Abstract
The Cancer Genome Atlas (TCGA) is a multidisciplinary, multi-institutional effort to characterize several types of cancer. Datasets from biomedical domains such as TCGA present a particularly challenging task for those interested in dynamically aggregating its results because the data sources are typically both heterogeneous and distributed. The Linked Data best practices offer a solution to integrate and discover data with those characteristics, namely through exposure of data as Web services supporting SPARQL, the Resource Description Framework query language. Most SPARQL endpoints, however, cannot easily be queried by data experts. Furthermore, exposing experimental data as SPARQL endpoints remains a challenging task because, in most cases, data must first be converted to Resource Description Framework triples. In line with those requirements, we have developed an infrastructure to expose clinical, demographic and molecular data elements generated by TCGA as a SPARQL endpoint by assigning elements to entities of the Simple Sloppy Semantic Database (S3DB) management model. All components of the infrastructure are available as independent Representational State Transfer (REST) Web services to encourage reusability, and a simple interface was developed to automatically assemble SPARQL queries by navigating a representation of the TCGA domain. A key feature of the proposed solution that greatly facilitates assembly of SPARQL queries is the distinction between the TCGA domain descriptors and data elements. Furthermore, the use of the S3DB management model as a mediator enables queries to both public and protected data without the need for prior submission to a single data source.
Keywords: TCGA, SPARQL, RDF, Linked Data, Data integration
1. Introduction
The Cancer Genome Atlas (TCGA) is a multi-institutional, cross-discipline effort led by the National Cancer Institute to characterize and sequence 20 cancer types at the molecular level [1]. The results, such as the discovery of new oncogenic mutations, come with the promise of clinically relevant population stratification and have recently been widened to form a coordinated international network of similarly minded initiatives [2]. TCGA is also a valuable resource for those interested in hypothesis-driven translational research as the bulk of its data results from direct experimental evidence. The level of complexity and detail of TCGA presents both an opportunity to statistically integrate the data [3] and a challenge in its representation. Heterogeneity and distribution of data sources are characteristics almost ubiquitous in biomedical datasets, which are often made available as data services without consistent data retrieval mechanisms and formats [4]. As such, advances in translational research often require complex infrastructures to integrate data from various autonomous sources and transverse several scientific domains [5]. Even when biomedical data are exposed as Web services, these tend to reflect the heterogeneity of the data, creating a challenge for its analysis with automated tools [6]. The communities of those producing and consuming biomedical data sources have mostly agreed that wide adoption of Web services that share common protocols can greatly improve data reuse and integration without the need to locally store large quantities of data [7,8]. The Linked Data best practices [9] include a collection of standards for publishing and connecting structured data on the Web that have matured to the point of providing a practical solution for the life sciences [10], namely through use of Resource Description Framework (RDF) as a data representation formalism and SPARQL as its query language [11–13].
1.1. Resource Description Framework
RDF is a generic model that relies on two key assertions: (a) that everything is a resource referenced by a Universal Resource Identifier (URI) and (b) that every resource is part of a triple [11]. A key feature of RDF is the separation between content and presentation, which makes it useful for transversing a variety of domains, organizations and data structures [14,15]. Datasets may be converted to RDF by identifying their data elements, which are assigned to URIs, and formalizing their relationships as triples of URIs. Common vocabularies and terminologies, such as those made available by the National Center for Biomedical Ontology [16], are often used to link different datasets. Projects such as Dbpedia [17], Bio2RDF [18], Neurocommons [19], Diseasome [20,21] and others already provide a large amount of linked biomedical data available as RDF [22].
1.2. Sparql
SPARQL, the schema-free RDF query language, was designed to allow queries to be expressed across diverse data sources based on data properties and the relationships established with other data elements rather than on the physical location of the data [23]. SPARQL queries are constructs of one or more three-element graph patterns, such as “?Person :hasName ?Name.”, each including a subject as the first element (?Person), a predicate as the second element (:hasName) and an object as the third element (?Name). SPARQL graph patterns support both variable elements (for example ?Person and ?Name) and non-variable elements (:hasName), where the prefix “:” indicates the Universal Resource Locator (URL) portion of a URI. The elements specific to the domain of discourse are typically the predicates (:hasName), which provide an anchor for the query. The solution to a SPARQL query is a directed labeled graph reusable in future queries. These properties make SPARQL endpoints, particularly those available as Web services, a very attractive solution for biomedical data services [22] given their recurrent need for data integration methodologies and shared queries [24]. Experimental biomedical data exposed as SPARQL endpoints can greatly facilitate discovery in the life sciences as each data source can be re-used as part of query federation approaches [25].
However, several problems have been identified that hamper exposure and query of data through SPARQL endpoints without extensive technical knowledge of RDF. Notably, SPARQL is a schema-free protocol; as such formulating a query usually requires some level of eye-parsing of the data, which hinders automation [26]. Tools such as MashQL [26] or Exhibit [27] have been developed to aid in the assembly of SPARQL queries by using the underlying RDF dataset structure.
1.3. Services for an integrative infrastructure
In this report we describe an infrastructure to expose the experimental data collected by the TCGA initiative as a programmatically accessible SPARQL endpoint. TCGA experimental datasets were broken into their fundamental data elements and assigned to entities of the Simple Sloppy Semantic Database (S3DB) management model [28]. S3DB defines entities and relationships using an RDF schema (RDFS) core model that enables encapsulation of RDF triples as part of a domain description, also represented as RDF triples [29]. This solution allows both the data elements and the description of the domain to have a representation in RDF, thereby supporting SPARQL queries formulated using the domain descriptors while targeting the data elements. It is worth noting that the processing of queries in the infrastructure developed overcomes the problems associated with a static RDF representation of the data by serializing SPARQL to S3DB's protocol and query language (S3QL). A graphical tool was developed that automatically assembles SPARQL queries while navigating the description of the domain and probing the properties of its instantiation. The intended end users of the system are researchers interested in biomarker discovery that require access to both molecular raw data and clinical covariates or researchers interested in linking their own datasets to TCGA. Usage is illustrated with a case study in which biomarker identification and its biological annotation are integrated with the Diseasome dataset [20,21]. The various components of the infrastructure are made available as Representational State Transfer (REST) Web services such that each component may be re-used independently.
2. Materials and methods
The Cancer Genome Atlas is a cancer genome characterization and sequencing project generating high-throughput molecular biology data about clinical samples. That data needs to be organized, integrated and analyzed in order to identify and characterize the genomic changes in 20 cancer types. A total of 500 samples from each type of tumor were collected, along with clinical and demographic covariates. Experiments were performed by 11 distinct genomic and sequencing characterization centers (GSCCs) to obtain data regarding miRNA expression, single nucleotide polymorphisms, exon expression, DNA methylation, copy number, trace-gene-sample relationships and somatic mutations. The publicly available TCGA datasets are deposited by individual genomic characterization and sequencing centers into a shared File Transfer Protocol (FTP) location (ftp1.nci.nih.gov).
2.1. S3DB core model URIs
The S3DB engine (http://s3db.org) was used to reassemble data elements from the TCGA initiative as RDF triples. The organizational model of S3DB defines a total of seven entities that define relationships between data elements. These are: Deployment, an entity representing an instance of an S3DB engine; User, an authenticated entity of any S3DB Deployment; Project, an entity that represents a specific domain by aggregating its entities and attributes; Collection, any entity associated with a domain that may be instantiated; Rule, the association between two Collections or between a Collection and a literal attribute; Item, an instance of a Collection; Statement, the relationship between two Items or between an Item and a literal value (see Fig. 4 in [28]). By design, each instance of an S3DB entity is automatically associated with a URI that consists of a URL (identifying the S3DB deployment in which the data are kept) concatenated with an alphanumeric identifier composed of the first character of the entity name (D, U, P, C, I, R or S) and a numeric component unique for each deployment of S3DB. The TCGA domain descriptors and their relationships, i.e. the metadata describing the data, were assigned to S3DB Collections and Rules, whereas the TCGA data elements and their attributes were assigned to S3DB Items and Statements. All assignment steps were performed using the S3DB protocol (S3QL), which supports select, insert, update and delete operations.
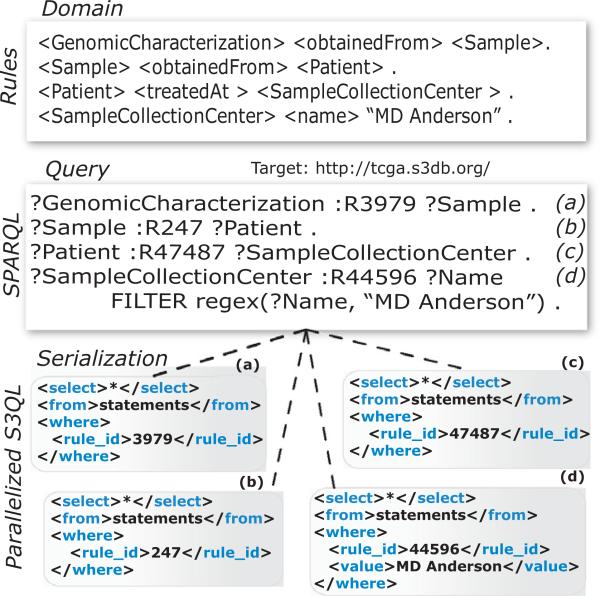
Fig. 4.
Serialization and parallelization of a SPARQL query. The description of the domain as S3DB Rules is mapped into a SPARQL query by replacing the predicate of each rule with its identifier. Each graph pattern is then serialized to its equivalent S3QL query; for example, “?GenomicCharacterization :R3979 ?Sample” is translated into http://ibl.mdanderson.org/TCGA/S3QL.php?query=<S3QL><select>*</select><from>statement</from><where><rule_id>3979</rule_id></where></S3QL>&format=rdf, equivalent to http://ibl.mdanderson.org/TCGA/S3QL/statement/rule_id=3979, and executed in parallel, with the results intersected to produce a solution.
2.2. TCGA data structures
The TCGA datasets are made available through the TCGA portal (http://cancergenome.nih.gov/) as compact assemblies of data elements with various degrees of structure: as FTP directory structures, as eXtended Markup Language (XML) and as Microarray and Gene Expression Tabular (MAGE-tab) format. MAGE-tab is a spreadsheet-based, standard format for microarray data that includes an investigation description format (IDF) file, which contains details about each experiment; a sample to data relationship format (SDRF) file, which describes the association of each sample with raw and processed data files and several files (.CEL or .txt) containing the experimental or analytical results [30]. Each format was handled separately during the process of assignment of TCGA data elements to S3DB entities. The data primer document [1] released by the TCGA consortia was used to assist in the interpretation of each archive name and code.
2.2.1. Formal representation of the TCGA workflow
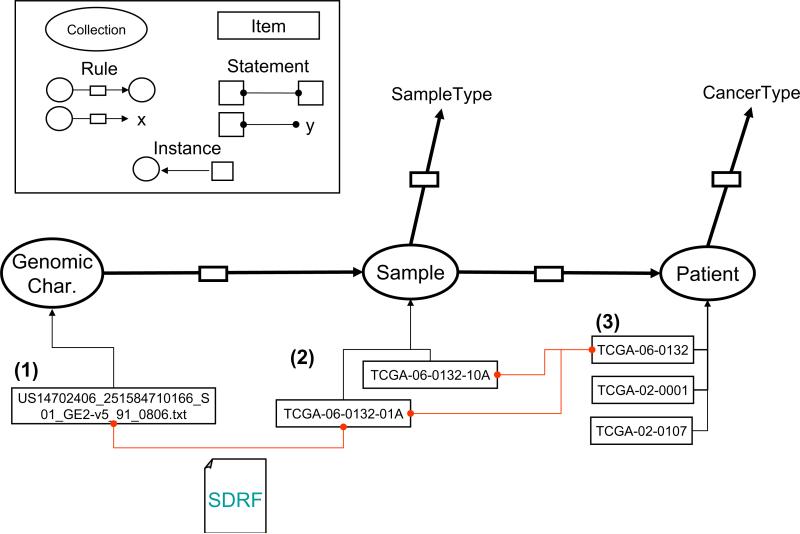
The 3 TCGA dataset formats described are generated during the course of an experimental workflow to produce genomic characterization files (Fig. 1). The workflow consists of obtaining a genomic characterization element, corresponding to the latest revision of a raw data file, from a Sample, which in turn was collected from a Patient. Data elements involved in this workflow (Genomic Characterization, Sample and Patient) were assigned to S3DB Collections whereas the relationships established between them were assigned to S3DB Rules.
Fig. 1.
Mapping the TCGA experimental workflow to S3DB entities. A Genomic Characterization element links a raw array data file containing either copy number or expression to a patient's clinical information (1–3). The filename syntax “US14702406_251584710166_S01_GE2-v5_91_0806.txt” (1) was used to link the raw data to the patient indirectly using the information in the SDRF file. In the example, the raw data is obtained from Sample “TCGA-06-0132-01A” (2), which was collected from a tumor (as indicated by “01A”) of Patient “TCGA-06-0132” (3). Each of these links was assigned to an S3DB Statement whereas the links between domain descriptors “GenomicCharacterization”, “Sample” and “Patient” were assigned to S3DB Rules.
2.2.2. Semantic caching
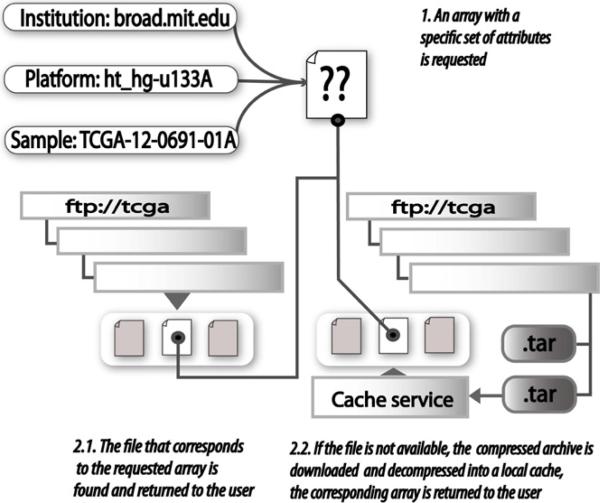
Raw TCGA genomic characterization files are compacted and distributed as compressed archives through the TCGA initiative FTP server at ftp1.nci.nih.gov. It is not uncommon for a specific revision of a file to be requested more than once in order to replicate an analysis. To avoid the need for creating a local copy and reprocessing the same large TCGA archives each time a revision is required, a caching Web service was developed as part of the infrastructure described here so that each compressed archive is downloaded only once, uncompressed and stored locally. This procedure, illustrated in Fig. 2, dynamically iterates through the FTP directory structure to discover the appropriate file associated with a specific sample given a platform, an institution and a cancer type. Each raw data file was assigned to an S3DB Statement as a symbolic link (including the required attributes) in the form http://tcga.s3db.org/TCGA-sync.php?institution=[institution_url]&platform=[platform_code]&sample_id=[sample_id]&cancer_type=[cancer_type].
Fig. 2.
A caching service for TCGA archives overcomes the need for bulk downloads. The caching service finds and retrieves the latest revision of a raw data file from the TCGA archives given an institution, a platform and a sample (1); the caching service will also retrieve a specific file revision if requested. The dynamic link generator, available at http://tcga.s3db.org/TCGAsync.php, recursively browses the TCGA datasets to return the raw data file corresponding to the requested array (2.1). If the archive has been compressed and the raw data file is not available as a symbolic link (2.2), then the TCGA archive is downloaded to the caching server, the archive is decompressed and the data file is returned. If the file has been requested previously, it is retrieved from the caching service.
2.2.3. Mapping between the TCGA datasets and S3DB entities
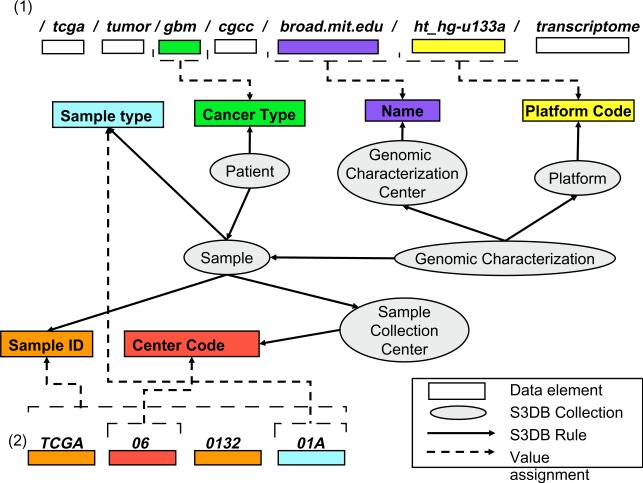
Attributes associated with each TCGA data file are obtained by recursively navigating the FTP directory structure. The symbolic directory paths that terminate in files containing data are used to retrieve attribute-values for data elements concerning the genomic characterization center, array platform, data type and archive serial index (Fig. 3.1), which are assigned to values of S3DB Statements. For example, the symbolic directory path /tcga/tumor/gbm/cgcc/broad.mit.edu/ht_hg-u133a/transcriptome/ describes the content as originating from the “tcga” initiative, specifically from a “tumor” study in which the cancer type was glioblastoma multiforme (“gbm”), the sample was collected at the Broad Institute (“broad.mit.edu”) Cancer Genomic Characterization Center (“cgcc”) and the analytical platform Affymetrix HT Human Genome U133 Array Plate Set (“ht_hg-u133a”) was used to generate “transcriptome” data.
Fig. 3.
Breaking TCGA data structures and assigning the data elements to S3DB statements. The path to raw data files (1) is separated into its constituent slash-separated portions, and the resulting data elements are assigned to values of S3DB Statements according to their position in the path. For example, the string “ht_hg-u133a” is assigned as the value for the “Platform Code” of an S3DB Statement concerning a specific Item of the Collection “Platform”. Similarly, each sample barcode (2) is broken down into its constituent elements to retrieve values for Sample ID, Sample Type and Sample Collection Center Code, among others (not shown).
Data generated by each of the participant genomic characterization centers for a given batch of patient samples and a given analytic platform are described in the MAGE-tab SDRF files, where a detailed listing of all the data files within an FTP archive can be found along with the associated sample barcodes. This index was used to establish a relationship between each raw data file and the corresponding sample (Fig. 3.2). The sample barcodes were separated into their constituent parts and used to characterize patient and sample metadata; as an example, the identifier of a sample is a 16-character barcode such as “TCGA-06-0132-01A,” where the second alphanumeric portion, “06,” was used to assign the appropriate sample collection center (“06” corresponds to the Henry Ford Hospital) and the last portion of the sample identifier, “01A,” was used to assign the tumor type of the sample (“01” corresponds to a solid tumor, “A” indicates the first vial).
Patient clinical data for TCGA samples were retrieved from the XML data files, when available, to assign clinical parameters, including biospecimen collection center barcodes, tumor tissue site, sample primary metastatic status, histological type and tumor sample anatomic location.
2.3. Availability and documentation
The complete set of S3DB Rules describing the TCGA domain is available at http://ibl.mdanderson.org/TCGA/S3QL/rule/project_id=126 and graphically at http://tcga.s3db.org/map. TCGA data elements may be browsed through the S3DB graphical interface at http://tcga.s3db.org/login by using “public” as both the user-name and password.
The element assignment procedure described here (by recursively browsing the TCGA archives and extracting information from both the FTP directory structure, the SDRF files and the clinical data XML files) was developed in PHP and is available for download, along with documentation at http://code.google.com/p/tcga2s3db/.
PHP and an S3DB deployment are necessary to execute the application. S3DB may be downloaded from http://s3db.org, having as dependencies a Web server, mySQL and PHP. Documentation for the S3QL protocol used to assign data elements to the S3DB core model is available at http://s3db.org/documentation/s3qlsyntax.
All infrastructure components are available as REST-compliant Web services: the SPARQL endpoint is available at http://tcga.s3db.org/sparql.php and the semantic caching utility is available at http://tcga.s3db.org/TCGAsync.php. A graphic user interface for SPARQL assembly is available at http://tcga.s3db.org and the resulting RDFS document is available at http://tcga.s3db.org/rdf. Two third party tools were also configured for visualizing the TCGA RDF representation: an Exhibit [31] representation is available at http://tcga.s3db.org/exhibit and Allegrograph compatibility is demonstrated in the screencast at http://www.youtube.com/watch?v=BI5bf-taGU4.
3. Results
3.1. Semantic Web services
The S3DB engine was used to mediate the exposure of experimental data from TCGA to Web services by assigning granular TCGA data elements to S3DB entities. As a consequence of this approach, two methodologies were made available that mediate the exposure of TCGA as a SPARQL endpoint: direct use of the exported data as an RDF document and serialization of SPARQL into S3DB's native query language and protocol (S3QL). Using the first method, all data that are part of an S3DB project are exported as an RDF document and queried using a SPARQL service, such as http://www.sparql.org/sparql.html with the URL for the RDF exported document (available at http://tcga.s3db.org/TCGA.rdf) in the “FROM” clause. However, Web-based repositories of experimental data, such as the TCGA datasets, are typically subject to updates, both in the amount of data and in their representation. The second method, in which the exposure of TCGA experimental data as a SPARQL endpoint is mediated by serializing SPARQL queries to S3QL, offers a flexible solution whereby data always reflect the latest state. For example, S3DB Statements that have been assigned to S3DB Rule R44596, which correspond to the “Name” attribute of a TCGA Sample Collection Center, are retrieved by the S3QL query http://ibl.mdanderson.org/TCGA/S3QL/statement/rule_id=44596. The resulting S3DB Statements are, essentially, triples in the form [Item-Rule-Item] or [Item-Rule-Literal]. Consequently, the S3QL query represented above has a SPARQL equivalent: ?SampleCollectionCenter :R44596 ?Name ., which is formulated directly from the description of the S3DB Rule R44596 used in the predicate : [SampleCollectionCenter hasName Name .]. Note that S3DB Rules are typically defined by the researchers producing the data through S3DB-associated interfaces and are not necessarily knowledgeable about Semantic Web technologies [29,32].
Fig. 4 illustrates a SPARQL query in which all genomic characterization arrays and corresponding Samples from patients treated at “MD Anderson” are retrieved. The query is assembled by mapping S3DB rules from the TCGA representation to SPARQL graph patterns and is executed through the SPARQL serialization Web service at http://tcga.s3db.org/sparql.php. The SPARQL serialization engine optimizes the time to a result by parallelizing and executing S3QL queries in stages, according to the amount of data that is expectable. For example, SPARQL patterns in which two of the three elements (subject, predicate and object) are constant, such as ?SampleCollectionCenter :R44596 ‘Henry Ford Hospital’, are executed first as they return a small number of results. Furthermore, both the computed SPARQL query result and each serialized S3QL result are cached in order to improve query performance. This cache may be deleted by indicating “&clean=1” in the URL. More SPARQL queries on the TCGA domain are available at http://tcga.s3db.org and http://s3db.org/documentation/sparql.
Whenever applicable, URIs created by assignment of TCGA domain descriptors and data elements to S3DB entities were mapped to terms from widely used controlled terminologies such as MGED Ontology [33], OBI [34] and NCI thesaurus [35]; Bioportal [36] was used to discover the appropriate terminology equivalents and a specialized extension of the S3QL protocol was devised to support the mapping of TCGA URIs to controlled terminologies (see http://s3db.org/documentation/s3qlsyntax/#TOC-Dictionary).
3.2. SPARQL endpoint interface
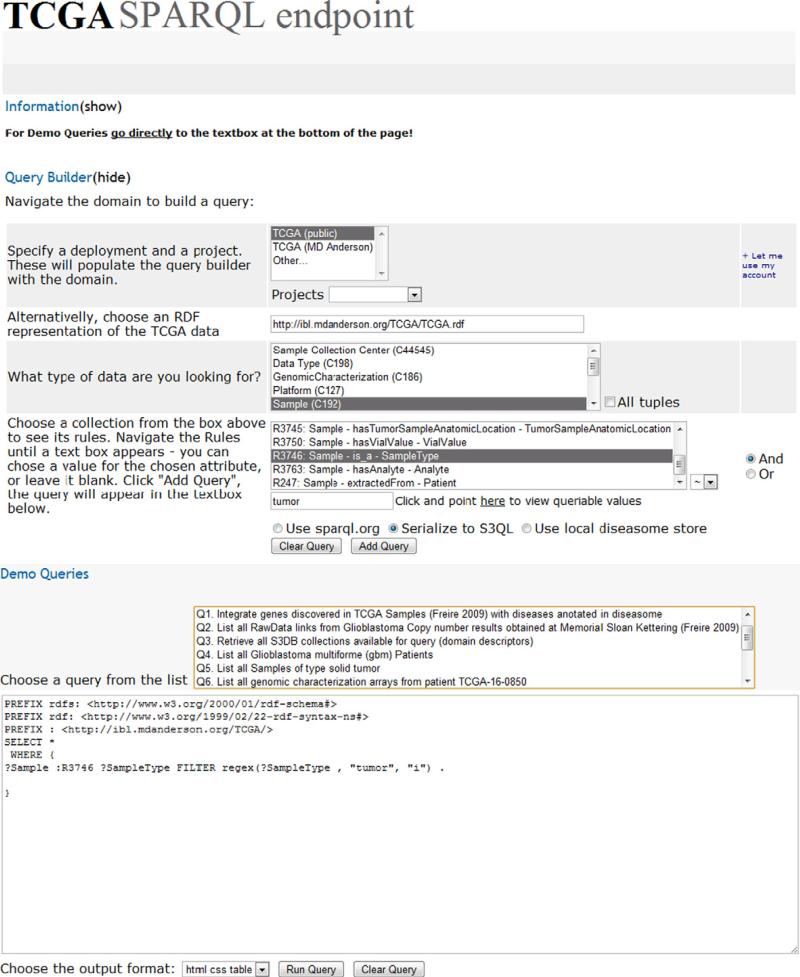
An interface to support the use of the TCGA SPARQL endpoint was developed (Fig. 5, http://tcga.s3db.org/). It relies on the navigation of the TCGA domain rule set to facilitate the construction of SPARQL queries. The interface is populated directly from S3DB Rules, therefore changes in the description of the domain are immediately reflected in the interface. The action of selecting a Collection will display the attributes available for query in the “Rules” box. A “FROM” or “GRAPH” clause may be added to the SPARQL query, as described by [23], to integrate the results with data sources external to S3DB.
Fig. 5.
A snapshot of the SPARQL interface for TCGA that automatically writes SPARQL queries by navigating the domain. Once an S3DB Collection is chosen, the Rules available for query will be displayed. Whenever an S3DB rule in which the object corresponds to an S3DB collection is chosen (for example, [Sample extractedFrom Patient]), a new rule menu is displayed. When the object of the rule is literal (for example, [Sample is_a SampleType]) a text box will appear in which a value for the chosen attribute may be fully matched (using “=”) or partially matched (using “~”). The TCGA SPARQL endpoint is available at http://tcga.s3db.org.
3.3. A copy number analysis use case
The advantage of exposing TCGA data to a SPARQL endpoint can be illustrated with the exploration of DNA copy number variation (CNV) in glioblastoma multiforme. The work presented in [3] presents a stand-alone tool that traverses, represents and analyzes CNV data for multiple tumor samples in real time using the TCGA data representation described in this report. The collection of data to perform such analysis in real time requires quick transversal of the TCGA datasets, an operation that would be challenging to integrate and correlate with clinical variables if data were retrieved directly from the TCGA FTP server. Alternatively, the data could be collected by performing a single SPARQL query in which the required parameters were configured, namely by setting the data type as “Copy Number results”, the genomic characterization center as “mskcc” and the cancer type as “gbm”. This query is illustrated in demo query Q2 at http://tcga.s3db.org. Performing the CNV analysis generated a list of 146 genes in aberrant regions, which were assigned to S3DB Statements in a related S3DB project termed TCGA analytics linked to the TCGA Samples described above (Fig. 3.2). This project includes as the main element an instance of Analysis, described as Exploratory Analysis of the Copy Number Alterations in Glioblastoma Multiforme, which functions as the intermediate between two lists: TCGA Samples that were analyzed and Genes that were found in aberrant regions. The TCGA analytics project can be visually explored at http://tcga.s3db.org/login.
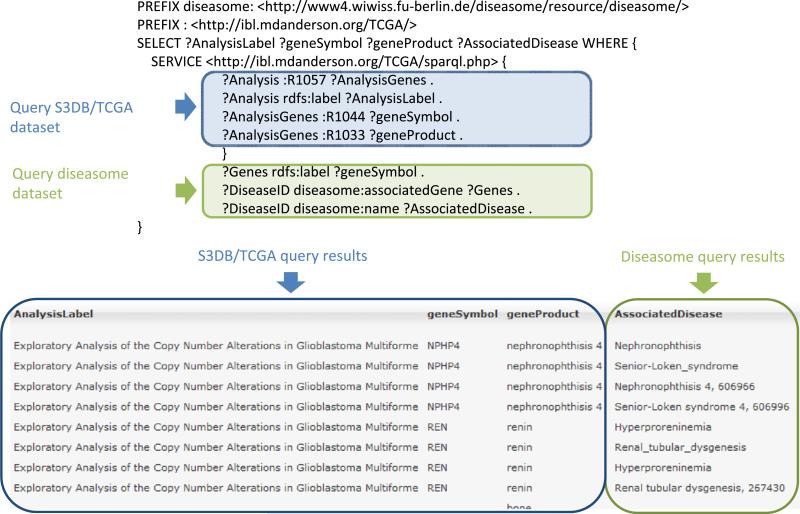
Using the retrieved gene list, a SPARQL query was devised to discover associated diseases using data from the Diseasome data-set [20] (Fig. 6). Because the SPARQL endpoint service from the Diseasome project was found to be down with some frequency, for logistic reasons we have also downloaded the Diseasome N-triple statements into an RDF store using the ARC library for PHP. The query illustrated in Fig. 6 combines the Diseasome and the TCGA datasets by making use of the ‘SERVICE’ tag [37] to retrieve a source RDF graph from a remote SPARQL endpoint. This avoids the need to locally store each data source to be queried thereby enabling SPARQL federation without forcing a static RDF representation of the data. Data from the two datasets is linked by means of the National Center for Biotechnology Information (NCBI) gene symbol. From the resulting integrated data it can be observed that a total of 72 diseases are associated with the same genes discovered in aberrant regions of glioblastoma multiforme, the most common being leukemia and melanoma with 3 concurrent observations.
Fig. 6.
A query to integrate TCGA derived gene list and the Diseasome dataset. The SPARQL ‘SERVICE’ tag is used to retrieve data from the TCGA SPARQL endpoint in real time without the need to create a local representation of the complete RDF graph. Illustrating the effective aggregation of the two distinct data sources, the output of the query includes both data collected from the S3DB/TCGA source and data from the diseasome project. Complete query results are available at http://tcga.s3db.org (demo query Q1).
3.4. System architecture
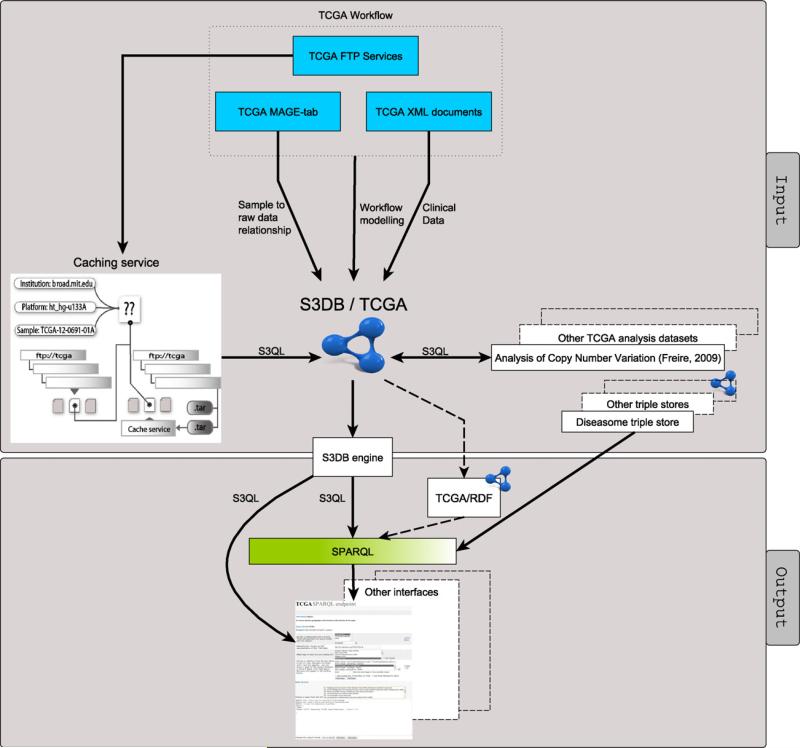
The overall architecture of the infrastructure developed including input and output components is depicted in Fig. 7. TCGA domain descriptors were manually assigned to S3DB Rules using S3DB-associated interfaces, and individual data elements were programmatically retrieved from 3 types of TCGA data structures (FTP directory, XML files and MAGE-tab format). The caching service, described in Fig. 2, was successfully tested as a buffer for scalability problems caused by the need to transfer large archives and frequent modifications in the structure of the FTP directory. The analytical results of the copy number variation use case were linked to the original samples providing the data by assignment to S3DB entities. It is worth noting that the Diseasome dataset used to discover diseases associated with aberrant genes in glioblastoma multiforme, described in Fig. 6, is not a component of the S3DB/TCGA system. Integration with the Diseasome dataset is achieved by using the proposed W3C standard “SERVICE” tag in the SPARQL query.
Fig. 7.
Overview of the infrastructure developed to expose experimental data from The Cancer Genome Atlas as a SPARQL endpoint. The system components are divided into input and output components. The input components are directed mainly at breaking three types of TCGA data structures into their data elements while assigning them to S3DB entities. The output components are aimed at providing application programming interfaces for extracting data assigned to S3DB entities.
Integrated data may be retrieved from the S3DB engine either by obtaining the complete TCGA/RDF representation or through a SPARQL endpoint in which the query is primarily serialized into its S3QL equivalent (see Fig. 4). Additionally, data may be retrieved by direct use of S3QL (see http://s3db.org/documentation/s3qlsyntax for documentation on S3QL).
4. Discussion
An infrastructure has been developed to programmatically expose clinical and molecular data generated by The Cancer Genome Atlas project to Linked Data Web best practices, in particular as a SPARQL endpoint. The proposed solution makes use of the S3DB management model by assigning TCGA data elements and domain descriptors to entities of the S3DB RDF Schema core model. Specifically, the TCGA data elements were assigned to S3DB Statements, which in turn instantiate a separate set of domain descriptors, the S3DB Rules (Figs. 1 and 3). Resulting domain descriptors were mapped to terms from widely used controlled vocabularies and a Web application was developed to assemble REST-full SPARQL calls by navigating the description of the TCGA domain (Fig. 5).
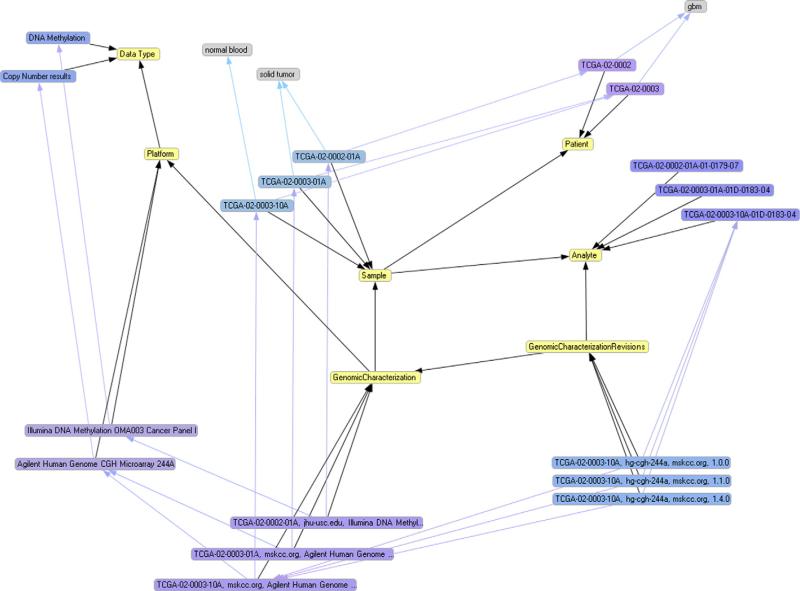
The formal representation of experimental data from TCGA using the RDF model facilitates the task of data integration at various levels when compared to current data integration practices. One of the key benefits of the RDF model for data discovery is the reorganization of data according to its relevance in the domain rather than by content management needs. As an example, the most relevant data elements in TCGA, the raw data files that represent the outcome of a genomic characterization experiment, are retrieved by assembling a single intuitive SPARQL query such as the one in Fig. 4. Linked Data queries may thus be formulated in terms of the workflow pursued to collect the data, i.e. using SPARQL variables such as “?Sample” or “?Patient”, rather than in terms of the infrastructure used to represent it. To retrieve those same data elements from the original portal, it would be necessary to browse through several data structures in several data processing steps. A second key benefit of using the RDF model is its flexibility in establishing links with datasets that were generated with different purposes. For example, integration of the TCGA experimental dataset and the Diseasome dataset collected by the Human Disease Network includes a list of 72 diseases that could potentially be related to glioblastoma multiforme because they share the same aberrant genes. A common impediment to the adoption of RDF in information management systems derives from the same decoupling of content and presentation that grant flexibility to RDF, which results in the absence of a clearly defined data schema to be used as an anchor for queries. Often some ‘eye-parsing’ of the data is required in order to formulate a query [26]. A key feature of the solution proposed here is the separation of the domain descriptors from their instantiation, provided by the assignment of data elements to the S3DB management model [28,29]. Asan illustration, a graphical RDF representation tool, Sentient Knowledge Explorer, was used to generate Fig. 8 from the complete RDF graph of the TCGA datasets; the domain descriptors (yellow nodes) are clearly separate from the data elements (blue and grey nodes such as “Affymetrix HT Human Genome U133 Array Plate Set”) because they are assigned to entities from the S3DB core model.
Fig. 8.
A fragment of the RDF graph representation of TCGA created by the software tool Sentient Knowledge Explorer. Yellow nodes represent S3DB Collections, blue and purple nodes represent S3DB Items and grey nodes represent values of properties of the items. Light blue lines represent the relationships between items; black lines represent the connections between the elements of the core. Assignment of TCGA data elements to S3DB core elements results in a directed labeled graph in which the domain (yellow nodes) is clearly separate from its instantiation (blue and grey nodes). The separation of the domain facilitates the assembly of SPARQL queries, as it separates what is an actual data element (for example, sample “TCGA-02-0002-01A”) from what is a domain descriptor (for example, the yellow node “Sample”).
The S3DB constraint that requires S3DB statements (blue lines) to be instances of S3DB rules (black lines) is an intermediate step that greatly facilitates the relational algebra exercise of assembling SPARQL to retrieve TCGA contents. As an example, the SPARQL query presented in Fig. 4 has an intuitive syntax that is built from the description of the domain; that is, it emerges from mapping a user-defined Rule such as [Genomic Characterization obtained From Sample] (identified as R3979) to the SPARQL triple [?GenomicCharacterization :R3979 ?Sample]. The development of Web applications that generate SPARQL queries based on user-defined domains, an example of which is available at http://tcga.s3db.org/ (Fig. 5), therefore becomes an exercise of mapping the description of the domain, assigned to S3DB Rules, to SPARQL graph patterns.
An additional outcome of annotating TCGA domain descriptors to S3DB Rules is the creation of an intermediate layer between analytical applications and raw data. This prevents changes in the original TCGA FTP structure, such as compressing archives, from affecting data retrieval. An extreme example of rewiring an FTP directory would force only a small change in the automated assignment procedure; however, it would not affect query functionality. The description of S3DB Rules can also be freely edited, because the relationship between the domain descriptors and their data elements are established using alphanumeric identifiers rather than descriptive terms [29].
Finally, it is worth noting that by developing the Web service at http://tcga.s3db.org/ based on S3DB, an open-source biological management tool [38], it benefits not only from the REST protocols described but also from code portability, distribution with peer-to-peer interoperability and the availability of queries on protected data given the appropriate credentials (username and password), requirements that data management systems for biomedical domains must be able to juggle. Because the S3DB engine is distributed as open source, users of the system may choose to replace the default TCGA S3DB deployment with any S3DB deployment of their choice, where the domain may be freely configured and extended. Changes to S3DB Rules are immediately reflected in the SPARQL configuration tool, allowing researchers with a focus on TCGA dataset analysis to attempt alternative configurations that may be useful for their own purposes. Users of the system may opt to simultaneously query the public TCGA data and protected data that would otherwise not be exposed to the Linked Data Web. These features have made the S3DB system and its SPARQL endpoint an attractive data service solution for client-side analytical platforms such as the Cancer Genome Browser [3]. The availability of a SPARQL serialization engine enables data annotated to S3DB to be immediately available for query without the need for generating an RDF document. Researchers interested in integrating their own gene list results with the Diseasome dataset using the SPARQL service provided with every S3DB deployment need only assign their genes to S3DB statements.
4.1. System scalability and query performance
We have evaluated query performance of the system with a SPARQL query that was tested on TCGA data using both the SPARQL serialization engine described (see Section 3, Fig. 4) and a SPARQL engine without serialization using the complete TCGA/RDF representation as the data source. A screencast with these results is available at: http://www.youtube.com/watch?v=yrkA4uAT5GY. When the queries were executed simultaneously, the SPARQL serialization engine returned a result in approximately half the time required for the SPARQL engine without serialization.
4.2. Combining approaches to query federation
The cancer Biomedical Informatics Grid (caBIG®) is a project aimed at enabling the sharing of cancer-related data using a federated query model, whereby different institutions working on related problems can share data either by adapting their local repositories to a set of data models provided by caBIG or by adopting one of the caBIG applications [39]. The various data sources in caBIG can then be queried simultaneously using the caGrid query language [40]. The caBIG approach offers the advantage of facilitating query assembly because it relies on a set of common data structures; this approach is therefore indicated for knowledge fields that are very well established. The linked data approach [9] does not impose a data model before integration is possible; instead data can be integrated using SPARQL queries as soon as an RDF representation is available. Although the latter approach requires that datasets be linked through the use of common terminologies before the assembly of SPARQL queries, it is better suited for knowledge areas that evolve quickly as it benefits from having novel data immediately available for integration. The two approaches could therefore greatly benefit from each other; indeed, a call for Semantic Web opportunities has been launched by the caBIG community [41]. The semCDI [42] and Corvus [43] projects, for example, have already developed extensive work towards modeling and integrating the various data models available at caBIG including the availability of SPARQL engines. The architecture described in this report could thus be easily integrated with caBIG datasets that are made available as RDF or if a SPARQL endpoint is provided. Indeed, one of the steps in that direction was mapping the terms within TCGA S3DB representation to NCI Thesaurus, also widely used by the caBIG community [44,45].
4.3. Limitations to the proposed solution
The work presented here attempts to provide a formal linked representation of the TCGA datasets that can be queried using SPARQL. However, datasets that are unavailable to the public may not be included in the RDF representation due to the academic nature of this work.
TCGA domain elements were mapped to biomedical ontologies whenever possible using RDF Schema and Web Ontology Language. We believe that this mapping is necessarily incomplete because it can only reflect a snapshot of currently available ontologies and will most likely not address all the needs of application development. As a consequence, the dictionary extension to the S3QL protocol was devised to enable extending the mapping beyond the S3DB schema, while still enabling mapped resources to be queried through the SPARQL serialization engine.
Acknowledgments
We thankfully acknowledge IO-informatics for providing a Sentient Knowledge Explorer license to the Integrative Bioinformatics Laboratory of The University of Texas M. D. Anderson Cancer Center. We thank Rebecca Partida for reviewing the manuscript. We would also like to acknowledge two anonymous reviewers, who provided valuable insights towards the improvement of this report.
This work was funded by the Fundação para a Ciência e Tecnologia and the Center for Clinical and Translational Sciences under contracts SFRH/BD/45963/2008 and 1UL1RR024148, respectively. The work was also supported in part by the National Heart, Lung and Blood Institute and by the National Cancer Institute of the US National Institutes of Health under contracts N01-HV-28181 and P50 CA70907, respectively.
References
- 1.TCGA Data Primer Version 1.0. < http://tcga-data.nci.nih.gov/docs/TCGA_Data_Primer.pdf>.
- 2.The International Cancer Genome Consortium International network of cancer genome projects. Nature. 2010;464:993–8. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freire P, Vilela M, Deus H, Kim Y-W, Koul D, Colman H, et al. Exploratory analysis of the copy number alterations in glioblastoma multiforme. PLoS ONE. 2008;3:e4076. doi: 10.1371/journal.pone.0004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephens SM, Rung J. Advances in systems biology: measurement, modeling and representation. Curr Opin Drug Discov Devel. 2006;9:240–50. [PubMed] [Google Scholar]
- 5.Ruttenberg A, Clark T, Bug W, Samwald M, Bodenreider O, Chen H, et al. Advancing translational research with the Semantic Web. BMC Bioinformatics. 2007;8(Suppl. 3):S2. doi: 10.1186/1471-2105-8-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandervalk BP, McCarthy EL, Wilkinson MD. Moby and Moby 2: creatures of the deep (web). Brief Bioinform. 2009;10:114–28. doi: 10.1093/bib/bbn051. [DOI] [PubMed] [Google Scholar]
- 7.Baker CJO, Cheung K-H. Semantic web: revolutionizing knowledge discovery in the life sciences. Springer; New York: 2007. [Google Scholar]
- 8.Goble C, Stevens R, Hull D, Wolstencroft K, Lopez R. Data curation + process curation=data integration + science. Brief Bioinform. 2008;9:506–17. doi: 10.1093/bib/bbn034. [DOI] [PubMed] [Google Scholar]
- 9. Linked Data. < http://www.w3.org/DesignIssues/LinkedData.html>.
- 10.Stephens S, Morales A, Quinlan M. Applying Semantic Web technologies to drug safety determination. Ieee Intell Syst. 2006;21:82–6. [Google Scholar]
- 11. Semantic Web Roadmap. < http://www.w3.org/DesignIssues/Semantic.html>.
- 12. W3C. Semantic Web Best Practices and Deployment Working Group. < http://www.w3.org/2001/sw/BestPractices/>.
- 13.Wang X, Gorlitsky R, Almeida JS. From XML to RDF: how semantic web technologies will change the design of ‘omic’ standards. Nat Biotechnol. 2005;23:1099–103. doi: 10.1038/nbt1139. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez MA, Watkins JH, Bollen J, Gershenson C. Using RDF to model the structure and process of systems. InterJ Complex Sys. 2007:2131. [Google Scholar]
- 15.Powers S. Practical RDF: solving problems with the resource description framework. O'Reilly & Associates Inc.; 2003. [Google Scholar]
- 16.Rubin DL, Lewis SE, Mungall CJ, Misra S, Westerfield M, Ashburner M, et al. National Center for Biomedical Ontology: advancing biomedicine through structured organization of scientific knowledge. Omics. 2006;10:185–98. doi: 10.1089/omi.2006.10.185. [DOI] [PubMed] [Google Scholar]
- 17.Auer S, Bizer C, Kobilarov G, Lehmann J, Cyganiak R, Ives Z. DBpedia: a nucleus for a web of open data. Semantic Web, Proc. 2007;4825:722–35. [Google Scholar]
- 18.Belleau F, Nolin MA, Tourigny N, Rigault P, Morissette J. Bio2RDF: towards a mashup to build bioinformatics knowledge systems. J Biomed Inform. 2008;41:706–16. doi: 10.1016/j.jbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Ruttenberg A, Rees JA, Samwald M, Marshall MS. Life sciences on the Semantic Web: the neurocommons and beyond. Brief Bioinform. 2009;10:193–204. doi: 10.1093/bib/bbp004. [DOI] [PubMed] [Google Scholar]
- 20. D2R Server publishing the Diseasome Dataset. < http://www4.wiwiss.fu-berlin.de/diseasome/>.
- 21.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci USA. 2007;104:8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goble C, Stevens R. State of the nation in data integration for bioinformatics. J Biomed Inform. 2008;41:687–93. doi: 10.1016/j.jbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 23. SPARQL Query Language for RDF – W3C Recommendation. < http://www.w3.org/TR/rdf-sparql-query/>.
- 24.Cheung KH, Prud'hommeaux E, Wang Y, Stephens S. Semantic Web for Health Care and Life Sciences: a review of the state of the art. Brief Bioinform. 2009;10:111–3. doi: 10.1093/bib/bbp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung KH, Frost HR, Marshall MS, Prud'hommeaux E, Samwald M, Zhao J, et al. A journey to Semantic Web query federation in the life sciences. BMC Bioinformatics. 2009;10(Suppl. 10):S10. doi: 10.1186/1471-2105-10-S10-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarrar M, Dikaiakos MD. MashQL: a query-by-diagram topping SPARQL.. Proceeding of the 2nd international workshop on ontologies and information systems for the semantic web.; Napa Valley, California, USA: ACM. 2008. [Google Scholar]
- 27. Exhibit: Publishing Framework for Data-Rich Interactive Web Pages. < http://www.simile-widgets.org/exhibit/>.
- 28.Deus HF, Stanislaus R, Veiga DF, Behrens C, Wistuba II, Minna JD, et al. A semantic web management model for integrative biomedical informatics. PLoS ONE. 2008;3:e2946. doi: 10.1371/journal.pone.0002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almeida JS, Chen C, Gorlitsky R, Stanislaus R, Aires-de-Sousa M, Eleuterio P, et al. Data integration gets ‘Sloppy’. Nat Biotechnol. 2006;24:1070–1. doi: 10.1038/nbt0906-1070. [DOI] [PubMed] [Google Scholar]
- 30.Rayner TF, Rocca-Serra P, Spellman PT, Causton HC, Farne A, Holloway E, et al. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huynh DF, Karger DR, Miller RC. Exhibit: lightweight structured data publishing.. Proceedings of the 16th international conference on World Wide Web.; Banff, Alberta, Canada: ACM. 2007. [Google Scholar]
- 32. S3DB documentation: webS3DB. < http://s3db.org/documentation/webs3db>.
- 33.Stoeckert CJ, Parkinson H. The MGED ontology: a framework for describing functional genomics experiments. Comp Funct Genomics. 2003;4:127–32. doi: 10.1002/cfg.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. The ontology for biomedical investigations. < http://obi-ontology.org/page/Main_Page>.
- 35.Sioutos N, de Coronado S, Haber MW, Hartel FW, Shaiu WL, Wright LW. NCI thesaurus: a semantic model integrating cancer-related clinical and molecular information. J Biomed Inform. 2007;40:30–43. doi: 10.1016/j.jbi.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith N, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009;37:W170–3. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. SPARQL 1.1 Federation Extensions. < http://www.w3.org/2009/sparql/docs/fed/service#introduction>.
- 38.Antezana E, Kuiper M, Mironov V. Biological knowledge management: the emerging role of the Semantic Web technologies. Brief Bioinform. 2009;10:392–407. doi: 10.1093/bib/bbp024. [DOI] [PubMed] [Google Scholar]
- 39.caBIG-Community. An Introduction to caGrid Technologies and Data Sharing In.
- 40. caGrid Query Language (CQL) documentation. < http://cagrid.org/display/dataservices/CQL>.
- 41. caBIG Semantic Web Opportunities. < https://wiki.nci.nih.gov/display/VCDE/caBIG+Semantic+Web+Opportunities>.
- 42.Shironoshita EP, Jean-Mary YR, Bradley RM, Kabuka MR. SemCDI: a query formulation for semantic data integration in caBIG. J Am Med Inform Assoc. 2008;15:559–68. doi: 10.1197/jamia.M2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCusker JP, Phillips JA, Gonzalez Beltran A, Finkelstein A, Krauthammer M. Semantic web data warehousing for caGrid. BMC Bioinformatics. 2009;10(Suppl. 10):S2. doi: 10.1186/1471-2105-10-S10-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cimino JJ, Hayamizu TF, Bodenreider O, Davis B, Stafford GA, Ringwald M. The caBIG terminology review process. J Biomed Inform. 2009;42:571–80. doi: 10.1016/j.jbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. caBIG Enterprise Vocabulary Services (EVS). < https://cabig.nci.nih.gov/concepts/EVS/>.