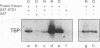
Abstract
STD1 (MSN3) was isolated independently as a multicopy suppressor of mutations in the TATA-binding protein and in SNF4, suggesting that STD1 might couple the SNF1 kinase signaling pathway to the transcriptional machinery. We report here a direct physical interaction between STD1 and the TATA-binding protein (TBP), observed in vivo by the two-hybrid system and in vitro by binding studies. STD1 bound both native TBP in yeast cell-free extracts and purified recombinant TBP. This interaction was altered when TBP delta 57 was used, suggesting a role for the non-conserved N-terminal domain of TBP in mediating protein-protein interactions. We also show that perturbation of STD1-TBP stoichiometry alters SUC2 expression in vivo and that this effect is dependent on the N-terminal domain of TBP. The activation of SUC2 expression by increased copy number of STD1 occurs at the level of mRNA accumulation and it requires the same TATA element and uses the same transcription start site as does activation of SUC2 by glucose limitation. Taken together, these results suggest that STD1 modulates SUC2 transcription through direct interactions with TBP.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartel P., Chien C. T., Sternglanz R., Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993 Jun;14(6):920–924. [PubMed] [Google Scholar]
- Carlson M., Taussig R., Kustu S., Botstein D. The secreted form of invertase in Saccharomyces cerevisiae is synthesized from mRNA encoding a signal sequence. Mol Cell Biol. 1983 Mar;3(3):439–447. doi: 10.1128/mcb.3.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J. L., Carlson M. Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol Cell Biol. 1989 Nov;9(11):5034–5044. doi: 10.1128/mcb.9.11.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J. L., Eng F. J., Carlson M. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol Cell Biol. 1989 Nov;9(11):5045–5054. doi: 10.1128/mcb.9.11.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C. T., Bartel P. L., Sternglanz R., Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriacy M. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol Gen Genet. 1977 Jul 20;154(2):213–220. doi: 10.1007/BF00330840. [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Strubin M., Ponticelli A. S., Struhl K. Functional differences between yeast and human TFIID are localized to the highly conserved region. Cell. 1991 Apr 19;65(2):341–348. doi: 10.1016/0092-8674(91)90167-w. [DOI] [PubMed] [Google Scholar]
- Durfee T., Becherer K., Chen P. L., Yeh S. H., Yang Y., Kilburn A. E., Lee W. H., Elledge S. J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993 Apr;7(4):555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Chapon C., Roberts S. M., Dollard C., Winston F. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics. 1994 Jul;137(3):647–657. doi: 10.1093/genetics/137.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D., Zimmermann F. K. New genes involved in carbon catabolite repression and derepression in the yeast Saccharomyces cerevisiae. J Bacteriol. 1982 Sep;151(3):1123–1128. doi: 10.1128/jb.151.3.1123-1128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989 Jul 20;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Ganster R. W., Shen W., Schmidt M. C. Isolation of STD1, a high-copy-number suppressor of a dominant negative mutation in the yeast TATA-binding protein. Mol Cell Biol. 1993 Jun;13(6):3650–3659. doi: 10.1128/mcb.13.6.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G., Tjian R. A highly conserved domain of TFIID displays species specificity in vivo. Cell. 1991 Apr 19;65(2):333–340. doi: 10.1016/0092-8674(91)90166-v. [DOI] [PubMed] [Google Scholar]
- Goldstein A., Lampen J. O. Beta-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Hagemeier C., Bannister A. J., Cook A., Kouzarides T. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1580–1584. doi: 10.1073/pnas.90.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hateboer G., Timmers H. T., Rustgi A. K., Billaud M., van 't Veer L. J., Bernards R. TATA-binding protein and the retinoblastoma gene product bind to overlapping epitopes on c-Myc and adenovirus E1A protein. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8489–8493. doi: 10.1073/pnas.90.18.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993 Jul;7(7B):1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hirschhorn J. N., Brown S. A., Clark C. D., Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992 Dec;6(12A):2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Yamamoto T., Ohkuma Y., Weil P. A., Roeder R. G. Analysis of structure-function relationships of yeast TATA box binding factor TFIID. Cell. 1990 Jun 29;61(7):1171–1178. doi: 10.1016/0092-8674(90)90681-4. [DOI] [PubMed] [Google Scholar]
- Horikoshi N., Maguire K., Kralli A., Maldonado E., Reinberg D., Weinmann R. Direct interaction between adenovirus E1A protein and the TATA box binding transcription factor IID. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5124–5128. doi: 10.1073/pnas.88.12.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard E. J., Jiang R., Carlson M. Dosage-dependent modulation of glucose repression by MSN3 (STD1) in Saccharomyces cerevisiae. Mol Cell Biol. 1994 Mar;14(3):1972–1978. doi: 10.1128/mcb.14.3.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles C. J., Shales M., Cress W. D., Triezenberg S. J., Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991 Jun 13;351(6327):588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- Kuddus R., Schmidt M. C. Effect of the non-conserved N-terminus on the DNA binding activity of the yeast TATA binding protein. Nucleic Acids Res. 1993 Apr 25;21(8):1789–1796. doi: 10.1093/nar/21.8.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrer K., Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- Lai J. S., Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. K., DeJong J., Hashimoto S., Horikoshi M., Roeder R. G. TFIIA induces conformational changes in TFIID via interactions with the basic repeat. Mol Cell Biol. 1992 Nov;12(11):5189–5196. doi: 10.1128/mcb.12.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. S., Kao C. C., Bryant G. O., Liu X., Berk A. J. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991 Oct 18;67(2):365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- Lieberman P. M., Berk A. J. The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev. 1991 Dec;5(12B):2441–2454. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- Lieberman P. M., Schmidt M. C., Kao C. C., Berk A. J. Two distinct domains in the yeast transcription factor IID and evidence for a TATA box-induced conformational change. Mol Cell Biol. 1991 Jan;11(1):63–74. doi: 10.1128/mcb.11.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Miller C. W., Koeffler P. H., Berk A. J. The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol Cell Biol. 1993 Jun;13(6):3291–3300. doi: 10.1128/mcb.13.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. W., Subler M. A., Muñoz R. M., Brown D. R., Deb S. P., Deb S. p53 and SV40 T antigen bind to the same region overlapping the conserved domain of the TATA-binding protein. Biochem Biophys Res Commun. 1993 Aug 31;195(1):428–434. doi: 10.1006/bbrc.1993.2061. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Neigeborn L., Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984 Dec;108(4):845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon D., Schroeder S., Wang C. K., Yamamoto T., Horikoshi M., Roeder R. G., Weil P. A. The conserved carboxy-terminal domain of Saccharomyces cerevisiae TFIID is sufficient to support normal cell growth. Mol Cell Biol. 1991 Oct;11(10):4809–4821. doi: 10.1128/mcb.11.10.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransone L. J., Kerr L. D., Schmitt M. J., Wamsley P., Verma I. M. The bZIP domains of Fos and Jun mediate a physical association with the TATA box-binding protein. Gene Expr. 1993;3(1):37–48. [PMC free article] [PubMed] [Google Scholar]
- Reddy P., Hahn S. Dominant negative mutations in yeast TFIID define a bipartite DNA-binding region. Cell. 1991 Apr 19;65(2):349–357. doi: 10.1016/0092-8674(91)90168-x. [DOI] [PubMed] [Google Scholar]
- Rigby P. W. Three in one and one in three: it all depends on TBP. Cell. 1993 Jan 15;72(1):7–10. doi: 10.1016/0092-8674(93)90042-o. [DOI] [PubMed] [Google Scholar]
- Ruden D. M., Ma J., Li Y., Wood K., Ptashne M. Generating yeast transcriptional activators containing no yeast protein sequences. Nature. 1991 Mar 21;350(6315):250–252. doi: 10.1038/350250a0. [DOI] [PubMed] [Google Scholar]
- Schüller H. J., Entian K. D. Molecular characterization of yeast regulatory gene CAT3 necessary for glucose derepression and nuclear localization of its product. Gene. 1988 Jul 30;67(2):247–257. doi: 10.1016/0378-1119(88)90401-5. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. TATA-binding protein is a classless factor. Cell. 1992 Mar 6;68(5):819–821. doi: 10.1016/0092-8674(92)90023-6. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Thompson-Jaeger S., François J., Gaughran J. P., Tatchell K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991 Nov;129(3):697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. J., Jackson S. P., Rigby P. W. A role for the TATA-box-binding protein component of the transcription factor IID complex as a general RNA polymerase III transcription factor. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1949–1953. doi: 10.1073/pnas.89.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. J., Jackson S. P. The TATA-binding protein: a central role in transcription by RNA polymerases I, II and III. Trends Genet. 1992 Aug;8(8):284–288. doi: 10.1016/0168-9525(92)90255-3. [DOI] [PubMed] [Google Scholar]
- Yang X., Hubbard E. J., Carlson M. A protein kinase substrate identified by the two-hybrid system. Science. 1992 Jul 31;257(5070):680–682. doi: 10.1126/science.1496382. [DOI] [PubMed] [Google Scholar]
- Zhou Q. A., Schmidt M. C., Berk A. J. Requirement for acidic amino acid residues immediately N-terminal to the conserved domain of Saccharomyces cerevisiae TFIID. EMBO J. 1991 Jul;10(7):1843–1852. doi: 10.1002/j.1460-2075.1991.tb07710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Berk A. J. The yeast TATA-binding protein (TBP) core domain assembles with human TBP-associated factors into a functional TFIID complex. Mol Cell Biol. 1995 Jan;15(1):534–539. doi: 10.1128/mcb.15.1.534. [DOI] [PMC free article] [PubMed] [Google Scholar]