final command stage in a chain of cells extending from the brain to the spinal cord. They trigger activity in the autonomic (sympathetic) nervous system, such as sweating under stressful conditions, and they send instructions to muscles in the form of the neurotransmitter molecule acetylcholine that cause the muscles to contract. Motor coordination is achieved by the precise spatial and temporal control of hundreds of muscles. Consequently, the complexity of the musculoskeletal system is mirrored by a similarly complex arrangement of motor neurons in the spinal cord.
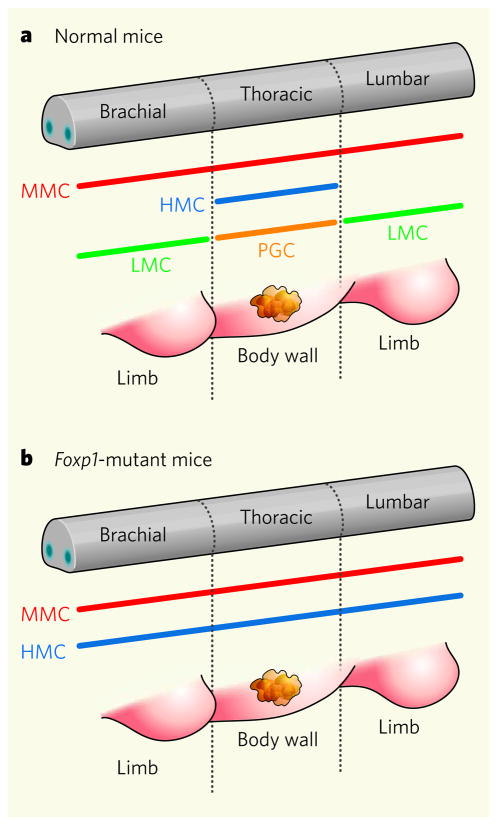
During embryo formation, motor neurons grow axons to their appropriate muscles in the periphery — the equivalent in IT terms of threading Internet cable to its destination. Among the motor-neuron subclasses examined by Dasen et al.3 and Rousso et al.4 were the median motor column (MMC) neurons that innervate axial muscles supporting posture; the hypaxial motor column (HMC) neurons that innervate body-wall muscles around the rib cage; the preganglionic motor column (PGC) neurons that innervate peripheral ganglia of the autonomic nervous system; and the lateral motor column (LMC) neurons that innervate muscles in the arms and legs. The motor neurons in the spinal cord and the muscles they control are typically in register with one another (Fig. 1a). LMC neurons occur only in the brachial and lumbar spinal cord, aligned with the arms and legs respectively (or wings/legs in birds and fins and fish), whereas HMC neurons are located at thoracic levels where the rib cage is present.
Figure 1. Neuron specification in the spinal cord of embryonic mice3,4.
a, In normal mice, Hox genes and Foxp1 interact to generate the correct subclass of motor neuron at the brachial, thoracic and lumbar levels of the spinal cord. The cell bodies of the neurons align into longitudinal columns called the LMC, which innervates limbs; MMC, which innervates axial muscles; HMC, which innervates body-wall muscles; and PGC, which innervates ganglia of the sympathetic nervous system. b, Mutations in Foxp1 disrupt Hox activity involved in specifying the LMC and PGC. The spinal cord of Foxp1 mutant mice defaults to generating motor neurons such as those of more primitive vertebrates, for example the lamprey. (Based on Fig. 1 of ref. 3.)
The spinal cord is derived from undifferentiated cells that comprise the embryo’s neural tube. The generation of specific subtypes of motor neuron is initiated by cell–cell interactions mediated by different secreted factors that in effect create an x, y, z coordinate system1,3. In general, these signals activate intracellular pathways that eventually culminate in the transcription or repression of genes needed for the specialized properties of each type of motor neuron. Multiple layers of transcription factors are then involved in establishing motor-neuron identity1,5, with the Hox family lying at the core of the circuitry. Hox genes have undergone several duplications in more highly evolved vertebrates, presumably to expand their repertoire of functions. In mice, for example, Hox6 proteins contribute to the specification of brachial LMC motor neurons, Hox9 specify PGC neurons, and Hox10 specify lumbar LMC neurons.
In some cases, however, Hox expression imprecisely correlates with motor-neuron subtype. Dasen et al.3 and Rousso et al.4 set out to understand why, starting with the knowledge that Hox proteins typically function with cofactors such as Meis, Pbx/Prep, Engrailed and Fox6,7. Meis and Pbx/Prep are not restricted to particular subtypes, and Engrailed is not expressed at all by motor neurons, so these were poor candidates for further study. In contrast, FoxP1 looked promising, being expressed at low levels in PGC motor neurons and at high levels in LMC cells.
Both groups3,4 found that mutation of the mouse Foxp1 gene does not interfere with motor-neuron generation as such, but that PGC and LMC cells fail to emerge and motor neurons seem instead to remain stuck in their ‘ground state’ and resemble HMC cells. From studies with the fruitfly Drosophila, we can infer that Hox proteins functionally interact with the Fox homologue called sloppy paired (Slp)6,7, which could modify their DNA-binding and/or transcriptional activity. Nevertheless, Hox proteins also seem to have FoxP1-independent functions3, and therefore individual genes in each motor-neuron class could contain either Hox-only or Hox–FoxP1 complexes, which as a composite might specify subtype identity.
Furthermore, the concentration of FoxP1 seems to be significant: misexpression of high levels favours LMC development, whereas low levels trigger PGC specification3. The mechanistic basis for this observation remains unknown, but one possibility is that ternary Hox complexes form with different amounts of FoxP1 and have unique activities. Understanding these details might help us to understand how FoxP1 also functions in non-neuronal contexts such as cardiac- and blood-cell development.
Mice lacking Foxp1 are not viable3,4 and have a strange type of spinal cord that in some ways resembles the spinal cord of more primitive animals (Fig. 1b). Ancient vertebrates such as lamprey and hagfish swim using axial and hypaxial muscles, and their spinal cords seem to have neurons of MMC and HMC character but not PGC and LMC cells8. The appearance of LMC and PGC neurons is linked to the formation of lateral fins and limbs and a sympathetic nervous system — features that emerged later in vertebrate evolution. Although the Hox and Fox genes can be traced back to invertebrates, evolutionary pressure evidently exploited them to expand the repertoire of functions mediated by the CNS. Comparisons of the promoters of Fox genes and of Fox protein structures from different animals should help us to understand how genes can be co-opted from one context to be used in another9.
References
- 1.Jessell TM. Nature Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 2.Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Dasen JS, De Camilli A, Wang B, Tucker PW, Jessell TM. Cell. 2008;134:304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 4.Rousso DL, Gaber ZB, Wellik D, Morrisey EE, Novitch BG. Neuron. 2008;59:226–240. doi: 10.1016/j.neuron.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shirasaki R, Pfaff SL. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- 6.Mann RS, Affolter M. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- 7.Moens CB, Selleri L. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 8.Fetcho JR. Brain Behav Evol. 1992;40:82–97. doi: 10.1159/000113905. [DOI] [PubMed] [Google Scholar]
- 9.True JR, Carroll SB. Annu Rev Cell Dev Biol. 2002;18:53–80. doi: 10.1146/annurev.cellbio.18.020402.140619. [DOI] [PubMed] [Google Scholar]