Abstract
Heparan sulfate proteoglycans (HSPGs) participate in a wide range of biological processes through interactions with a number of ligand proteins. The nature of these interactions largely depends on the heparan sulfate (HS) moiety of HSPGs, which undergoes a series of modifications by various HS-modifying enzymes (HSMEs). Although the effects of alterations in a single HSME on physiological processes have started to be studied, it remains elusive how a combination of these molecules control the structure and function of HS. Here we systematically manipulated the HS structures and analyzed their effect on morphogenesis and signaling, using the genetically tractable model organism, Drosophila. We generated transgenic fly strains overexpressing HSMEs alone or in combination. Unsaturated disaccharide analyses of HS showed that expression of various HSMEs generates distinct HS structures, and the enzymatic activities of HSMEs are influenced by coexpression of other HSMEs. Furthermore, these transgenic HSME animals showed a different extent of lethality, and a subset of HSMEs caused specific morphological defects due to defective activities of Wnt and bone morphogenetic protein signaling. There is no obvious relationship between HS unsaturated disaccharide composition and developmental defects in HSME animals, suggesting that other structural factors, such as domain organization or sulfation sequence, might regulate the function of HS.
Keywords: Decapentaplegic, Drosophila, heparan sulfate proteoglycan, heparan sulfate-modifying enzyme, Wingless
Introduction
Heparan sulfate proteoglycans (HSPGs), composed of a core protein and heparan sulfate (HS) chains, are major constituents of the extracellular matrix and cell surface. HSPGs regulate a wide spectrum of developmental and physiological events by regulating the activities of various proteins, such as growth factors, cell adhesion molecules, proteases and lipoproteins. These interactions largely, but not entirely, depend on the HS moiety of HSPGs, which has highly heterogeneous structures resulting from complex, multistep modification processes in the Golgi network (Esko and Selleck 2002; Nakato and Kimata 2002; Kirkpatrick and Selleck 2007). Biosynthesis of HS begins from the formation of a tetrasaccharide linkage attached to the serine residues of the core protein. Subsequently, EXT proteins, which encode HS copolymerases, add repeating disaccharides composed of N-acetylglucosamine (GlcNAc) and glucuronic acid (GlcA) to polymerize the HS chain. As the chain is extending, N-deacetylase/N-sulfotransferase (NDST) removes the acetyl groups from some of the GlcNAc residues and replaces them with sulfate groups. After N-sulfation, heparan sulfate C5-epimerase (Hsepi) converts GlcA to iduronic acid (IdoA), and 2-O, 6-O and 3-O sulfotransferases (Hs2st, Hs6st and Hs3st, respectively) add sulfate groups on specific ring positions of the HS chain. The epimerization and sulfation by these HS modification enzymes (HSMEs) contribute to the structural complexity of HS, which is thought to allow the selective binding to a variety of ligand proteins (Esko and Selleck 2002; Nakato and Kimata 2002). It has recently been proposed that some HSMEs are assembled into a physical complex called a “gagosome”, and the composition of the gagosome affects the structure of HS (Esko and Selleck 2002). Supporting this concept, previous studies have identified a physical association between Hsepi and Hs2st (Pinhal et al. 2001) and between NDST1 and EXT2 (Presto et al. 2008).
Mutations in HSME genes induce specific developmental defects by interfering with growth factor signaling (Bullock et al. 1998; Li et al. 2003; Habuchi et al. 2007). Although these studies unambiguously highlighted the roles of HSME genes during development, it remains elusive which structural alterations of HS contributed to the observed defects. Further complexity comes from HS sulfation compensation. Previous studies have shown that the loss of particular HSME genes induces a compensatory increase in sulfation at other position on HS (Merry et al. 2001). In Drosophila, an increase of 6-O sulfation can compensate for losses of 2-O sulfation, and vice versa, thus maintaining growth factor signaling essential for normal development in both Hs2st and Hs6st mutants (Kamimura et al. 2006). Similar compensation of HS sulfation has also been observed in Hs2st and Hs6st mutant mice (Merry et al. 2001; Sugaya et al. 2008). Although the mechanism for sulfation compensation is unknown, it suggests the existence of a complex regulatory network that controls the activity of the HS modification machinery.
The Drosophila model provides an excellent system to study the mechanisms of HS modification and its biological significance. First, Drosophila has a complete set of HSMEs found in mammalian species. Using these molecules, Drosophila produces complex HS structures that are equivalent to mammalian HS (Toyoda et al. 2000). Second, Drosophila has only one gene for most of the HSMEs. Therefore, there is no genetic redundancy, which could hamper the genetic analyses of these molecules in mammalian systems. Furthermore, in this model organism, signaling pathways and gene regulatory networks controlling patterning and morphogenesis have been extensively characterized. This enables us to identify the molecular foundation underlying the developmental processes controlled by HS. Finally, a number of genetic tools, including mutations, RNA interference knock-down transgenic animals and gain-of-function strains bearing overexpression constructs for almost all the known genes of the HS-modifying machinery are available (Nakato et al. 1995; Tsuda et al. 1999; Kirkpatrick et al. 2004; Kreuger et al. 2004; Takeo et al. 2005; Kleinschmit et al. 2010). With sophisticated molecular genetic technologies available in this model system, we are able to manipulate HS structures in vivo.
In this study, to elucidate the effects of HSMEs on HS structures, morphogenesis and growth factor signaling, we performed a comprehensive in vivo gain-of-function analysis of HSMEs using Drosophila. We generated HSME transgenic flies, which allow the overexpression of a single HSME or a combination of multiple HSMEs. Disaccharide analyses of HS isolated from HSME-overexpressing animals showed that expression of different HSMEs generates distinct HS structures. We also found that the activities of HSMEs are affected by coexpression of other HSMEs. Furthermore, expression of HSMEs induced different levels of lethality, and a subset of HSMEs caused specific defects in the adult wing structures as well as in the signaling activities of Wingless (Wg, a Drosophila member of the Wnt family) and Decapentaplegic (Dpp, a member of the bone morphogenetic protein family). We found no obvious relationship between HS unsaturated disaccharide compositions and developmental abnormalities in HSME animals, suggesting that other structural factors, such as domain organization or sulfation sequence, may regulate HS function.
Results
Structural changes of HS induced by overexpression of HSME genes
To manipulate HS structures in vivo, we generated transgenic flies bearing constructs to express four HS-modifying enzymes (HSMEs): sulfateless (sfl) which encodes NDST, C5-epimerase (Hsepi), 2-O sulfotransferase (Hs2st) and 6-O sulfotransferase (Hs6st). Each gene was inserted downstream of the Gal4-responsive elements, upstream activating sequences (UAS) (Brand and Perrimon 1993). The activities of some HSMEs are dependent on the reactions of other enzymes. For example, the HS modifications by Hsepi, Hs2st and Hs6st mostly occur on the specific HS structures, which are formerly catalyzed by NDST and/or Hsepi (Esko and Selleck 2002). Therefore, in order to promote the epimerization and O-sulfation events on HS by these enzymes, we also produced five transgenic flies simultaneously bearing two HSME transgenes (sfl–Hsepi, sfl–Hs2st, sfl–Hs6st, Hsepi–Hs2st and Hsepi–Hs6st). By crossing these nine HSME transgenic strains to Gal4 drivers, we combinatorially overexpressed the HSME genes under temporal and spatial control and examined their effects on HS structure and developmental events. Since we focused on the effects of overexpression of HSMEs in this study, HSME-expressing animals are simply referred to “HSME animals” in this paper.
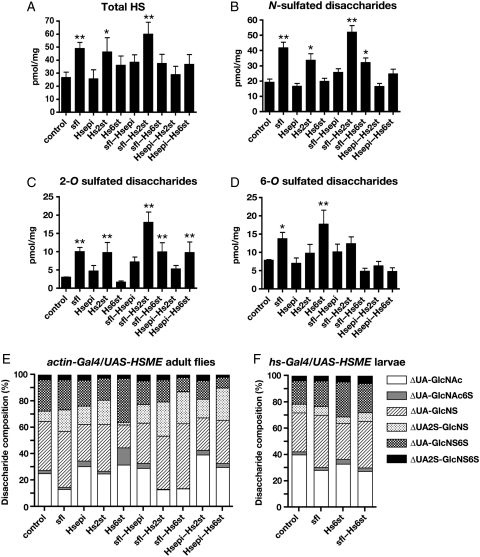
We first examined whether expression of HSME genes results in structural alteration of HS. The HSME genes were induced by a ubiquitous actin-Gal4 driver and the unsaturated disaccharide profiles of HS extracted from adult flies were determined (Kinoshita and Sugahara 1999). Notably, the HSME manipulations not only affected the HS unsaturated disaccharide composition, but also led to an increase in the total level of HS (Figure 1A and Table I). A significant increase in HS was observed in sfl and Hs2st animals, and this effect was enhanced by coexpression of these two genes. Although the mechanisms for this increase in HS levels are unknown, one explanation is that sfl and Hs2st affect the activities of Drosophila EXTs to enhance the HS polymerizing reaction. Alternatively, the HS structures modified by these HSMEs may influence the stability and metabolism of HSPGs.
Fig. 1.
Structural analyses of HS from HSME-expressing animals. Structural analyses of HS isolated from actin-Gal4/UAS-HSME adult flies (A–E). (A) The HS levels were increased in sfl, Hs2st and sfl–Hs2st animals. The levels of N- (B), 2-O (C) and 6-O (D) sulfated disaccharide units for each genotype. The values (pmol/mg dry whole adult flies) for all graphs represent mean and standard deviations based on three independent experiments. Disaccharide composition of HS from actin-Gal4/UAS-HSME adult flies (E) and hs-Gal4/UAS-HSME larvae (F). The values are given as mol% of total disaccharides. ΔUA, unsaturated uronic acid; ΔUA2S, 2-O sulfated unsaturated uronic acid; GlcNAc, N-acetylglucosamine; GlcNAc6S, 6-O sulfated N-acetylglucosamine; GlcNS, N-sulfated glucosamine; GlcNS6S, N- and 6-O sulfated glucosamine. *Statistically different from control (actin-Gal4/UAS-GFP). *P < 0.05; **P < 0.01.
Table I.
Disaccharide analyses of HS from actin-Gal4/UAS-HSME adult flies
| ΔUA-GlcNAc | ΔUA-GlcNAc6S | ΔUA-GlcNS | ΔUA2S-GlcNS | ΔUA-GlcNS6S | ΔUA2S-GlcNS6S | Total | Sulfate/dimer | |
|---|---|---|---|---|---|---|---|---|
| Control | 6.8 ± 2.0 (25.1) | 0.64 ± 0.08 (2.4) | 10.0 ± 2.4 (37.1) | 2.0 ± 0.1 (7.8) | 6.2 ± 0.3 (23.9) | 0.97 ± 0.06 (3.7) | 26.6 ± 4.2 | 1.13 ± 0.11 |
| sfl | 6.4 ± 0.7 (13.1) | 0.81 ± 0.44 (1.6) | **20.8 ± 1.9 (42.4) | **8.1 ± 1.0 (16.5) | **11.0 ± 1.7 (22.4) | **1.93 ± 0.17 (3.9) | **49.0 ± 4.6 | **1.34 ± 0.03 |
| Hsepi | 7.9 ± 3.3 (30.4) | 1.06 ± 0.37 (4.1) | 7.0 ± 1.3 (27.8) | 3.6 ± 1.1 (14.1) | 4.9 ± 0.7 (19.5) | 1.08 ± 0.47 (4.1) | 25.5 ± 7.1 | 1.11 ± 0.07 |
| Hs2st | 11.6 ± 3.5 (24.9) | 0.94 ± 0.15 (2.1) | *16.2 ± 2.9 (35.4) | **8.6 ± 2.4 (18.5) | 7.7 ± 2.0 (16.6) | 1.17 ± 0.31 (2.5) | *46.2 ± 11.1 | 1.15 ± 0.03 |
| Hs6st | 11.4 ± 2.1 (31.6) | **4.79 ± 1.65 (13.0) | 6.3 ± 1.0 (17.4) | 0.7 ± 0.2 (2.0) | **12.0 ± 2.0 (33.3) | 0.95 ± 0.15 (2.7) | 36.1 ± 7.1 | 1.09 ± 0.01 |
| sfl–Hsepi | 11.1 ± 1.4 (29.1) | 1.49 ± 0.49 (3.8) | 11.6 ± 1.7 (30.4) | 5.4 ± 1.0 (14.1) | 6.9 ± 1.3 (17.9) | *1.78 ± 0.44 (4.6) | 38.3 ± 5.9 | 1.12 ± 0.05 |
| sfl–Hs2st | 7.8 ± 1.6 (12.9) | 0.20 ± 0.01 (0.3) | **24.1 ± 3.3 (40.3) | **15.7 ± 2.5 (26.2) | *9.9 ± 1.5 (16.4) | **2.30 ± 0.37 (3.9) | **60.0 ± 9.1 | **1.37 ± 0.01 |
| sfl–Hs6st | 5.2 ± 1.9 (13,6) | 0.06 ± 0.04 (0.2) | **18.3 ± 2.3 (49.2) | **9.2 ± 2.3 (24.3) | 3.9 ± 0.7 (10.6) | 0.80 ± 0.17 (2.1) | 37.4 ± 7.1 | 1.26 ± 0.03 |
| Hsepi–Hs2st | 11.4 ± 3.1 (39.2) | 1.05 ± 0.25 (3.6) | 7.1 ± 1.8 (24.5) | 4.1 ± 0.9 (14.3) | 4.0 ± 0.9 (13.9) | 1.23 ± 0.14 (4.3) | 28.9 ± 6.5 | *0.98 ± 0.06 |
| Hsepi–Hs6st | 11.0 ± 3.2 (29.8) | 1.13 ± 0.42 (3.0) | 12.0 ± 2.1 (32.7) | **9.1 ± 2.7 (24.5) | *2.9 ± 0.5 (8.1) | 0.70 ± 0.19 (1.9) | 36.8 ± 7.5 | 1.07 ± 0.10 |
Values represent mean ± standard deviations (n = 3) of the level of each disaccharide unit and total HS (pmol/mg dry whole adult flies), and the number of sulfate groups per disaccharide. The values in parentheses are given as mol% of total disaccharides. Statistically different from control (*P < 0.05; **P < 0.01).
The unsaturated disaccharide composition of HS in HSME animals revealed that expression of distinct HSMEs differentially affects the structure of HS (Figure 1B–E, and Table I). We first analyzed the disaccharide compositions from animals expressing a single HSME. Consistent with the enzymatic activity of each HSME, expression of these enzymes significantly increased the number of corresponding sulfate groups: sfl increased N-sulfation, and Hs2st and Hs6st increased the 2-O and 6-O sulfate groups, respectively, confirming that our overexpression system is functionally effective. In contrast, little change in the sulfation level was observed in Hsepi-expressing animals. Interestingly, expression of sfl also increased 2-O and 6-O sulfate groups, and expression of Hs2st increased N- but not 6-O sulfate groups. These results suggested that expression of these HSMEs affects the activities of other HSMEs.
We next analyzed the structure of HS from animals expressing two HSMEs. Unexpectedly, these analyses showed that expression of particular HSMEs induced facilitatory or inhibitory effects on the HS structures modified by other HSMEs expressed simultaneously. The animals overexpressing both sfl and Hs2st (sfl–Hs2st animals) revealed a substantial increase in N- and 2-O sulfate groups compared with the animals singly overexpressing sfl or Hs2st, showing cooperative effects of the enzymatic activities of Sfl and Hs2st (Figure 1B and C). On the other hand, coexpression of other HSMEs did not generate HS structures reflecting each HSME activity. In particular, in sfl–Hs6st animals, the level of 6-O sulfated groups was not increased, despite the high levels of 6-O sulfation observed in sfl and Hs6st animals (Figure 1D). Similarly, sfl–Hsepi and Hsepi–Hs6st did not increase levels of N- and 6-O sulfation, respectively (Figure 1B–D). We also noticed that Hsepi–Hs6st increased the level of 2-O sulfate groups (Figure 1C). Although the precise molecular mechanism of these positive and negative effects is unknown, it is possible that overexpression of HSMEs may affect the gagosome composition, resulting in a change in the balance of each reaction. For the inhibitory effects, overexpression of specific sets of HSMEs may compete for binding to substrates, such as adenosine 3′-phosphate 5′-phosphosulfate and HS with specific fine structures.
As we discuss later, overexpression of HSME genes by actin-Gal4 affects the viability of adult flies. If overexpression of HSMEs causes cell death of a specific type of cells, it is possible that the changes in the structure of the HS isolated from whole animals could reflect changes in the proportions of HS derived from different cell sources. To address this point, we performed two experiments using sfl, Hs6st and sfl–Hs6st animals. First, we examined whether HSME overexpression by actin-Gal4 induced apoptosis. The staining of multiple tissues (wing disc, eye-antennal disc and central nervous system) with acridine orange showed no increase in cell death by HSME overexpression in survived animals (Supplementary data, Figure S1). Second, we expressed sfl, Hs6st or sfl–Hs6st transiently at lower levels using a heat shock-inducible GAL4 driver (hs-Gal4). The transgenic animals were cultured at 18°C until HSME expression was induced by a heat shock at 37°C for 30 min during the third-instar larval stage. In these conditions, no lethality or morphological abnormalities were induced (data not shown). HS was prepared at 4 h after the heat shock and subjected to disaccharide analysis. We observed that although less significant, HS structure shows a similar tendency to the results obtained from actin-Gal4 overexpression: In sfl and Hs6st animals, N- and 6-O sulfate groups were increased, respectively (Figure 1F and Table II). In addition, 6-O sulfation was not increased in sfl–Hs6st compared with that in Hs6st animals, but instead appears to be decreased (at a statistically insignificant level, P = 0.2). These results showed that HS structural changes observed in the HSME-overexpressing animals were mainly due to the altered levels of enzymatic activities during HS biosynthesis, not secondary effects such as changes in the cell populations from which HS is derived.
Table II.
Disaccharide composition of HS from hs-Gal4/UAS-HSME larvae
| ΔUA-GlcNAc | ΔUA-GlcNAc6S | ΔUA-GlcNS | ΔUA2S-GlcNS | ΔUA-GlcNS6S | ΔUA2S-GlcNS6S | |
|---|---|---|---|---|---|---|
| Control | 39.9 ± 4.0 | 2.3 ± 1.2 | 29.6 ± 1.5 | 6.6 ± 0.5 | 17.8 ± 2.4 | 3.7 ± 0.8 |
| sfl | 28.2 ± 4.3* | 2.1 ± 1.2 | 39.7 ± 3.1** | 6.6 ± 0.7 | 19.4 ± 1.6 | 3.9 ± 0.7 |
| Hs6st | 32.9 ± 4.1 | 3.7 ± 1.4 | 27.3 ± 2.5 | 5.1 ± 0.7* | 26.4 ± 3.4* | 4.6 ± 1.0 |
| sfl–Hs6st | 27.3 ± 4.7* | 2.7 ± 1.2 | 35.4 ± 3.2* | 6.8 ± 0.4 | 22.2 ± 3.0 | 5.7 ± 1.7 |
Values represent mean ± standard deviations (n = 3) of mol% of total disaccharides. Statistically different from control (*P < 0.05; **P < 0.01).
Thus, we established a model system in which we can manipulate HS structures in vivo in a systematic manner. This system provided evidence for the positive and negative functional interactions between HSMEs. It also enables us to assess the effects of structural changes of HS on morphogenesis and signaling.
Expression of HSMEs affects the viability of adult flies
The disaccharide analyses of HS from HSME animals showed that expression of distinct HSMEs induces different effects on the HS structures. A number of studies showed that loss of particular sulfate groups caused various morphological defects (Gorsi and Stringer 2007). On the other hand, recent studies showed that interactions between HS and proteins depend primarily on charge density rather than the precise positioning of various sulfate groups (Kreuger et al. 2006). To clarify the relationship between HS structures and function, we investigated which HSMEs induce developmental defects when overexpressed.
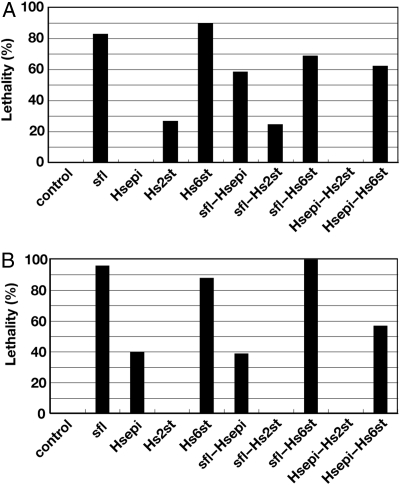
As the first step to elucidate the structural factors determining HS function, we examined the viability of HSME adult flies. To induce expression of HSME genes, we used actin-Gal4, which directs whole body gene expression, and hedgehog (hh)-Gal4, which directs expression in the posterior compartment of many epidermal tissues. Expression of the HSME genes by actin-Gal4 induced lethality at different levels (Figure 2A). The most prominent lethality was observed in flies expressing sfl, Hs6st and sfl–Hs6st. Expression of sfl–Hsepi and Hsepi–Hs6st also reduced their viability; however, no lethality was observed in Hsepi and Hsepi–Hs2st animals. Coexpression of Hsepi or Hs2st with sfl ameliorates the effects of sfl expression on lethality. The lethality of Hs6st animals was also decreased by coexpression of Hsepi.
Fig. 2.
Adult lethality caused by expression of HSME genes. (A) Lethality of actin-Gal4/UAS-HSME animals. The lethality and the number of scored animals for each genotype are: actin-Gal4/UAS-GFP (0%, n = 50), sfl (83%, n = 63), Hsepi (0%, n = 51), Hs2st (26%, n = 95), Hs6st (89%, n = 112), sfl–Hsepi (58%, n = 55), sfl–Hs2st (24%, n = 54), sfl–Hs6st (69%, n = 54), Hsepi–Hs2st (0%, n = 63) and Hsepi–Hs6st (62%, n = 50). (B) Lethality of hh-Gal4/UAS-HSME animals. The lethality and the number of scored animals for each genotype are: hh-Gal4/UAS-GFP (0%, n = 100), sfl (96%, n = 49), Hsepi (40%, n = 38), Hs2st (0%, n = 25), Hs6st (88%, n = 104), sfl–Hsepi (39%, n = 59), sfl–Hs2st (0%, n = 25), sfl–Hs6st (100%, n = 83), Hsepi–Hs2st (0%, n = 15) and Hsepi–Hs6st (57%, n = 30).
A similar pattern of the lethality was observed in animals overexpressing HSMEs by hh-Gal4 (Figure 2B). Interestingly, however, we noticed that some HSME genes show lethality uniquely induced by hh-Gal4. For example, the lethality by Hsepi expression was only observed in hh-Gal4/UAS-Hsepi animals. In contrast, no lethality was induced by the expression of Hs2st and sfl–Hs2st by hh-Gal4. These Gal4 driver-specific effects suggested that the lethality caused by some HSMEs is sensitive to spatial and temporal patterns of their expression. Collectively, our results indicated that expression of a specific subset of HSME genes affects the viability of adult flies.
Expression of HSMEs induces distinct effects on the patterning of adult wing structures
A number of studies have shown that HSPGs play critical roles in normal patterning of the Drosophila wing by regulating the activities of heparin-binding secreted growth factors such as Wg, Dpp and Hedgehog (Kirkpatrick and Selleck 2007). Impaired signaling activities of these molecules cause distinct defects in adult wing structures (Couso et al. 1994; De Celis 2003). To determine whether expression of HSME genes affects these signaling pathways, we examined the adult wing morphology of the HSME-expressing animals.
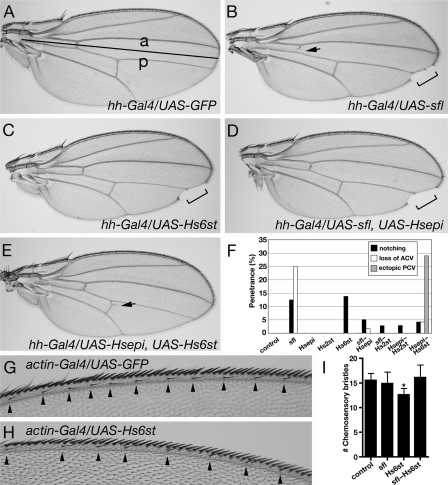
Expression of HSME genes was induced specifically in the posterior compartment by hh-Gal4. Wing phenotypes were examined for all genotypes used in Figure 2B except sfl–Hs6st animals which show fully penetrant lethality (Figure 2B). We found that expression of different HSME genes caused distinct defects in their adult wings (Figure 3B–F). These abnormalities include a deletion of the wing margin (notching), and the loss of ectopic wing cross veins. All these phenotypes were observed specifically in the posterior compartment. The notching phenotype was most obvious in sfl and Hs6st animals (Figure 3B, C and F). A small fraction of sfl–Hsepi, sfl–Hs2st, Hsepi–Hs2st and Hsepi–Hs6st animals also showed the wing margin defects (Figure 3D and F). Since formation of the wing margin is known to be controlled by Wg signaling, these results suggested that specific alterations of HS structures affect the activities of this pathway (Couso et al. 1994). Furthermore, development of chemosensory bristles at the wing margin, which also reflects Wg signaling activity, was impaired in actin-Gal4/UAS-Hs6st animals (Figure 3G–I). These results collectively support the idea that Wg signaling is affected in these animals.
Fig. 3.
Expression of HSME genes affects the morphology of adult wings. Adult wings of hh-Gal4/UAS-GFP (A), hh-Gal4/UAS-sfl (B), hh-Gal4/UAS-Hs6st (C), hh-Gal4/UAS-sfl, UAS-Hsepi (D) and hh-Gal4/UAS-Hsepi, UAS-Hs6st (E). Black line in (A) indicates the border between anterior (a) and posterior (p) compartments. Brackets in (B–D) show the wing margin defects. Arrows show the loss of anterior cross vein in the posterior compartment (B) and ectopic posterior cross vein (E). (F) The percentage of HSME-expressing animals showing the notching (black), loss of anterior cross vein (ACV, white) and ectopic posterior cross vein (PCV, gray) phenotypes. The penetrance of each defect (notching:loss of ACV:ectopic PCV) and the scored number for each genotype are: control (0:0:0, n = 45), sfl (13:25:0, n = 8), Hsepi (0:0:0, n = 27), Hs2st (0:0:0, n = 32), Hs6st (14:0:0, n = 29), sfl–Hsepi (5:2:0, n = 59), sfl–Hs2st (3:0:0, n = 36), Hsepi–Hs2st (3:0:0, n = 34) and Hsepi–Hs6st (4:4:29, n = 24). Anterior wing margin of actin-Gal4/UAS-GFP (G) and actin-Gal4/UAS-Hs6st (H) flies. Arrowheads indicate positions of chemosensory bristles. (I) Bar graph showing the average number of chemosensory bristles in control (actin-Gal4/UAS-GFP, 15.6), sfl (actin-Gal4/UAS-GFP, 14.9), Hs6st (actin-Gal4/UAS-Hs6st, 12.6) and sfl–Hs6st (actin-Gal4/UAS-sfl, UAS-Hs6st, 16.1) animals (n = 10 for each genotype). Error bars indicate the standard deviation. *Statistically different from control (P < 0.01).
In addition to the wing margin defect, we also observed cross vein defects in animals expressing sfl, sfl–Hsepi and Hsepi–Hs6st (Figure 3B, E and F). Interestingly, although expression of either sfl or Hs6st causes severe wing margin defects, only sfl but not Hs6st induced the loss of the anterior cross vein (Figure 3B, C and F). Furthermore, an ectopic posterior cross vein phenotype was observed only in the Hsepi–Hs6st animals (Figure 3E and F). The formation of wing veins is largely dependent on the activity of Dpp signaling (De Celis 2003). Therefore, expression of particular HSME genes appears to influence the sulfation patterns of HS, which is indispensable for the normal activity of Dpp signaling (Jackson et al. 1997; Fujise et al. 2003; Belenkaya et al. 2004; Akiyama et al. 2008). Taken together, our results revealed that expression of different HSME genes has distinct effects on the formation of adult wings, possibly by affecting the activities of several growth factor ligands.
Expression of HSMEs affects Wg signaling and extracellular distribution of Wg protein
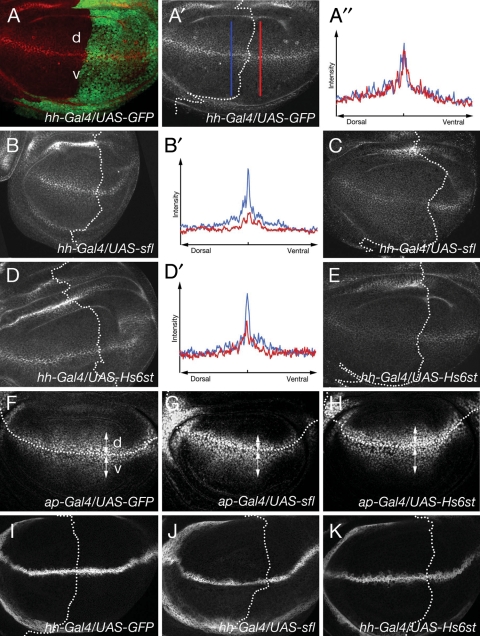
The notching phenotype of the adult wing margin in HSME-expressing animals suggested that these genes affect Wg signaling during wing development. In the larval wing disc, Wg is secreted from a few rows of cells at the dorsoventral (D/V) border and diffuses to form a concentration gradient. Previous studies have demonstrated that HS is required for the extracellular diffusion of Wg protein (Han et al. 2004; Kirkpatrick et al. 2004; Kreuger et al. 2004). Therefore, we investigated whether overexpression of HSME genes affects Wg localization in wing discs (Figure 4A–E). We expressed HSME genes using hh-Gal4 and monitored extracellular Wg using a protocol which specifically detects epitopes in the extracellular space (Strigini and Cohen 2000). Since the gradient of extracellular Wg is formed on the basolateral surface of the wing epithelium, we compared the Wg levels in the basolateral membranes between anterior and posterior compartments (Strigini and Cohen 2000). We found that the level of extracellular Wg in sfl and Hs6st animals was significantly reduced in the posterior compartment, which is in consistent with the severe notching phenotype (Figure 4B and D). Signal intensity plots showed that the Wg protein level was affected both near the D/V boundary and in the most of Wg-receiving cells of the wing discs (Figure 4B′ and D′). Expression of sfl–Hsepi also induced a modest reduction of Wg (data not shown). These results showed that expression of several HSME genes affects the formation of the Wg gradient. In addition to the decrease in the basolateral Wg protein levels in sfl and Hs6st animals, Wg levels on the apical surface of the Wg-producing cells in these animals was increased (Figure 4C and E). A recent report indicated that Wg is secreted from the apical surface of the producing cells and, subsequently, HSPGs regulate the internalization and transfer of Wg to the basolateral membrane which is required for the long-range distribution of Wg (Gallet et al. 2008). Therefore, our results suggest that overexpression of sfl and Hs6st interferes with the internalization and/or the transport of Wg protein near the Wg-expressing cells.
Fig. 4.
Expression of HSME genes affects the level of extracellular Wg protein. (A–E) Expression of HSME genes affects the level of extracellular Wg. Extracellular Wg staining in hh-Gal4/UAS-GFP (A, A′), hh-Gal4/UAS-sfl (B, C) and hh-Gal4/UAS-Hs6st (D, E) larval wing discs. (A) Wg (red) diffuses from D/V border in wing disc (d, dorsal; v, ventral). GFP signal (green) shows the posterior compartment. (A), (B) and (D) show the basolateral side, and (C) and (E) show the apical surface of their wing discs. Signal intensity of the basolateral Wg protein level was plotted in anterior and posterior compartments near the boundary (indicated by blue and red lines, respectively, in A′) for control (A″), sfl (B′) and Hs6st (D′) animals. (F–K) Expression of HSME compromises Wg signaling but not the transcriptional activity of wg. Anti-Dll staining of ap-Gal4/UAS-GFP (F), ap-Gal4/UAS-sfl (G) and ap-Gal4/UAS-Hs6st (H) wing discs. The level of Dll protein is significantly decreased in the dorsal compartment in which sfl or Hs6st was misexpressed. Arrows mark the domain with high levels of Dll staining in the dorsal (d) and ventral (v) compartments. Wing discs of hh-Gal4/UAS-GFP (I), hh-Gal4/UAS-sfl (J) and hh-Gal4/UAS-Hs6st (K) stained with anti-β-galactosidase antibody to mark wg-lacZ.
We next asked whether Wg signal transduction is affected by overexpression of sfl and Hs6st by monitoring expression of a downstream target of Wg signaling, Distal-less (Dll). Expression of these HSME genes was specifically induced in the dorsal compartment using apterous (ap)-Gal4. Anti-Dll antibody staining revealed that the level of Dll protein is significantly decreased in the dorsal compartment (Figure 4F–H). In contrast, overexpression of these genes did not affect the levels and patterns of a wg-lacZ reporter (Figure 4I–K), suggesting that sfl and Hs6st impaired Wg signaling without affecting wg transcription. Together, expression of several HSME genes affects Wg signaling by influencing the extracellular distribution of the Wg ligand in the developing wing.
Expression of specific HSMEs reduces cell proliferation by affecting Dpp signaling
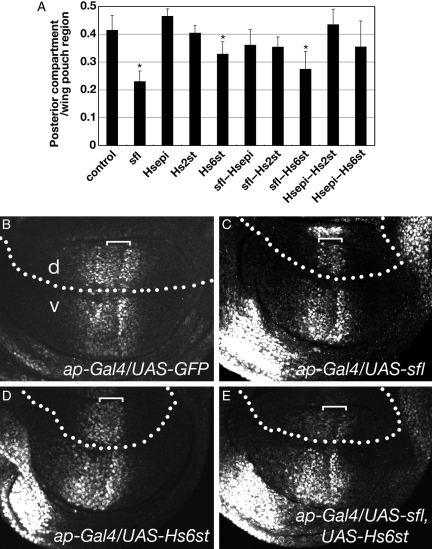
In addition to the patterning defects, the size of the posterior compartment was reduced in the wing discs of several hh-Gal4/UAS-HSME strains (Figure 5A). The areas expressing sfl and sfl–Hs6st were markedly affected (56 and 66% of those of hh-gal4/UAS-GFP animals, respectively). Expression of Hs6st also significantly decreased the size of the posterior compartment. The reduction in the size of the posterior compartment in these animals suggested that these HSME genes affect cell proliferation.
Fig. 5.
Expression of HSMEs affects cell proliferation and Dpp signaling in the developing wing. (A) Expression of HSME genes affects the size of the posterior compartments of the wing disc. The graph indicates the average ratio of the size of posterior compartment to the whole wing pouch region (n = 10 for each genotype). The ratio for each genotype is: 0.41 for control, 0.23 for sfl, 0.46 for Hsepi, 0.40 for Hs2st, 0.33 for Hs6st, 0.36 for sfl–Hspi, 0.35 for sfl–Hs2st, 0.27 for sfl–Hs6st, 0.43 for Hsepi–Hs2st and 0.35 for Hsepi–Hs6st. Expression of GFP and all HSME genes was induced by hh-Gal4. Error bars indicate the standard deviations. *Statistically different from control (P < 0.01). Wing discs of ap-Gal4/UAS-GFP (B), ap-Gal4/UAS-sfl (C), ap-Gal4/UAS-Hs6st (D) and ap-Gal4/UAS-sfl, UAS-Hs6st (E) were stained with anti-pMad antibody. In the dorsal compartment, pMad levels were significantly reduced in the Dpp-receiving cells of sfl, Hs6st or sfl–Hs6st discs (d, dorsal; v, ventral). Brackets indicate the Dpp-expressing domain.
It has been shown that Dpp signaling regulates cell proliferation in the wing disc (Burke and Basler 1996; Martin-Castellanos and Edgar 2002). To determine whether HSME expression affects Dpp signaling, phosphorylation of Mothers against dpp (Mad), a direct readout of Dpp signal transduction, was monitored in the wing discs expressing sfl, Hs6st or sfl–Hs6st (Figure 5B–E). Staining the discs with an antibody against phosphorylated form of Mad protein (pMad) showed that expression of these HSME genes significantly reduced the level of pMad in the Dpp-receiving cells. Interestingly, the pMad levels were not decreased in the Dpp-expressing cells of the sfl and Hs6st discs (brackets in Figure 5C and D). In fact, this pattern of pMad resembles that observed in the discs mutant for dally, which encodes a Drosophila glypican (Fujise et al. 2003). These results suggest that specific HSME expression interferes with Dpp signaling, leading to the reduced cell proliferation, probably by disrupting normal function of a Dpp co-receptor, Dally.
Discussion
A number of in vivo studies have shown the importance of specific sulfate groups on HS during development (Gorsi and Stringer 2007). These analyses have focused on the effects of alterations of a single HSME on developmental processes, and it remains elusive how a combination of these molecules controls the structure and function of HS. Recent models of gagosome assembly and function (Pinhal et al. 2001; Presto et al. 2008) and findings of the HS sulfation compensation system (Merry et al. 2001; Kamimura et al. 2006; Sugaya et al. 2008) suggested that there are various networks controlling the activities of HSMEs in the HS modification machinery. It is now important to understand how the activity of each HSME is controlled in such a regulatory system and how it affects biological processes. In this study, we established a model system in which we can manipulate HS structures in vivo in a systematic manner using Drosophila and evaluate their effects on morphogenesis and signaling.
Our disaccharide analyses of HS from various HSME animals demonstrated several novel features of these enzymes. First, we noticed that expression of sfl, Hs2st and sfl–Hs2st affects the total level of HS. Although the mechanisms for this increase in HS are unknown, one possibility is that Sfl and Hs2st recruit EXTs into the HS biosynthetic machinery and/or increase their activities, promoting HS polymerization. Alternatively, expression of these HSMEs may generate HS structures, which affect the stability and turnover rate of HSPGs. The other unexpected aspect of HSMEs emerged from analyses of animals coexpressing two HSMEs. Although expression of a single HSME increases the corresponding sulfate groups, in many cases these effects were impaired by coexpression of other HSMEs. For example, the increase in the level of 6-O sulfation in Hs6st animals was disrupted by coexpression of sfl or Hsepi. One exception was observed in sfl–Hs2st animals, which possess more N- and 2-O sulfate groups than sfl and Hs2st animals. Thus, Sfl and Hs2st show a cooperative relationship, while Sfl is inhibitory to overexpressed Hs6st. It was previously reported that in mouse embryonic stem cells deficient for NDST1 and NDST2, HS does not contain 2-O sulfate groups but 6-O sulfation occurs without N-sulfation (Holmborn et al. 2004), suggesting that the activity of Hs2st, but not Hs6st, entirely depends on N-sulfation. Such distinct relationships between NDST–Hs2st and NDST–Hs6st appear to contribute to the facilitatory and inhibitory effects of these enzymes on HS sulfation in sfl–Hs2st and sfl–Hs6st animals. For example, Hs6st may influence the initiation process of HS modification by NDST by antagonizing NDST's activity. Briefly, our results as well as previous studies suggested that the enzymatic activities of HSMEs are significantly affected by the levels of other HSMEs. Although the precise molecular mechanism of this phenomenon remains to be clarified, our findings suggest the existence of a regulatory network controlling HSME function during the biosynthesis and modification of HS in consistent with the gagosome model.
Morphological analyses of HSME animals revealed that expression of a subset of HSMEs induces distinct defects in adult wing structures. Expression of sfl, Hs6st, sfl–Hsepi, sfl–Hs2st, Hsepi–Hs2st and Hsepi–Hs6st caused wing margin defects, which are characteristic phenotypes of Wg signaling mutants (Couso et al. 1994). In consistent with this, we found that distribution of Wg protein is disrupted in sfl and Hs6st animals, which displayed the most severe notching phenotype in HSME animals. These results indicated that structural changes of HS by overexpression of these HSMEs compromised the activity of Wg signaling. We also observed cross vein defects in sfl, sfl–Hsepi and Hsepi–Hs6st animals, and impaired cell proliferation in sfl, Hs6st and sfl–Hs6st. Previous studies indicated that the formation of the cross vein and cell proliferation are controlled by the Dpp pathway, suggesting that expression of these HSME genes also affects Dpp signaling (Burke and Basler 1996; Martin-Castellanos and Edgar 2002; De Celis 2003). Indeed, this was the case: Mad phosphorylation was disrupted by overexpression of sfl, Hs6st and sfl–Hs6st. Thus, our analyses on adult phenotypes as well as signaling markers showed that overexpression of various HSMEs caused distinct morphological defects by affecting different signaling pathways. Particularly, sfl–Hs2st and Hsepi–Hs2st showed defects in Wg-mediated wing margin formation but not in Dpp-mediated cross vein formation. These results strongly suggest that distinct fine structures of HS differentially affect binding of specific growth factor ligands.
Our recent analysis of Sulf1, a Drosophila HS 6-O endosulfatase, showed that this enzyme is involved in the formation of the wing margin by regulating Wg signaling (Kleinschmit et al. 2010). Overexpression of Sulf1 also affects Dpp signaling, leading to reduced proliferation of wing cells. HS disaccharide analysis revealed that Sulf1 mutants have abnormally high levels of tri-S disaccharide unit (ΔUA2S-GlcNS6S), indicating that Sulf1 selectively removes 6-O sulfate group from tri-S disaccharide unit. These observations suggest the pivotal role of this HS structure in Wg and Dpp signaling. Interestingly, the effects of Sulf1 overexpression on developmental events and signaling were similar to, not opposite to, those observed in HSME animals. In our morphological analyses of HSME animals, the most severe defects were observed in sfl, Hs6st and sfl–Hs6st animals. These animals showed a reduced viability, and defects in wing margin formation and the cell proliferation. However, we did not see a simple correlation between the severity of morphological defects and the degree of HS structural change (Table III). sfl showed an increase in total HS, the total number of sulfate groups and the density of sulfate groups on HS, and Hs6st and sfl–Hs6st showed elevated levels of particular sulfate groups (Figure 1 and Table I). However, these effects may not necessarily account for their severe defects since expression of sfl–Hs2st caused only modest morphological defects despite the high levels of HS, sulfate groups and sulfation density as well as alteration of unsaturated disaccharide composition (Figure 1 and Table I). Furthermore, the effect of sfl–Hsepi on the levels of sulfation was moderate, but it caused obvious morphological defects such as the loss of the wing margin and cross vein. These results indicated that changes in charge density and unsaturated disaccharide composition are not directly associated with morphological defects and signaling phenotypes. Since unsaturated disaccharide analysis provides a limited amount of information on HS structure, it is possible that other structural alterations of HS may contribute to the defects. For example, the occurrence of C5-epimerization and 3-O sulfation on HS was not detected in this analysis. Also, previous studies showed that HS structures involving N-sulfated domains separated by N-acetylated domain mediate interactions with many proteins (Kreuger et al. 2006). Furthermore, more specific sulfation sequences could be responsible for selective ligand binding. Thus, our HSME expression system, in combination with further detailed studies on HS structures, will be useful to understand the structure–function relationship of HS.
Table III.
Structural alterations of HS and morphological defects in HSME animals
| Total HS | N-sulfation | 2-O sulfation | 6-O sulfation | Wing margin defects | Cross vein defects | Cell proliferation defects | Wg distribution defects | Dpp signaling defects | |
|---|---|---|---|---|---|---|---|---|---|
| sfl | + | + (+) | + (+) | + (N.S.) | ++ | ++ | ++ | ++ | ++ |
| Hsepi | N.S. | N.S. (N.S.) | N.S. (+) | N.S. (N.S.) | N.S. | N.S. | N.S. | N.S. | * |
| Hs2st | + | + (N.S.) | + (+) | N.S. (−) | N.S. | N.S. | N.S. | N.S. | * |
| Hs6st | N.S. | N.S. (−) | N.S.(−) | ++ (++) | ++ | N.S. | + | ++ | ++ |
| sfl–Hsepi | N.S. | N.S. (N.S) | N.S. (+) | N.S. (N.S.) | + | + | N.S. | + | * |
| sfl–Hs2st | ++ | ++ (+) | ++ (++) | N.S. (−) | + | N.S. | N.S. | N.S. | * |
| sfl–Hs6st | N.S. | + (+) | + (++) | N.S. (−) | * | * | + | N.S. | ++ |
| Hsepi–Hs2st | N.S. | N.S. (−) | N.S. (+) | N.S. (−) | + | N.S. | N.S. | N.S. | * |
| Hsepi–Hs6st | N.S. | N.S. (N.S) | + (++) | N.S. (−) | + | + (extra vein) | N.S. | N.S. | * |
Summary obtained from this study. + represents increased levels of total HS, sulfate groups and the severity of morphological defects (++, dramatically increased). The change in the proportion of disaccharides that contains each sulfate group is shown in parentheses. N.S., not significantly affected; *, not determined.
Materials and methods
Fly stocks
The detailed information for fly strains used is described in Flybase (http://flybase.bio.indiana.edu/). All flies were maintained at 25°C. UAS-HSME transgenic strains were generated as follows. Total RNA was extracted from third-instar larvae, and the cDNA for each HSME gene was synthesized and amplified by reverse transcription–polymerase chain reaction using standard protocols. The PCR fragments were sequenced, recombined with the pDONR 221 vectors (Invitrogen, CA, USA) and thereafter recombined into pUAST destination vectors (Drosophila Genomics Resource Center, IN, USA) using Gateway technology (Invitrogen, CA, USA). P-element-mediated germ-line transformation was performed by Genetic Service, Inc. Other transgenic animals used were: winglessen11 (wg-lacZ), UAS-GFP, actin-Gal4, hh-Gal4, ap-Gal4 and hh-Gal4. In the Gal4/UAS system, expression level of each transgene in animals bearing two UAS transgenes might be lower than that in animals bearing one transgene, since the level of Gal4 protein is invariant in these animals. To avoid this possibility, in our phenotypic analyses of UAS-HSME/actin (or hh)-Gal4 animals, we adjusted the induction level of each HSME by introducing UAS-GFP and equalizing the number of UAS transgene in all animals. UAS-GFP was also used as a control for UAS-HSME.
To determine the relative viability of actin (or hh)-Gal4/UAS-HSME flies, UAS-HSME flies were crossed to actin (or hh)-Gal4/CyO (or TM6B), and adult progenies with the balancer and the non-balancer chromosomes were counted.
Disaccharide analyses of HS
Thirty actin-Gal4/UAS-HSME adult flies were used for disaccharide analysis. For hs-Gal4/UAS-HSME animals, animals were reared at 18°C until the third-instar larval stage. After a heat shock at 37°C for 30 min, the larvae were incubated at 25°C for 4 h before collection. Thirty animals were used for disaccharide analysis. The collected animals were lyophilized to dryness, and homogenized with 1 mL of ice-cold acetone. The homogenates were stirred for 1 h at 4°C and centrifuged at 15,000 × g for 10 min. The pelleted samples were homogenized again with 1 mL of acetone, and stirred and centrifuged as above. The resulting pellets were air-dried, and re-suspended in 0.2 mL of 0.2 M NaOH, 0.5% SDS. The samples were stirred overnight at room temperature. After neutralization with 1.5 μL of acetic acid, the solutions were diluted with 0.3 mL of 50 mM Tris–HCl, pH 8.0. The samples were then mixed with 2.5 μL of 2 mg/mL Actinase E (Sigma, MO, USA), and were incubated overnight at 37°C. The solutions were heated at 100°C for 10 min, and centrifuged at 15,000 × g for 10 min. The supernatants were mixed with 3 vol. of 95% ethanol, 1.3% potassium acetate and incubated for 30 min on ice (ethanol precipitation). The samples were centrifuged at 15,000 × g for 10 min, and the resultant pellets were dissolved in 300 μL of distilled water. Ethanol precipitation was further repeated twice. The final pellets were dissolved in 30 μL of 50 mM ammonium acetate and 1 mM calcium acetate. Six microliters of heparin lyase mixture containing 0.17 U/mL each of heparitinase I, heparitinase II and heparinase (Seikagaku, Tokyo, Japan) was added to the samples, which were incubated overnight at 37°C. Subsequently, the samples were mixed with 3 μL of heparin lyase mixture and were incubated for further 3 h at 37°C. The samples were mixed with 117 μL of ethanol and incubated for 30 min on ice. After centrifugation at 15,000 × g for 10 min, the supernatants were dried by SpeedVac lyophilization. The dried materials were treated with 5 μL of 0.35 M 2-aminobenzamide, 1 M sodium cyanoborohydride in 30% acetic acid and 70% dimethyl sulfoxide for 2 h at 65°C. After the removal of excess 2-aminobenzamide by paper chromatography, the fluorescently labeled unsaturated disaccharides were analyzed by high-performance liquid chromatography using a YMC pack PA-03 column (YMC, Kyoto, Japan) according to the method described by Kinoshita and Sugahara (1999). We analyzed the HS structures three times for each genotype, and statistical analysis was performed using unpaired t-test and analysis of variance (ANOVA) to compare each disaccharide component for all genotypes to control, followed by Dunnett's test.
Preparation of adult wings
The adult wings were dehydrated in ethanol and subsequently with xylene. After the wings were mounted in EUKITT (Takahashi Giken Glass Co., Tokyo, Japan), phenotypes were observed and photographed using a Nikon Eclipse E800 microscope.
Immunohistochemistry
Conventional antibody staining was performed as described (Fujise et al. 2003). The following antibodies were used: mouse anti-Distal-less (Dll, 1:500, a gift from D. Duncan), rabbit anti-β-galactosidase (1:500, Cappel, PA, USA) and rabbit anti-pSMAD3 (1:1000, Epitomics, CA, USA). Extracellular Wg staining was performed as described (Strigini and Cohen 2000). Mouse anti-Wg antibody was used at 1:3 dilution (4D4, Developmental Studies Hybridoma Bank, IA, USA). The primary antibody was detected by Alexa Fluor 568-conjugated goat anti-mouse IgG antibody (Jackson Immunoresearch, PA, USA). Images were captured with a Nikon C1 confocal microscope, and the signal intensity of Wg and the size of posterior compartments in HSME animals were analyzed with ImageJ 1.33u. Statistical analysis was performed using ANOVA to compare different genotypes to control, followed by Dunnett's test. Acridine orange staining was performed as previously described (Abrams et al. 1993).
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Conflict of interest statement
None declared.
Funding
This work was partly supported by the National Institute of Health (R01 HD042769 to H.N.) and Grant-in-Aid for Young Scientists (No. 21770154 to K.K.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Abbreviations
ACV, anterior cross vein; ANOVA, analysis of variance; Ap, apterous; Dll, Distal-less; Dpp, Decapentaplegic; ΔUA-GlcNAc, Δ4,5 unsaturated hexuronate-N-acetyl glucosamine; ΔUA-GlcNAc6S, ΔUA-6-O sulfated GlcNAc; ΔUA-GlcNS, ΔUA-N-sulfated glucosamine; ΔUA-GlcNS6S, ΔUA6-O sulfated GlcNS; ΔUA2S-GlcNS, 2-O sulfated ΔUA-GlcNS; Hh, Hedgehog; HS, heparan sulfate; Hs3st, heparan sulfate 3-O sulfotransferase; Hs6st, heparan sulfate 6-O sulfotransferase; Hsepi, heparan sulfate C5-epimerase; Hs2st, heparan sulfate 2-O sulfotransferase; HSME, heparan sulfate-modifying enzyme; HSPG, heparan sulfate proteoglycan; IdoA, iduronic acid; Mad, Mother against dpp; NDST, N-deacetylase/N-sulfotransferase; PCV, posterior cross vein; Sfl, sulfateless; UAS, upstream activating sequence; Wg, Wingless.
Acknowledgements
We are grateful to D. Duncan, the Developmental Studies Hybridoma Bank and the Bloomington Stock Center for fly stocks and reagents.
References
- Abrams JM, White K, Fessler LI, Steller H. Programmed cell death during Drosophila embryogenesis. Development. 1993;117:29–43. doi: 10.1242/dev.117.1.29. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Kamimura K, Firkus C, Takeo S, Shimmi O, Nakato H. Dally regulates Dpp morphogen gradient formation by stabilizing Dpp on the cell surface. Dev Biol. 2008;313:408–419. doi: 10.1016/j.ydbio.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenkaya TY, Han C, Yan D, Opoka RJ, Khodoun M, Liu H, Lin X. Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell. 2004;119:231–244. doi: 10.1016/j.cell.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Bullock SL, Fletcher JM, Beddington RS, Wilson VA. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev. 1998;12:1894–1906. doi: 10.1101/gad.12.12.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R, Basler K. Dpp receptors are autonomously required for cell proliferation in the entire developing Drosophila wing. Development. 1996;122:2261–2269. doi: 10.1242/dev.122.7.2261. [DOI] [PubMed] [Google Scholar]
- Couso JP, Bishop SA, Martinez Arias A. The wingless signalling pathway and the patterning of the wing margin in Drosophila. Development. 1994;120:621–636. doi: 10.1242/dev.120.3.621. [DOI] [PubMed] [Google Scholar]
- De Celis JF. Pattern formation in the Drosophila wing: The development of the veins. Bioessays. 2003;25:443–451. doi: 10.1002/bies.10258. [DOI] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Fujise M, Takeo S, Kamimura K, Matsuo T, Aigaki T, Izumi S, Nakato H. Dally regulates Dpp morphogen gradient formation in the Drosophila wing. Development. 2003;130:1515–1522. doi: 10.1242/dev.00379. [DOI] [PubMed] [Google Scholar]
- Gallet A, Staccini-Lavenant L, Therond PP. Cellular trafficking of the glypican Dally-like is required for full-strength Hedgehog signaling and wingless transcytosis. Dev Cell. 2008;14:712–725. doi: 10.1016/j.devcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Gorsi B, Stringer SE. Tinkering with heparan sulfate sulfation to steer development. Trends Cell Biol. 2007;17:173–177. doi: 10.1016/j.tcb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Habuchi H, Nagai N, Sugaya N, Atsumi F, Stevens RL, Kimata K. Mice deficient in heparan sulfate 6-O-sulfotransferase-1 exhibit defective heparan sulfate biosynthesis, abnormal placentation, and late embryonic lethality. J Biol Chem. 2007;282:15578–15588. doi: 10.1074/jbc.M607434200. [DOI] [PubMed] [Google Scholar]
- Han C, Belenkaya TY, Khodoun M, Tauchi M, Lin X, Lin X. Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signalling and gradient formation. Development. 2004;131:1563–1575. doi: 10.1242/dev.01051. [DOI] [PubMed] [Google Scholar]
- Holmborn K, Ledin J, Smeds E, Eriksson I, Kusche-Gullberg M, Kjellen L. Heparan sulfate synthesized by mouse embryonic stem cells deficient in NDST1 and NDST2 is 6-O-sulfated but contains no N-sulfate groups. J Biol Chem. 2004;279:42355–42358. doi: 10.1074/jbc.C400373200. [DOI] [PubMed] [Google Scholar]
- Jackson SM, Nakato H, Sugiura M, Jannuzi A, Oakes R, Kaluza V, Golden C, Selleck SB. dally, a Drosophila glypican, controls cellular responses to the TGF-beta-related morphogen, Dpp. Development. 1997;124:4113–4120. doi: 10.1242/dev.124.20.4113. [DOI] [PubMed] [Google Scholar]
- Kamimura K, Koyama T, Habuchi H, Ueda R, Masu M, Kimata K, Nakato H. Specific and flexible roles of heparan sulfate modifications in Drosophila FGF signaling. J Cell Biol. 2006;174:773–778. doi: 10.1083/jcb.200603129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Sugahara K. Microanalysis of glycosaminoglycan-derived oligosaccharides labeled with a fluorophore 2-aminobenzamide by high-performance liquid chromatography: application to disaccharide composition analysis and exosequencing of oligosaccharides. Anal Biochem. 1999;269:367–378. doi: 10.1006/abio.1999.4027. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CA, Dimitroff BD, Rawson JM, Selleck SB. Spatial regulation of Wingless morphogen distribution and signaling by Dally-like protein. Dev Cell. 2004;7:513–523. doi: 10.1016/j.devcel.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CA, Selleck SB. Heparan sulfate proteoglycans at a glance. J Cell Sci. 2007;120:1829–1832. doi: 10.1242/jcs.03432. [DOI] [PubMed] [Google Scholar]
- Kleinschmit A, Koyama T, Dejima K, Hayashi Y, Kamimura K, Nakato H. Drosophila heparan sulfate 6-O endosulfatase regulates Wingless morphogen gradient formation. Dev Biol. 2010;345:204–214. doi: 10.1016/j.ydbio.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuger J, Perez L, Giraldez AJ, Cohen SM. Opposing activities of Dally-like glypican at high and low levels of Wingless morphogen activity. Dev Cell. 2004;7:503–512. doi: 10.1016/j.devcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Kreuger J, Spillmann D, Li JP, Lindahl U. Interactions between heparan sulfate and proteins: the concept of specificity. J Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JP, Gong F, Hagner-McWhirter A, Forsberg E, Abrink M, Kisilevsky R, Zhang X, Lindahl U. Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking L-iduronic acid and in neonatal lethality. J Biol Chem. 2003;278:28363–28366. doi: 10.1074/jbc.C300219200. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Edgar BA. A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development. 2002;129:1003–1013. doi: 10.1242/dev.129.4.1003. [DOI] [PubMed] [Google Scholar]
- Merry CL, Bullock SL, Swan DC, Backen AC, Lyon M, Beddington RS, Wilson VA, Gallagher JT. The molecular phenotype of heparan sulfate in the Hs2st-/- mutant mouse. J Biol Chem. 2001;276:35429–35434. doi: 10.1074/jbc.M100379200. [DOI] [PubMed] [Google Scholar]
- Nakato H, Futch TA, Selleck SB. The division abnormally delayed (dally) gene: a putative integral membrane proteoglycan required for cell division patterning during postembryonic development of the nervous system in Drosophila. Development. 1995;121:3687–3702. doi: 10.1242/dev.121.11.3687. [DOI] [PubMed] [Google Scholar]
- Nakato H, Kimata K. Heparan sulfate fine structure and specificity of proteoglycan functions. Biochim Biophys Acta. 2002;1573:312–318. doi: 10.1016/s0304-4165(02)00398-7. [DOI] [PubMed] [Google Scholar]
- Pinhal MA, Smith B, Olson S, Aikawa J, Kimata K, Esko JD. Enzyme interactions in heparan sulfate biosynthesis: uronosyl 5-epimerase and 2-O-sulfotransferase interact in vivo. Proc Natl Acad Sci U S A. 2001;98:12984–12989. doi: 10.1073/pnas.241175798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presto J, Thuveson M, Carlsson P, Busse M, Wilen M, Eriksson I, Kusche-Gullberg M, Kjellen L. Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc Natl Acad Sci U S A. 2008;105:4751–4756. doi: 10.1073/pnas.0705807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strigini M, Cohen SM. Wingless gradient formation in the Drosophila wing. Curr Biol. 2000;10:293–300. doi: 10.1016/s0960-9822(00)00378-x. [DOI] [PubMed] [Google Scholar]
- Sugaya N, Habuchi H, Nagai N, Ashikari-Hada S, Kimata K. 6-O-sulfation of heparan sulfate differentially regulates various fibroblast growth factor-dependent signalings in culture. J Biol Chem. 2008;283:10366–10376. doi: 10.1074/jbc.M705948200. [DOI] [PubMed] [Google Scholar]
- Takeo S, Akiyama T, Firkus C, Aigaki T, Nakato H. Expression of a secreted form of Dally, a Drosophila glypican, induces overgrowth phenotype by affecting action range of Hedgehog. Dev Biol. 2005;284:204–218. doi: 10.1016/j.ydbio.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Kinoshita-Toyoda A, Selleck SB. Structural analysis of glycosaminoglycans in Drosophila and Caenorhabditis elegans and demonstration that tout-velu, a Drosophila gene related to EXT tumor suppressors, affects heparan sulfate in vivo. J Biol Chem. 2000;275:2269–2275. doi: 10.1074/jbc.275.4.2269. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Kamimura K, Nakato H, Archer M, Staatz W, Fox B, Humphrey M, Olson S, Futch T, Kaluza V, Siegfried E, Stam L, Selleck SB. The cell-surface proteoglycan Dally regulates Wingless signalling in Drosophila. Nature. 1999;400:276–280. doi: 10.1038/22336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.