Abstract
DREB (dehydration-responsive element-binding protein) transcription factors have important roles in the stress-related regulation network in plants. A DREB orthologue, GmDREB3, belonging to the A-5 subgroup of the DREB subfamily, was isolated from soybean using the RACE (rapid amplification of cDNA ends) method. Northern blot analysis showed that expression of GmDREB3 in soybean seedlings was induced following cold stress treatment for 0.5 h and was not detected after 3 h. However, it was not induced by drought and high salt stresses or by abscisic acid (ABA) treatment. This response was similar to those of members in the A-1 subgroup and different from those of other members in the A-5 subgroup, suggesting that the GmDREB3 gene was involved in an ABA-independent cold stress-responsive signal pathway. Furthermore, analysis of the GmDREB3 promoter elucidated its cold-induced modulation. A promoter fragment containing bases −1058 to −664 was involved in response to cold stress, and its effect was detected for 1 h after treatment, but a transcriptional repressor appeared to impair this response by binding to a cis-element in the region −1403 to −1058 at 24 h after the beginning of cold stress. Moreover, the GmDREB3 protein could specifically bind to the DRE element in vitro, and activated expression of downstream reporter genes in yeast cells. In addition, overexpression of GmDREB3 enhanced tolerance to cold, drought, and high salt stresses in transgenic Arabidopsis. Physiological analyses indicated that the fresh weight and osmolality of GmDREB3 transgenic Arabidopsis under cold stress were higher than those of wild-type controls. GmDREB3 transgenic tobacco accumulated higher levels of free proline under drought stress and retained higher leaf chlorophyll levels under high salt stress than wild-type tobacco. In addition, constitutive expression of GmDREB3 in transgenic Arabidopsis caused growth retardation, whereas its expression under control of the stress-inducible Rd29A promoter minimized negative effects on plant growth under normal growth conditions, indicating that a combination of the Rd29A promoter and GmDREB3 might be useful for improving tolerance to environmental stresses in crop plants.
Keywords: Abiotic stress, DREB transcription factor, drought tolerance, gene function, soybean
Introduction
Plants encounter variable environmental stresses such as cold and osmotic stresses that may affect normal growth and productivity. Investigations on physiological, biochemical, and molecular aspects of plant stress tolerance have unravelled aspects of the intrinsic mechanisms developed during evolution to mitigate against stresses (Vij and Tyagi, 2007). So far, 299 drought-inducible genes, 213 high-salt stress-inducible genes, and 54 cold-inducible genes have been identified in Arabidopsis (Seki et al., 2002; Shinozaki and Yamaguchi-Shinozaki, 2003). Among these genes, many are transcription factors involved in responses to drought, high salt, and cold stresses, and regulate the expression of downstream target genes through specific binding to cis-acting elements in the promoters of down-regulated genes (Liu et al., 1998; Bartels and Sunkar, 2005; Vinocur and Altman, 2005). These genes were classified into several large families, such as AP2/EREBP, bZIP, NAC, MYB, MYC, Cys2His2 zinc-finger, and WRKY (Umezawa et al., 2006).
AP2/EREBP is an important transcription factor family. In Arabidopsis, 145 AP2/EREBP transcription factors were classified into five subfamilies, including DREB(dehydration-responsive element-binding protein), ERF (ethylene-responsive transcription factor), AP2 (APETALA 2), RAV (related to ABI3/VP1), and one very specific gene, AL079349, based on the similarities of their DNA-binding domain (AP2 domain). Genes belonging to the DREB subfamily were thought to be important switches to regulate expression of many stress-inducible genes. The group was further divided into six subgroups (A-1–A-6), among which DREB1/CBF (C-repeat binding factor)-like genes, belonging to the A-1 subgroup, are induced by low temperature and activate the expression of many cold stress-responsive genes, whereas DREB2-like genes, belonging to the A-2 subgroup, are mainly involved in osmotic stress-responsive gene expression (Sakuma et al., 2002; Nakashima and Yamaguchi-Shinozaki, 2006). Currently, homologous DREB1/CBF genes have been identified in a variety of plants, such as Arabidopsis, common wheat (Triticum aestivum L.), rice (Oryza sativa L.), rye (Secale cereale L.), and maize (Zea mays L.), and overexpression of these genes in transgenic plants increases tolerance to drought, high salt, and freezing stresses (Dubouzet et al., 2003; Shen et al., 2003; Qin et al., 2004; Zhang et al., 2004; Hong and Kim, 2005; Benedict et al., 2006; Zhao et al., 2006). Thus, it is apparent that the DREB1/CBF regulon is conserved and plays a key role in regulating stress responses of higher plants, and that DREB1/CBF-like genes may be useful for improving the stress tolerance of crops (Yamaguchi-Shinozaki and Shinozaki, 2006). Except for DREB1/CBF-like and DREB2-like genes, the characteristics and functions of members of the other subgroups in the DREB subfamily remain to be studied. Novel members belonging to the A-5 subgroup, such as PpDBF1, GmDREB2, and GhDBF1, have been identified (Huang and Liu, 2006; Chen et al., 2007; Liu et al., 2007). For example, PpDBF1 from the moss Physcomitrella patens was induced by drought, high salt, cold stresses, and abscisic acid (ABA) treatment, and overexpression of this gene enhanced tolerance of transgenic plants to drought, high salt, and cold stresses, but did not cause growth retardation (Liu et al., 2007). A soybean GmDREB2 gene, whose expression and function under various abiotic stresses were similar to those of PpDBF1 (Chen et al., 2007), were also isolated, suggesting that members of the A-5 subgroup, like those in A-1, are important genetic resources for improving stress tolerance in crop plants. However, more members of this subgroup need to be identified in order to elucidate their function and regulation mechanisms in plants tolerant to various stresses.
The DREB pathway plays an important role in the stress-responsive regulation network of plants (Cook et al., 2004), and important insights about the regulatory mechanisms are beginning to emerge. A transcription factor gene, Inducer of CBF Expression 1 (ICE1), encoding a MYC-like basic helix–loop–helix (bHLH) protein, binds specifically to MYC sites in the DREB1A/CBF3 promoter region and increases the expression of DREB1A/CBF3, which in turn activates expression of many downstream genes, leading to a significantly enhanced tolerance to chilling and freezing (Chinnusamy et al., 2003). However, an ice1 mutation had little effect on cold-induced accumulation of DREB1C/CBF2 transcripts, and two sequences, designated as ICEr1 and ICEr2 (induction of CBF expression region 1 or 2), in the promoter of DREB1C/CBF2 stimulated transcription of DREB1C/CBF2 in response to cold stress (Zarka et al., 2003). In addition to MYC-like bHLH proteins, other proteins affecting the expression of DREB1/CBF-like genes were also identified. For example, the gene LOS4 encoding a DEAD-box RNA helicase had a positive role in regulating DREB1/CBF expression (Gong et al., 2002). In contrast, FRY2 (Xiong et al., 2002) and HOS1 (Lee et al., 2001) appeared to down-regulate DREB1/CBF expression. FRY2, encoding a transcriptional repressor, showed limited homologies to the genes of yeast and human C-terminal domain (CTD) phosphatases that were recently found to be involved in gene transcription and pre-mRNA processing, and HOS1, encoding a RING finger protein, might be an E3 ligase involved in ubiquitination and protein degradation of DREB1/CBF proteins (Lee et al., 2001; Xiong et al., 2002). Moreover, DREB1C/CBF2 negatively regulated the expression of DREB1B/CBF1 and DREB1A/CBF3 (Novillo et al., 2004). These results indicate that expression of DREB genes is tightly controlled by a complex gene network, which guarantees the correct induction of downstream genes and precise development of tolerance to freezing and other stresses. However, the regulatory mechanisms of most of the DREB genes remain unclear, probably due to the lack of focus on the promoters.
In this study, a DREB gene, GmDREB3, belonging to the A-5 subgroup, was isolated from soybean. Northern blot analysis showed that GmDREB3 was induced only by cold stress, and its overexpression in transgenic plants increased their tolerance to cold, drought, and high salt stresses, similar to DREB1/CBF-like genes, but obviously different from other members in the A-5 subgroup. In addition, promoter analysis of GmDREB3 showed that a promoter segment, from bp −1058 to −664, was sufficient to activate cold-responsive expression, but elements in region −1403 to −1058 might work with a transcriptional repressor to impair this activity, suggesting that both transcriptional activators and repressors are involved in fine-tuning expression of GmDREB3 in response to cold stress.
Materials and methods
Plant materials and growth conditions
Arabidopsis plants (genotype Colombia) used for transformation were grown in soil at 22 °C and ∼70% humidity under 14 h light and 10 h darkness. T1 seeds were surface-sterilized and planted on MS medium supplemented with 50 μg ml−1 kanamycin for the selection of transgenic plants. After emergence, seedlings of transgenic Arabidopsis plants were transferred to pots for further functional analyses. Tobacco (Nicotiana tabacum L., genotype W38) seedlings grown on MS medium were used for transformation. After transgenic plants were identified by PCR, seedlings were transferred to pots and grown under a 12 h light:12 h darkn:25 °C regime for further functional analyses.
Isolation of the GmDREB3 gene
In order to isolate the genes encoding DREB from soybean, an AP2 consensus peptide sequence was used as a query to search the expressed sequence tag (EST) database of soybean (http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=soybean). A total of 103 EST sequences containing AP2 domains were obtained and further systematic phylogenetic analyses of those sequences were carried out on the basis of homology of AP2 domains. In order to study the characteristics and functions of a member belonging to the A-5 subgroup of the DREB subfamily, an EST sequence belonging to the A-5 subgroup was chosen for further analyses (EST code in GenBank: BQ629398). Using the RACE (rapid amplification of cDNA ends) method, the full-length cDNA sequence, designated GmDREB3 (GenBank accession no. DQ208969), was isolated from total RNA of soybean cv. Tiefeng 8 (a salt-tolerant soybean cultivar). RACE was carried out as described in the Instruction Manual (Rapid Amplification of cDNA Ends System, Gibco-BRL, Rockville, MD, USA).
Application of abiotic stresses and northern blot analysis
Soybean cv. Tiefeng 8 plants were used for gene isolation and expression pattern analyses. Soybean seeds were planted in pots, irrigated with water, and subjected to a 12 h light:12 h dark:25 °C regime. To determine the expression pattern of GmDREB3 under high salt stress, some 2-week-old soybean plants were removed from the soil. An initial sample represented an untreated control (or high salt treatment at 0 h). Other soybean plants were soaked in a solution containing 250 mM NaCl for various time periods prior to sampling. For ABA treatment, leaf tissues of soybean plants were sprayed with 200 μM ABA solution and then sampled after different time intervals. For drought treatment, soybean plants were sampled at 0 h; other soybean plants were sampled after being placed on filter paper for various time periods. For cold treatment, soybean plants were placed in a refrigerator at 4 °C under dim light. After exposure to stresses, samples were immediately frozen in liquid nitrogen for later analysis of expression patterns. Total RNA was extracted from plant samples harvested after different time points as described by Zhang et al. (1996). About 30 μg of total RNA was fractionated in a 1% (w/v) agarose gel containing formaldehyde and then transferred onto a Hybond-N+ nylon membrane in 20× SSC. The GmDREB3 probe was labelled with [α-32P]dCTP and the Random Primer DNA Labeling Kit (TaKaRa Biotech, Dalian, China). Hybridization was performed as described in the Instruction Manual for the Hybond-N+ nylon membrane filter (Amersham Biosciences, Piscataway, NJ, USA).
Isolation and activity analysis of the GmDREB3 promoter
The promoter fragment of GmDREB3 was isolated from the soybean genome using the SiteFinding-PCR method (Tan et al. 2005). Promoter sequence analysis was performed using the PLACE Signal Scan Search Program (http://www.dna.affrc.go.jp/PLACE/signalscan.html). According to the predicted position of the cis-element in the GmDREB3 promoter, five fragments deleted from the 5′ end of the promoter containing bp −1587 to +41, −1403 to +41, −1058 to +41, −644 to +41, and −226 to +41 were amplified, and separately inserted into binary vector pBI121 (Clontech, Mountain View, CA, USA) to replace the CaMV 35S (cauliflower mosaic virus 35S) promoter upstream of the GUS (β-glucuronidase) reporter gene. These vectors and a positive control pBI121 were transferred into calli induced from mature embryos of wheat (cv. Yumai 66) using a biolistic particle acceleration device (PDS 1000/He, BioRad, Hercules, CA, USA) under a chamber pressure of 27 mmHg and 6 cm distance from the rupture disc to the microcarriers (Vasil et al., 1993). After transformation, calli were held on MS medium plus 100 μM ABA, 200 mM NaCl, or low temperature (4 °C) conditions for 1 h and 4 d, respectively. Treated calli were then stained with GUS staining solution (Jefferson et al., 1987) to identify GUS activity, which was quantified using a Lambda 35 UV/VIS Spectrometer (Perkin Elmer, Foster, City, CA, USA) as described previously (Facchini et al., 1996). The relative GUS activity was calculated as the ratio of GUS activities of the deleted GmDREB3 promoter series to that of the CaMV 35S promoter (pBI121) under the same stress treatments. This transient expression assay was repeated three times for each construct within each treatment.
Preparation and electrophoretic mobility shift assays (EMSA) of glutathione S-transferase (GST) fusion proteins
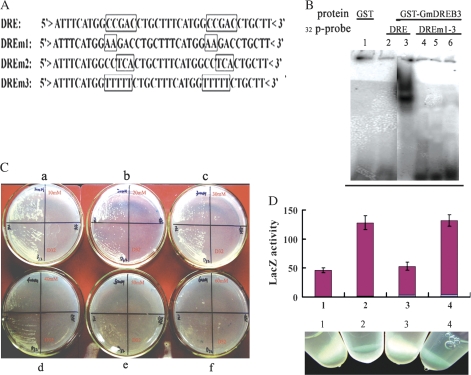
The 528 bp fragment of GmDREB3 containing the DNA-binding domain was amplified using the primer pair: GmDREB3PP-1, 5′-CCCTCTAGAGAATTCATGGCGAAACCCAGCAGC-3′ (forward); and GmDREB3PP-2, 5′-CCCCTCGAGCGGCATTT CCGGCACATA-3′ (reverse). This fragment was cloned into the EcoRI–XhoI site of the pGEX4T-1 vector (Amersham Biosciences) and transferred into Escherichia coli BL21 cells (Amersham Biosciences) to produce a GST fusion protein. The GST fusion protein was purified using a glutathione–Sepharose 4B column (Amersham Biosciences) according to the manufacturer's instructions. EMSA was completed as described previously (Liu et al., 1998). The 37 bp DNA fragment containing two copies of the wild-type or mutant DRE elements was synthesized (Fig. 3A). The DNA fragment was labelled by filling in 5′ overhangs with [γ-32P]dCTP (25 μCi μl−1; Amersham Biosciences). The DNA-binding reaction was allowed to proceed for 30 min at 25 °C in 20 μl of binding buffer [25 mM HEPES/KOH, pH 7.9, 50 mM KCl, 0.5 mM dithiothreitol (DTT), 0.5 mM EDTA, 5% (w/v) glycerol, 5 μg μl−1 bovine serum albumin (BSA)] that contained 20 000 dpm of the 32P-labelled probe, 2 μl of glycerine, and purified fusion protein as described previously (Liu et al., 1998). The resulting DNA–protein complexes were loaded on 0.5× Tris-borate-EDTA, 6% polyacylamide gels. After eletrophoresis, gels were dried and visualized by autoradiography.
Fig. 3.
EMSA and transcriptional activation analyses of GmDREB3 proteins. (A) Nucleotide sequences of DRE (DRE) and mutated DRE (DREm1–m3) probes. The nucleotide mutation in the DRE core motif of each probe is boxed. (B) EMSA was performed using 0.2 μg of GmDREB3 protein, and radiolabelled DRE or mutant DRE probes. Lanes: 1, GST proteins control; 2, free labelled DRE probe; 3, GST–GmDREB3 fusion plus labelled wild-type DRE element; 4–6, GST–GmDREB3 fusion proteins plus labelled mutated DRE (DREm1–m3) probes. (C) Recombinant plasmids containing GmDREB3 were transferred into yeast cells carrying the dual reporter genes under the control of the 71 bp promoter region containing the wild-type DRE (left half of each plate) or the mutant DRE core sequence (right half of every plate). Transferred yeast cells were examined for growth on the medium in the presence of 10, 20, 30, 40, 50, and 60 mM 3-AT (a–f). (D) LacZ activity of yeast cells transformed with GmDREB3. 1 and 3, stain intensities of yeast cells carrying mutant DRE elements; 2 and 4, stain intensities of yeast cells carrying the wild-type DRE element. LacZ activities of the transformed yeast cells were semi-quantitated. The results are shown above the photographs.
Transcriptional activation assay of GmDREB3 protein
The coding region of the GmDREB3 gene was cloned into YepGAP, a yeast expression vector with the promoter of the glyceraldehyde 3-phosphate dehydrogenase gene and terminator of the ADH1 gene (Liu et al., 1998). The recombinant plasmid was then transferred into yeast strain YM4271 carrying the reporter genes His3 and LacZ with four copies of a tandemly repeated 71 bp DNA fragment containing the DRE core sequence (TACCGACAT) or a mutated DRE (mDRE) core sequence (TATTTTCAT) upstream of a TATA element (Liu et al., 1998). Both the yeast expression vector YepGAP and yeast strain YM4271 were kindly provided by Professor Qiang Liu, Tsinghua University, Beijing. The yeast strain expressing the His gene at the basal level grows on synthetic dextrose (SD) medium lacking histidine, but cannot grow in the presence of 10 mM 3-AT (3-aminotriazole; Sigma, St Louis, MO, USA), a competitive inhibitor of the His3 gene product, and cannot induce LacZ (β-galactosidase) activity. The growth status of the yeast cells transferred with the recombinant plasmids was compared on SD medium without His plus different concentrations of 3-AT (10, 20, 30, 40, 50, and 60 mM). LacZ activity was assayed as described by Chen et al. (2003) and was semi-quantified by comparing fluorescence intensities of stained yeast cells using Quantity One software (Bio-Rad, Hercules, CA, USA).
Transformation and stress tolerance analyses of transgenic Arabidopsis plants
The coding region of GmDREB3 was amplified using the primer pair: GmDREB3FP-1, 5′-TATCCCGGGATGGCGAAACCCAGCAGCGA-3′ (forward); and GmDREB3FP-2, 5′-GCAGAGCTCGCGGCCGCTCAAAAATTCCACAAGAAAGAC-3′ (reverse), and inserted into the MCS (multiple cloning site) of two binary vectors, p35S and pRd29A, derived from pBI121, to produce vectors p35S-GmD3 and pRd29A-GmD3, respectively. For the construction of the pRd29A vector, pBI121 was digested with HindIII and SacI to delete the GUS gene and CaMV 35S promoter, and ligated with a fragment containing the Rd29A promoter and MCS of the pBluescript SK vector (Clontech, Mountain View, CA, USA). To construct the p35S vector, the pRd29A vector was digested with HindIII and SmaI to delete the Rd29A promoter and ligated with the CaMV 35S promoter. To construct the p35S-GmD3 and pRd29A-GmD3 vectors, the amplified fragments of the GmDREB3 cDNA were inserted into the XbaI–SacI site of the p35S and pRd29A vectors, with GmDREB3 being driven by the constitutive CaMV 35S and stress-inducible Rd29A promoters in vectors p35S-GmD3 and pRd29A-GmD3, respectively.
Two plasmids, p35S-GmD3 and pRd29A-GmD3, were introduced into Agrobacterium tumefaciens strain C58C1. The Arabidopsis plants used for transformation were grown in 8 cm pots at 25 °C for 5 weeks and transferred by vacuum infiltration as described by Bechtold and Pelletier (1998). T2 transgenic Arabidopsis plants were identified by selection for kanamycin resistance and used for further functional analysis. For drought tolerance analyses, the wild-type, and 35S:GmDREB3 and Rd29A:GmDREB3 transgenic Arabidopsis plants grown in pots were treated without watering for 19 d, followed by rewatering. Survival rates of the wild-type and transgenic plants were evaluated 8 d later. For high salt tolerance analyses, seeds of wild-type and 35S:GmDREB3 plants were germinated on normal MS medium and then moved onto MS medium in the presence of 160 mM NaCl for 18 d at 22 °C. For cold tolerance analysis, 2-week-old wild-type and 35S:GmDREB3 transgenic plants were removed from the agar plates and put on filter paper saturated with water exposed to −6 °C for 1 h, and allowed to recover at 25 °C for 24 h, prior to evaluation of survival rates. Two plasmids, p35S-GmD3 and pRd29A-GmD3, were transferred into tobacco calli by Agrobacterium tumefaciens strain EH105. T0 transgenic tobacco plants were selected on MS medium containing 100 μg ml−1 kanamycin and further identified using PCR with the specific primers GmDREB3FP-1 and GmDREB3FP-2. All transgenic tobacco genotypes were transformed to pots for functional analyses.
Analysis of osmolality, free proline, and chlorophyll in GmDREB3 transgenic plants
Before and after cold stress, the fresh weights and osmolalities of wild-type and 35S:GmDREB3 transgenic Arabidopsis plants were measured with a vapour pressure osmometer (VAPRO™ 5520, Wescor, Logan, UT, USA) (1–3 replicates per genotype). Average osmolality values for each line were used for statistical analysis. For measurement of free proline content, wild-type and transgenic tobacco plants were grown in pots under a 12 h light:12 h dark:25 °C regime. Eight-weeks-old plants were treated without watering for 16 d and leaves were harvested at 0 d and 16 d. Free proline contents of harvested leaves were measured as described previously by Zhang et al. (1990). To determine the salt tolerance of transgenic tobacco, leaf discs of 1 cm diameter were cut from healthy and fully expanded leaves of wild-type and GmDREB3 transgenic tobacco plants, and floated on MS liquid medium containing 400 mM NaCl for 5 d (12 h of white light:12 h of darkness:25 °C). Chlorophyll contents were determined following Aono et al. (1993). Data for osmolality values, free proline, and chlorophyll contents were analysed by SAS software (SAS Corporation, Cory, NC, USA) using t-tests to test the significance of differences between means.
Results
The phylogenetic analysis of the GmDREB3
The full length of the GmDREB3 gene was 597 bp, encoding a protein of 199 amino acids. Further analysis of its deduced amino acid sequence using the SMART program (http://smart.embl-heidelberg.de) revealed that this protein contained a conserved AP2 domain of 58 amino acids and a putative NLS (nuclear localization signal sequence) (Supplementary Fig. S1A available at JXB online). Systematic phylogenetic analysis was carried out on the basis of the similarities of the AP2 domains in AP2/EREBP proteins isolated from soybean Arabidopsis, maize, rice, tomato (Solanum lycopersicum L.), barley (Hordeum vulgare L.), cotton (Gossypium hirsutum L.), tobacco (N. tabacum L.), and canola (Brassica napus L.) using CLUSTAL W software (Sakuma et al. 2002). The results showed that the GmDREB3 gene was classified into the A-5 subgroup of the DREB subfamily (data not shown), which was according to the study of Liu et al. (2007). In addition, GmDREB1 and GmDREB2 from soybean, RAP2.1 and RAP2.10 from Arabidopsis, GhDBF1 from cotton, and PpDBF1 from moss (P. patens) were also classified into the A-5 subgroup. In order to investigate the relationship between GmDREB3 and the other members in the A-5 subgroup, the deduced full-length amino acid and AP2 domain sequence of GmDREB3 was compared with that of the six DREB proteins from soybean, moss, cotton, and Arabidopsis (Okamuro et al., 1997; Huang and Liu, 2006; Chen et al., 2007; Liu et al., 2007) (Supplementary Fig. S1B, S1C). GmDREB3 had high similarity to other members of the A-5 subgroup in the AP2 domain (>68%, Supplementary Fig. S1C) and low similarity outside of the AP2 domain. Homologies of the full-length amino acid sequences were low (<38%, Supplementary Fig. S1B). In the AP2 domain, the GmDREB3 protein had the same 14th valine as other members in the A-5 subgroup, but contained a 19th leucine instead of a glutamic acid (Supplementary Fig. S1A). Wang et al. (2005) reported that the C-terminal 98 amino acids of CBF1 function in transactivation. The LWSY domain in the C-terminus is the characteristic of CBF proteins (Jaglo et al., 2001). In the present work, the C-terminus of GmDREB3 shared a similar LWSY domain with PpDBF1, whereas it shared low similarity with GmDREB2 in the C-terminus (Supplementary Fig. S1A), suggesting that the transactivation activity of GmDREB3 might be different from that of GmDREB2.
Expression pattern of the GmDREB3 gene in soybean
Northern blots showed that GmDREB3 was responsive to cold stress, and not to high salt and drought stresses, or to ABA treatment. With cold stress treatment, GmDREB3 mRNA began to accumulate after 0.5 h and reached a maximum at 1 h after treatment, after which it was not detectable (Supplementary Fig. S2 at JXB online). The GmDREB3 gene was not responsive to ABA treatment or osmotic stresses, suggesting that this gene might be involved in an ABA-independent cold stress-responsive signal pathway.
Isolation and activity analysis of the GmDREB3 promoter
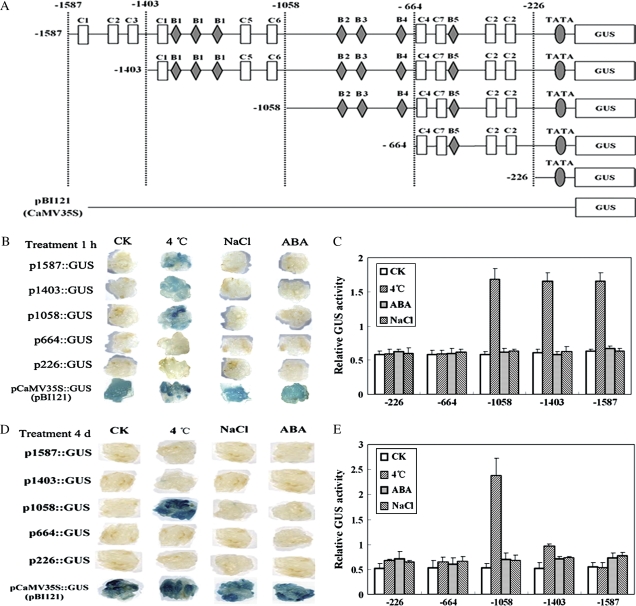
The promoter sequence of the GmDREB3 with a length of 1587 bp was isolated from the soybean genome. Using the PLACE Signal Scan Search Program, several cis-elements were predicted to be involved in transcriptional regulation; among them were one typical TATA-box located about −35 upstream of the transcription initiation point, ten MYC transcription factor recognition sites belonging to seven types (MC-1 to MC-7), and seven MYB transcription factor recognition sites belonging to five types (MB-1 to MB-5) (Fig. 1). Among the MYC recognition sites, MC-1, MC-4, and MC-7 shared the same consensus sequence with MYC-2, MYC-3, and MYC-4, respectively, in the DREB1A/CBF3 promoter (Chinnusamy et al., 2003). Deletion analysis from the 5′ end of the promoter showed that the GmDREB3 promoter was responsive only to cold stress (4 °C), and not to normal conditions, high salt stress, and ABA treatment (Fig. 2). However, the GmDREB3 promoter showed different regulation patterns after cold treatments for 1 h and 4 d. After treatment for 1 h, 5′ deletions to −1403 and −1058 did not impair cold responsiveness, whereas deletion to −664 resulted in almost complete elimination of cold responsiveness (Fig. 2B, C), indicating that part of the sequence between −1058 and −664 had an important role in the cold responsiveness of GmDREB3. After cold stress for 4 d, the 5′ deletion to −1058 showed a higher level of GUS activity (2.38±0.34) than after 1 h (1.96±0.16), whereas fragments containing −1587 to +41 and −1403 to +41, which include the region from −1403 to –1058, impaired the cold responsiveness (Fig. 2D, E). This indicated that the region from −1403 to −1058 was sufficient to impair the cold responsiveness of the GmDREB3 promoter and that the region might contain cold-responsive transcriptional repressor elements. The overall results suggested that a transcriptional activator might activate GUS expression after cold stress for 1 h by binding to cold-responsive elements in the region −1058 to −664, and that a transcriptional repressor impaired this activity by competitive binding to elements in region −1403 to −1058 after cold stress for 4 d. This probably also explained the greatly decreased expression of the GmDREB3 after cold stress for 3 h (Supplementary Fig. S2 at JXB online).
Fig. 1.
Nucleotide sequence of the promoter region of GmDREB3 including some predicted known cis-elements. The sequences homologous to MYC (MC) and MYB (MB) recognition sites are highlighted. A putative TATA box is underlined. An arrow shows the initiation point (+1) of transcription. Downward pointing arrows show the 5′ and 3′ ends of five deleted fragments of the GmDREB3 promoter.
Fig. 2.
Activity analysis of the GmDREB3 promoter. (A) Five fragments that were deleted from the 5′ end of the GmDREB3 promoter were inserted into a binary vector (pBI121). The locations of the 5′ ends of the five fragments of the GmDREB3 promoter are indicated; ‘C’ and ‘B’ represent MYC and MYB recognition sites, respectively. Binary vector pBI121 was used as the positive control. (B and D) After transformation with vectors containing the five fragments of the GmDREB3 promoter, calli were treated on MS medium plus 100 μM ABA (ABA), plus 200 mM NaCl (NaCl), and under low temperature (4 °C) stress and normal conditions (CK) for 1 h and 4 d. The histochemical staining results (GUS) are shown. (C and E) Quantitative results of GUS activity after treatments for 1 h and 4 d. The relative GUS activity was calculated as the ratio of GUS activity of a mutant GmDREB3 promoter to that of the CaMV 35S promoter (pBI121) under the same stress treatments.
DNA-binding activity and transcriptional activation analysis of GmDREB3
The wild-type DRE motif interacted with the GmDREB3–GST fusion protein and was retarded on SDS–PAGE, but the DRE mutation motifs with a two-base substitution at the 5′ end or a three-base substitution at the 3′ end, or a completely mutant DRE core sequence, severely inhibited interaction with the GmDREB3–GST fusion protein (Fig. 3B), suggesting that GmDREB3 protein could bind specifically to the DRE CCGAC core sequence in vitro.
Yeast cells with the recombinant plasmid harboring GmDREB3 grew on media lacking histidine in the presence of 10, 20, 30, and 40 mM 3-AT (Fig. 3C), and stained blue in X-gal solution after 4 h (Fig. 3D). However, when the recombinant plasmid containing GmDREB3 was transferred into yeast carrying the dual reporter genes fused to a 71 bp DNA fragment with base substitutions in the DRE core sequence, the yeast strain neither grew on media lacking histidine in the presence of all concentrations of 3-AT, nor stained blue (Fig. 3C, D), indicating that GmDREB3 protein bound specifically to the DRE element in order to activate transcription of the dual reporter genes in yeast cells.
Overexpression of GmDREB3 improved drought, high salt, and cold stress tolerance of transgenic Arabidopsis lines
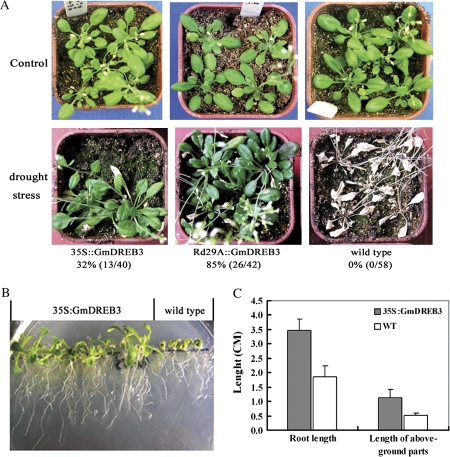
In drought tolerance analyses, all wild-type plants were dead (0/58), whereas 32% (14/30) of the 35S:GmDREB3 and 85% (26/42) of the Rd29A:GmDREB3 transgenic plants survived (Fig. 4A). This indicated that overexpression of GmDREB3 improved the drought tolerance of the transgenic plants.
Fig. 4.
Drought and high salt stress tolerance analyses of GmDREB3 transgenic Arabidopsis plants. (A) Drought tolerance analysis of GmDREB3 transgenic Arabidopsis plants. Control: 3-week-old plants grown under normal conditions. Drought stress: after 3 weeks wild-type and transgenic plants were treated without watering for 19 d, followed by rewatering. The growth status of treated plants 8 d after watering is shown. Percentage survival is indicated. (B) High salt tolerance analysis of 35S::GmDREB3 transgenic Arabidopsis plants. (C) Average lengths of roots and aboveground parts of 35S:GmDREB3 transgenics and controls shown in B.
High salt tolerance analyses showed that the growth of wild-type plants was severely inhibited, whereas the transgenics maintained normal growth (Fig. 4B). The average lengths of roots and aboveground parts of the transgenics were 3.48±0.38 cm and 1.13±0.29 cm, respectively, 1.88-fold (P <0.01) and 2.17-fold (P <0.01) greater than the wild-type controls (1.85±0.38 cm and 0.52±0.06 cm) (Fig. 4C). Thus GmDREB3 enhanced tolerance to high salt stress in transgenic plants.
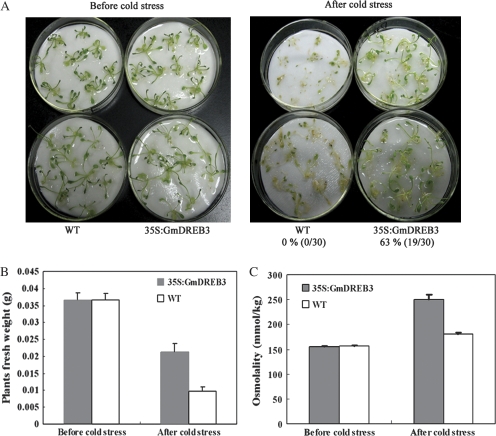
Cold tolerance analyses showed that all of the wild-type plants died (0/30), whereas 63% (19/30) of the 35S:GmDREB3 transgenic plants survived (Fig. 5A), indicating that overexpression of GmDREB3 improved the cold tolerance of transgenic Arabidopsis plants.
Fig. 5.
Evaluations of phenotypic and physiological effects of cold stresses on GmDREB3 transgenic Arabidopsis. (A) Cold stress tolerance analysis of GmDREB3 transgenic Arabidopsis plants. Two-week-old wild-type (WT) and transgenic (35S:GmDREB3) plants were removed from agar plates and exposed to –6° C for 1 h and then allowed to recover at 25 °C for 24 h, prior to scoring. Survival rates are shown below the photographs. (B and C) Average fresh weights and the osmolalities of transgenic (35S:GmDREB3) and wild-type (WT) plants before and after cold stress.
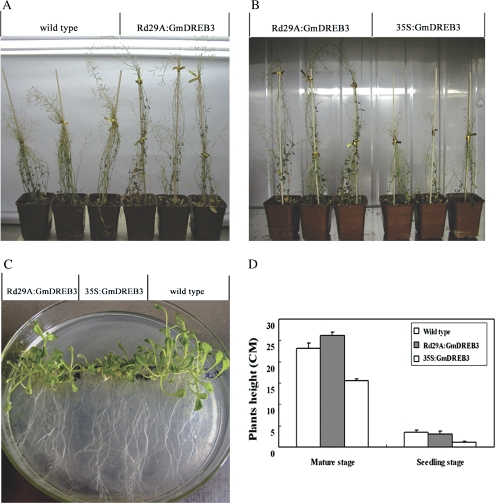
To evaluate phenotypic changes, wild-type and 35S:GmDREB3 and Rd29A:GmDREB3 transgenic Arabidopsis plants were grown on MS medium during the seedling stage (Fig. 6C) and then transformed to pots (Fig. 6A, B). At maturity, the average height of Rd29A:GmDREB3 transgenics (26.16±0.76 cm) was similar to that of the wild type (23.16±1.25 cm), but significantly higher (P <0.001) than that of the 35S:GmDREB3 transgenics (15.50±0.50 cm). Likewise, the average heights of the Rd29A:GmDREB3 transgenics (3.02±0.68 cm) and wild type (3.37±0.56 cm) seedlings were greater than that of the 35S:GmDREB3 transgenics (1.05±0.26 cm) (P <0.025) (Fig. 6D). Thus constitutive expression of GmDREB3 in Arabidopsis induced undesirable dwarfing, whereas the expression of GmDREB3 controlled by the stress-inducible Rd29A promoter minimized negative effects on growth under normal conditions.
Fig. 6.
Phenotypic evaluation of GmDREB3 transgenic Arabidopsis. (A and B) Growth retardation was observed in 35S:GmDREB3 transgenic Arabidopsis grown in pots after 70 d under normal conditions, whereas Rd29A:GmDREB3 transgenics were similar to the wild type. (C) After 22 d, the phenotype of Rd29A:GmDREB3 transgenics grown on MS medium was similar to that of the wild type, whereas the growth of 35S:GmDREB3 transgenics was obviously retarded. (D) Average seedling and mature plant heights of transgenics are compared with those of wild-type plants on MS medium and in pots.
Overexpression of GmDREB3 led to physiological changes in Arabidopsis and tobacco transgenic lines under cold, drought, and high salt stresses
To evaluate physiological changes in transgenic plants after cold stress, plant fresh weights and osmolalities were compared. Before cold stress, the fresh weights of the wild type (0.036667±0.001761 g) and 35S:GmDREB3 transgenics (0.036663±0.002002 g) were similar, whereas after cold stress (−6 °C for 1 h) 35S:GmDREB3 plants (0.02115±0.002527 g) were 2.2-fold (P <0.005) heavier than the wild type (0.00958±0.001249 g) (Fig. 5B). Similarly, before cold stress, the osmolality of wild-type (156.4±2.0 mmol kg−1) and transgenic plants (155.5±1.3 mmol kg−1) were similar, whereas after cold stress, the osmolality of transgenic plants (250.5±9.7 mmol kg−1) was higher than that of the wild-type plants (180.3±3.7 mmol kg−1) (P <0.001) (Fig. 5C).
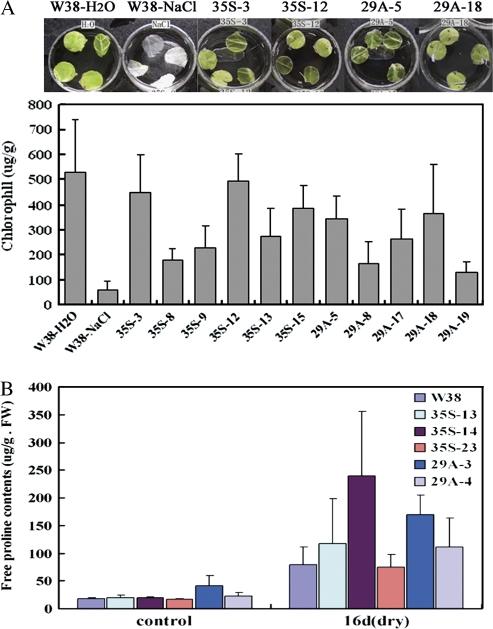
The chlorophyll contents in the leaves of wild-type and transgenic tobacco plants were measured following high salt treatment. Leaf discs from wild-type plants were bleached, whereas most of those from the transgenic plants remained green (Fig. 7A). The chlorophyll contents in the leaves of most of the transgenic tobacco plants were similar to those of wild-type discs floated in water, and higher than those of the wild type floated on 400 mM NaCl solution. Among the transgenic tobacco lines, 35S-3 and 35S-12 (35S:GmDREB3) at 449.33±125.65 μg g−1 and 494.79±110.72 μg g−1 were 7.6-fold (P <0.05) and 8.4-fold (P <0.01), and 29A-5 and 29A-18 (Rd29A:GmDREB3) at 343.76±89.11 μg g−1 and 364.86±197.78 μg g−1 were 5.8-fold (P <0.05) and 6.2-fold (P <0.05) higher in chlorophyll content compared with the wild-type tobacco (58.77±36.90 μg g−1) (Fig. 7A). These results indicated that overexpression of the GmDREB3 gene reduced the effects of high salt stress on chlorophyll formation and enhanced the salt tolerance of transgenic plants.
Fig. 7.
Chlorophyll and free proline content analyses of wild-type and transgenic tobacco plants under high salt and drought-stressed conditions. (A) Visual differences and chlorophyll contents (μg g−1 fresh weight) measured after 96 h of high salt treatment. The experiments were repeated three times, each replicate with four leaf discs. W38H2O and W38NaCl represent the wild type floated on H2O and high salt solution, respectively. 35S-3 to 35S-15 (35S:GmDREB3) and 29A-5 to 29A-15 (Rd29A:GmDREB3) are transgenics. (B) Free proline contents of wild-type and transgenic tobacco plants under normal or drought stress conditions. The wild-type (W38) and transgenic tobacco plants were grown in pots under normal conditions for 8 weeks when leaves were harvested as control samples. Both were then deprived of water for 16 d (dry); leaves were harvested at the 16th day for proline analyses.
The ability of transgenic tobacco to accumulate osmolytes, such as free proline, under normal and drought stress conditions was also investigated. Under normal conditions, free proline contents of transgenic plants were the same as those of wild-type plants (Fig. 7B). After drought treatment for 16 d, the free proline levels of the wild type and all transgenics were increased, and 35S-13 and 35S-14 (35S:GmDREB3) (117.79±80.89 μg g−1 and 239.73±115.66 μg g−1 FW) were 1.5-fold and 3-fold (P <0.2), and 29A-3 and 29A-4 (Rd29A:GmDREB3) (169.21±35.21 μg g−1 and 111.83±51.89 μg g−1 FW) were 2.1-fold (P <0.1) and 1.4-fold higher than wild-type tobacco (79.90±31.32 μg g−1 FW) (Fig. 7B). While none of these differences was significant, the consistent trend was that the transgenic tobacco plants accumulated higher levels of free proline than the wild type under drought stress, and that plants transferred with GmDREB3 are likely to be more tolerant to drought and high salt stresses.
Discussion
GmDREB3 belongs to the A-5 subgroup in the DREB subfamily and is a novel transcriptional activator
Most research on the DREB subfamily has focused on the A-1 and A-2 subgroups, and little is known about the characteristics and functions of members of the other subgroups. Studies on A-5 subgroup members, such as PpDBF1, GmDREB2, and GhDBF1 (Huang et al., 2006; Liu et al., 2007; Chen et al., 2007), suggested that the A-5 subgroup, like the A-1 subgroup, are important genetic resources, potentially useful for the improvement of crop stress tolerance. In this study, a novel DREB subfamily A-5 subgroup member, GmDREB3, was isolated from soybean. It was hypothesized that a gene transfer event might have introduced an AP2 gene from lower organisms to the common ancestor of the moss and plant, and then the AP2 genes began to spread in the genome by transposition and homing recombination (Magnani et al., 2004). During the course of evolution, AP2 genes diverged and acquired new functions by transposition and duplication events, and the DREB gene subfamily might have evolved from such events (Liu et al., 2007). During the course of the evolution of transcription factor genes in higher plants, the DNA-binding domain maintained a substantial selective constraint, whereas the non-DNA-binding region showed a higher level of evolutionary divergence due to relaxed constraint (Chang et al., 2005). In this study, the homology comparisons of DREB genes in the A-5 subgroup showed that GmDREB3 shared high similarity in the AP2 domain and low similarity in the full-length sequence with six other members, which might be due to the different rates of evolution within the AP2 conserved domain and other parts of the DREB proteins. Similar results for both the MYB family (Dias et al., 2003) and the bHLH family (Atchley et al., 1994) were also reported. In addition, the variation occurring in the promoter regions of different members, including insertions, deletions, transpositions, and substitutions of nucleotides, might result in the changes in expression pattern.
Expression of GmDREB3 might be co-regulated by different transcription factors
Among the members in the A-5 subgroup, PpDBF1 and GmDREB2 were induced by drought, high salt, cold stresses, and ABA treatment (Chen et al., 2007; Liu et al., 2007), and GhDBF1 was mainly induced by drought and high salt stress (Huang et al., 2006), whereas GmDREB3 was only responsive to cold stress (Supplementary Fig. S2 at JXB online). During the cold response, the transcription of GmDREB2 was induced after 3 h of treatment, and reached its maximum at 17 h (Chen et al., 2007), whereas the induction of GmDREB3 occurred much earlier than that of GmDREB2. Its expression had been restrained before 3 h, suggesting that GmDREB3 might be involved in the early cold response compared with GmDREB2 in soybean. In addition, the promoter sequence of GmDREB2 was obtained by searching Scaffold 26 in the Soybean genome project (www.phytozome.net/soybean.php). Sequence alignment indicated that the promoter sequence of GmDREB3 shared low identity (43.5%) with the promoter sequence of GmDREB2 (Supplementary Fig. S3 at JXB online), suggesting that although the two genes belonged to the same subgroup, they were involved in different stress-related signal pathways due to the different promoters controlling their expression.
Promoter activity analysis showed that a cold-responsive element located in region −1058 to −664 of the GmDREB3 promoter induced GUS expression after cold stress for 1 h. Following prolonged cold stress, it seemed that another transcriptional repressor could impair the activity of the GmDREB3 promoter by binding to an element in the region −1403 to −1058 (Fig. 2 and Supplementary Fig. S2 at JXB online). ICE1, a MYC-type bHLH transcription factor, regulates the expression of DREB1A, and there are five potential MYC sites (CANNTG) in the promoter region of DREB1A (Shinwari et al., 1998; Chinnusamy et al., 2003; Nakashima et al., 2006). Zarka et al. (2003) reported that a region in the DREB1C/CBF2 promoter, ICEr1, played a key role in cold-induced expression, and a MYC site (CACGTG) was detected in this ICEr1 region. However, in the present study, motif analysis indicated that there were no typical MYC recognition site in region −1058 to −664 of the GmDREB3 promoter, whereas three MYB recognition sites were in this region (MB2, MB3, and MB4) (Fig. 1). Overexpression of a rice MYB gene, Osmyb4, increased chilling and freezing tolerance of Arabidopsis plants, and its recognition sequence was mAC-II (AAGAAGGAAACC) (Vannini et al., 2004). Transcription factors belonging to the MYB family have binding specificity of either type I [CNGTT(A/G)] or type II [G(G/T)T(A/T)GTT(A/G)] and type IIG [G(G/T)T(A/T)GGT(A/G)], and members of different subgroups within the MYB family prefer different MYB recognition sequences (Romero et al., 1998). In region −1058 to −664 of the GmDREB3 promoter, three MYB sites (MB2, TAGTTA; MB3, TAGTAA; and MB4, TTGTTG) are more like type II MYB sites, which are more likely to be recognized by members belonging to subgroup B of the MYB family (Romero et al., 1998).
HOS1, a novel RING finger protein (Lee et al., 2001), and FRY2, a transcriptional repressor (Xiong et al., 2002), appear to be negative regulators of CBF expression. In addition, Agarwal et al. (2006) reported that a MYB transcription factor, MYB15, could bind to MYB recognition sequences in the promoters of CBF genes. Overexpression of MYB15 resulted in reduced expression of CBF genes, and its loss of function led to increased expression of CBF genes upon cold treatment. MYB15 preferentially binds to type II MYB or type IIG sites, and, to a much lesser extent, to the type I MYB recognition sequence (Romero et al., 1998). To date, due to lack of detection of negative regulatory elements related to these negative regulators of CBF, its cold-regulated mechanism in plants remains to be explained. In this study, in the region −1403 to −1058, three copies of consecutive MYB recognition site (TTGATA) were detected. This is similar to type II recognition sites, suggesting some MYB transcription factors might bind to these three MYB sites to regulate the expression of GmDREB3 negatively (Fig. 2D, E). Further identification of actively regulated elements in region −1058 to −664 and negatively regulated elements in region −1403 to −1058 will be important for elucidating the regulatory mechanism of GmDREB3 under cold stress.
Expression of GmDREB3 controlled by the stress-inducible Rd29A promoter increased tolerance to drought and high salt stresses, and minimized negative effects on plant growth under normal growth conditions
Overexpression of PpDBF1 in transgenic tobacco plants showed enhanced tolerance to salt, osmotic, and cold stresses, and, when grown on MS medium plus 200 mM NaCl and 250 mM sorbitol, root growth rates of transgenic tobacco plants were higher than those of the wild-type (Liu et al., 2007). Similarly, overexpression of GmDREB2 in transgenic plants also showed enhanced tolerance to salt and drought stresses (Chen et al., 2007). In the present work, GmDREB3 transgenic Arabidopsis plants were tolerant to drought, cold, and high salt stresses, similar to GmDREB2 transgenic plants. In addition, transcriptional activation analysis of GmDREB3 showed that yeast cells with the recombinant plasmid harbouring GmDREB3 grew on media lacking histidine in the presence of 10, 20, 30, and 40 mM 3-AT (Fig. 3C), whereas yeast cells with GmDREB2 grew only on media in the presence of 10 mM 3-AT (Chen et al., 2007), suggesting that under the same conditions (uniform plasmids were present in the yeast cells; data not shown), transcriptional activation of GmDREB3 was higher than that of GmDREB2. The C-terminal part of GmDREB3 shared low homology with GmDREB2 (Supplementary Fig. S1A at JXB online). Moreover, LWST domains, characteristic domains of CBF proteins (Jaglo et al., 2001), were detected only in GmDREB3 and PpDBF1. The structural differences might cause the different transcriptional activation characteristics of GmDREB3 and GmDREB2. Thus members of the A-5 subgroup might play key roles in regulating expression of stress-related genes, and, among them, GmDREB3, as an important member, might be useful for improving stress tolerance in crop plants.
In this study, overexpression of GmDREB3 in 35S:GmDREB3 transgenic plants led to reduced height of seedlings and mature plants (Fig. 6). Similar phenomena were observed with DREB1A, DREB1B, and DREB1C transgenic Arabidopsis (Liu et al., 1998; Kasuga et al., 1999; Gilmour et al., 2004). Recent studies showed that expression of some genes downstream of DREB1, such as the GA (gibberellin) biosynthesis gene and transcription repressor STZ, might be involved in growth retardation of DREB1 transgenic plants (Nakashima et al., 2006). To minimize the negative effects of DREB1 on plant growth, the stress-inducible Rd29A promoter, employed to control the expression of DREB1A, improved the drought, salt, and freezing stress tolerance of transgenic Arabidopsis (Yamaguchi-Shinozaki and Shinozaki, 1993), tobacco (Kasuga et al., 2004), rice (Dubouzet et al., 2003), wheat (Pellegrineschi et al., 2004), and potato (Behnam et al., 2006). Both the stress-inducible Rd29A and constitutive CaMV 35S promoters were used to control the expression of GmDREB3. Growth retardation of Rd29A:GmDREB3 transgenic plants was reduced (Fig. 6), and the survival rate of Rd29A:GmDREB3 transgenic Arabidopsis was high (Fig. 4A), providing hope that a combination of the Rd29A promoter and GmDREB3 might be useful for improving tolerance to environmental stresses in crop plants.
GmDREB3 transgenic plants accumulated higher levels of solute or free proline, enhancing the tolerance of transgenic plants to cold and drought stresses
The fresh weights of both wild-type and 35S:GmDREB3 transgenic plants were similar before cold stress, and both decreased after cold stress; however, the transgenics retained higher levels of water than wild-type plants (P <0.005) (Fig. 5B). Moreover, after cold stress, all wild-type plants withered and died, whereas most of the transgenics remained green (Fig. 5A). Thus 35S:GmDREB3 transgenics decreased water loss from cells, leading to enhanced tolerance to cold stress.
Many plants increase their tolerance to freezing when exposed to low, but non-freezing, temperatures in an adaptive process known as cold acclimation. During cold acclimation, the accumulation of compatible solutes, such as soluble sugars, protein, and betaine, occurs in many plant species (Guy, 1999). Yoshida et al. (1998) reported a positive correlation between freezing tolerance and soluble sugar content in 18 wheat cultivars. Uemura et al. (2003) reported that the freezing sensitivity of a cold-acclimated sfr4 mutant of Arabidopsis was due to its continued susceptibility to LOR (loss of osmotic responsiveness) and was associated with low sugar content in its cells. The level of freezing tolerance in canola was strongly dependent on the osmotic potential of the leaves, and the osmotic potential of acclimated leaves was lower than that of non-acclimated leaves under cold stress (−3 °C) (Gusta et al., 2004). The osmotic potential (ψS), as a function of the molal concentration of solute, was calculated using the van't Hoff relationship: ψS = –CiRT [where ψS =osmotic potential; C=osmolality of solute; i=ionization constant; R=gas constant (0.00831 kg MPa mol−1 K−1); T=absolute temperature (K)=+273 °C] (Bhatia et al., 2005). In this study, the osmolalies of wild-type and transgenic plants were similar before cold stress, whereas transgenics had significantly higher osmolalities than wild-type plants after cold stress (Fig. 5C), indicating that the osmotic potentials of transgenics were lower than those of wild-type plants after cold stress. Thus 35S:GmDREB3 transgenics are likely to accumulate higher levels of solutes, such as sugars, such that the osmotic potentials decline faster than in wild-type plants.
Many plants accumulate free proline under drought, high salt, and cold stress conditions, and this probably functions as an osmo-protectant in stressed plants, leading to tolerance (Igarashi et al., 1997). For example, transgenic Arabidopsis plants overexpressing DREB1A/CBF3 accumulate free proline under unstressed conditions (Gilmour et al., 2000). Free proline contents of GmDREB3 transgenic tobacco plants were compared with those of wild-type plants under drought stress conditions. Among the transgenic tobacco lines, 35S-14 and 29A-3 accumulated higher levels of free proline than wild-type plants after drought stress treatment for 16 d (Fig. 7B). These results suggested that overexpression of GmDREB3 in transgenic plants enhanced tolerance to drought stress by inducing expression of downstream genes involved in the synthesis and accumulation of higher levels of free proline.
The nucleotide sequence of the GmDREB3 gene reported in this article has been submitted to the GenBank database under the accession number DQ208969.
Supplementary material
The supplementary material is available at JXB online.
Figure S1. shows the amino acid sequence alignment of GmDREB3 and other members in the A-5 subgroup.
Figure S2. shows the expression pattern of the GmDREB3 gene in soybean under different abiotic stresses.
Figure S3. shows the promoter sequence alignment of GmDREB3 and GmDREB2 in the soybean genome.
Acknowledgments
The authors are grateful to Dr RA McIntosh (Plant Breeding Institute, University of Sydney, Australia) for a critical review of this manuscript. We thank Dr Tianfu Han (Institute of Crop Sciences, CAAS) for the soybean cultivars. This work was funded by the National 863 Project (#2006AA10A111 and #2008AA10Z124), the National Natural Science Foundation of China (#30700508 and #30700504), and the Science Foundation of Chinese Academy of Agricultural Sciences (#2060302-2-08 and #082060302-10), and the National Key Project for Research on Transgenic Plant (2008ZX08002-002).
References
- Agarwal M, Hao YJ, Kapoor A, Dong CH, Fujii H, Zheng XW, Zhu JK. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. Journal of Biological Chemistry. 2006;281:37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Aono M, Kubo A, Saji H, Tanaka K, Kondo N. Enhanced tolerance to photooxidative stress of transgenic Nicotiana tobaccum with high choroplastic glutathione reductase activity. Plant and Cell Physiology. 1993;34:129–136. [Google Scholar]
- Atchley W, Fitch W, Bronner-Fraser M. Molecular evolution of the MyoD family of transcription factor. Proceedings of the National Academy of Sciences, USA. 1994;91:11522–11526. doi: 10.1073/pnas.91.24.11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D, Sunkar R. Drought and salt tolerance in plants. Critical Reviews in Plant Sciences. 2005;24:23–58. [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods in Molecular Biology. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Behnam B, Kikuchi A, Celebi-Toprak F, Yamanaka S, Kasuga M, Yamaguchi-Shinozaki K, Watanabe KN. The Arabidopsis DREB1A gene driven by the stress-inducible Rd29A promoter increases salt-stress tolerance in proportion to its copy number in tetrasomic tetraploid potato (Solanum tuberosum) Plant Biotechnology Journal. 2006;23:169–177. [Google Scholar]
- Benedict C, Skinner JS, Meng R, Chang Y, Bhalerao R, Huner NA, Finn CE, Chen TH, Hurry V. The CBF1-dependent low temperature signaling pathway, regulon and increase in freeze tolerance are conserved in populus spp. Plant, Cell and Environment. 2006;29:1259–1272. doi: 10.1111/j.1365-3040.2006.01505.x. [DOI] [PubMed] [Google Scholar]
- Bhatia NP, Baker AJM, Walsh KB, Midmore DJ. A role for nickel in osmotic adjustment in drought-stressed pants of the nickel hyperaccumulator Stackhousia tryonii Bailey. Planta. 2005;223:134–139. doi: 10.1007/s00425-005-0133-8. [DOI] [PubMed] [Google Scholar]
- Chang SM, Lu YQ, Rausher MD. Neutral evolution of the nonbinding region of the anthocyanin regulatory gene Ipmyb1 in Ipomoea. Genetics. 2005;170:1967–1978. doi: 10.1534/genetics.104.034975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ, Dong Y, Wang YJ, Liu Q, Zhang JS. An AP2/EREBP-type transcript-factor gene from rice is cold-inducible and encodes a nuclear-localized protein. Theoretical and Applied Genetics. 2003;107:972–979. doi: 10.1007/s00122-003-1346-5. [DOI] [PubMed] [Google Scholar]
- Chen M, Wang QY, Cheng XG, Xu ZS, Li LC, Ye XG, Xia LQ, Ma YZ. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochemical and Biophysical Research Communications. 2007;353:299–305. doi: 10.1016/j.bbrc.2006.12.027. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes and Development. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:15243–15248. doi: 10.1073/pnas.0406069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias AP, Braun EL, McMullen MD, Grotewold E. Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiology. 2003;131:610–620. doi: 10.1104/pp.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high- salt- and cold-responsive gene expression. The Plant Journal. 2003;33:1–13. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Johnson AG, Poupart J, De Luca V. Uncoupled defense gene expression and antimicrobial alkaloid accumulation in elicited opium poppy cell cultures. Plant Physiology. 1996;111:687–697. doi: 10.1104/pp.111.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Molecular Biology. 2004;54:767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiology. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proceedings of the National Academy of Sciences, USA. 2002;99:11507–11512. doi: 10.1073/pnas.172399299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusta LV, Wisniewski M, Nesbitt NT, Gusta ML. The effect of water, sugars, and proteins on the pattern of ice nucleation and propagation in acclimated and nonacclimated Canola leaves. Plant Physiology. 2004;135:1642–1653. doi: 10.1104/pp.103.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CL. Cold acclimation and freezing stress tolerance: role of protein metabolism. Annual Review of Plant Physiology and Plant Molecular Biology. 1990;41:187–223. [Google Scholar]
- Hong JP, Kim WT. Isolation and functional characterization of the Ca-DREBLP1 gene encoding a dehydration-responsive element binding-factor-like protein 1 in hot pepper (Capsicum annuum L. cv. Pukang) Planta. 2005;220:875–888. doi: 10.1007/s00425-004-1412-5. [DOI] [PubMed] [Google Scholar]
- Huang B, Liu JY. A cotton dehydration responsive element binding protein functions as a transcriptional repressor of DRE element-mediated gene expression. Biochemical and Biophysical Research Communications. 2006;343:1023–1031. doi: 10.1016/j.bbrc.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Igarashi Y, Yoshiba Y, Sanada Y, Yamaguchi-Shinozaki K, Wada K, Shinozaki K. Characterization of the gene for delta l-pyrroline-5-carboxylate synthetase and correlation between the expression of the gene and salt tolerance in Oryza sativa L. Plant Molecular Biology. 1997;33:857–865. doi: 10.1023/a:1005702408601. [DOI] [PubMed] [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiology. 2001;127:910–917. [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnology. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. A combination of the Arabidopsis DREB1A gene and stress-inducible Rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant and Cell Physiology. 2004;45:346–350. doi: 10.1093/pcp/pch037. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Xiong LM, Gong ZZ, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes and Development. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhong NQ, Wang GL, Li LJ, Liu XL, He YK, Xia GX. Cloning and functional characterization of PpDBF1 gene encoding a DRE-binding transcription factor from Physcomitrella patens. Planta. 2007;226:827–838. doi: 10.1007/s00425-007-0529-8. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an AP2/EREBP DNA-binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression in Arabidopsis. The Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani E, Sjolander K, Hake S. From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. The Plant Cell. 2004;16:2265–2277. doi: 10.1105/tpc.104.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K. Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Plant Physiology. 2006;126:62–71. [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrineschi A, Reynolds M, Pacheco M, Brito RM, Almeraya R, Yamaguchi-Shinozaki K, Hoisington D. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome. 2004;47:493–500. doi: 10.1139/g03-140. [DOI] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Li J, Liu Q, Liu YQ, Shinozaki K, Yamaguchi-Shinozaki K. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant and Cell Physiology. 2004;45:1042–1052. doi: 10.1093/pcp/pch118. [DOI] [PubMed] [Google Scholar]
- Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J. More than 80R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. The Plant Journal. 1998;14:273–84. doi: 10.1046/j.1365-313x.1998.00113.x. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemical and Biophysical Research Communications. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold, and high-salinity stresses using a full-length cDNA microarry. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Shen YG, Zhang WK, He SJ, Zhang JS, Liu Q, Chen SY. An EREBP/AP2 type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theoretical and Applied Genetics. 2003;106:923–930. doi: 10.1007/s00122-002-1131-x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K. An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochemical and Biophysical Research Communications. 1998;250:161–170. doi: 10.1006/bbrc.1998.9267. [DOI] [PubMed] [Google Scholar]
- Tan GH, Gao Y, Shi M, Zhang XY, He SP, Chen ZL, An CC. SiteFinding-PCR: a simple and efficient PCR method for chromosome walking. Nucleic Acids Research. 2005;33:122–128. doi: 10.1093/nar/gni124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, Warren G, Steponkus PL. Freezing sensitivity in the sft4 mutant of Arabidopsis is due to low sugar content and is manifested by loss of osmotic responsiveness. Plant Physiology. 2003;131:1800–1807. doi: 10.1104/pp.102.013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Current Opinion in Biotechnology. 2006;17:113–122. doi: 10.1016/j.copbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. The Plant Journal. 2004;37:115–127. doi: 10.1046/j.1365-313x.2003.01938.x. [DOI] [PubMed] [Google Scholar]
- Vasil V, Srivastava V, Castillo AM. Rapid production of transgenic wheat plants by direct bombardment of cultured immature embryos. Bio/Technology. 1993;11:1553–1558. [Google Scholar]
- Vij S, Tyagi AK. Emerging trends in the functional genomics of the abiotic stress response in crop plants. Plant Biotechnology Journal. 2007;5:361–380. doi: 10.1111/j.1467-7652.2007.00239.x. [DOI] [PubMed] [Google Scholar]
- Vinocur B, Altman A. Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Current Opinion in Biotechnology. 2005;16:123–132. doi: 10.1016/j.copbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Wang Z, Triezenberg SJ, Thomashow MF, Stockinger EJ. Multiple hydrophobic motifs in Arabidopsis CBF1 COOH-terminus provide functional redundance in transactivation. Plant Molecular Biology. 2005;58:543–559. doi: 10.1007/s11103-005-6760-4. [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Tanaka Y, Stevenson B, Koiwa H, Bressan RA, Hasegawa PM, Zhu JK. Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2002;99:10899–10904. doi: 10.1073/pnas.162111599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation-responsive Rd29A gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Molecular and General Genetics. 1993;236:331–340. doi: 10.1007/BF00277130. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Physiology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Abe J, Moriyama M, Kuwabara T. Carbohydrate levels among winter wheat cultivars varying in freezing tolerance and snow mold resistance during autumn and winter. Plant Physiology. 1998;103:8–16. [Google Scholar]
- Zarka DG, Vogel JT, Cook D, Thomashow MF. Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiology. 2003;133:910–918. doi: 10.1104/pp.103.027169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DZ, Wang PH, Zhao HX. Determination of the content of free proline in wheat leaves. Plant Physiology Communications. 1990;4:62–65. [Google Scholar]
- Zhang JS, Zhou JM, Zhang C, Chen SY. Differential gene expression in a salt-tolerance rice mutant and its parental variety. Science in China Series C–Life Sciences. 1996;39:310–319. [Google Scholar]
- Zhang JZ, Creelman RA, Zhu JK. From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiology. 2004;135:615–621. doi: 10.1104/pp.104.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao TJ, Sun S, Liu Y, Liu JM, Liu Q, Yan YB, Zhou HM. Regulating the drought-responsive element (DRE)-mediated signaling pathway by synergic functions of trans-active and trans-inactive DRE binding factors in Brassia napus. Journal of Biological Chemistry. 2006;281:10752–10759. doi: 10.1074/jbc.M510535200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.