Abstract
A 1.8 kb 5′-flanking region of the large subunit of ADP-glucose pyrophosphorylase, isolated from watermelon (Citrullus vulgaris S.), has fruit-specific promoter activity in transgenic tomato plants. Two negative regulatory regions, from –986 to –959 and from –472 to –424, were identified in this promoter region by fine deletion analyses. Removal of both regions led to constitutive expression in epidermal cells. Gain-of-function experiments showed that these two regions were sufficient to inhibit RFP (red fluorescent protein) expression in transformed epidermal cells when fused to the cauliflower mosaic virus (CaMV) 35S minimal promoter. Gel mobility shift experiments demonstrated the presence of leaf nuclear factors that interact with these two elements. A TCCAAAA motif was identified in these two regions, as well as one in the reverse orientation, which was confirmed to be a novel specific cis-element. A quantitative β-glucuronidase (GUS) activity assay of stable transgenic tomato plants showed that the activities of chimeric promoters harbouring only one of the two cis-elements, or both, were ∼10-fold higher in fruits than in leaves. These data confirm that the TCCAAAA motif functions as a fruit-specific element by inhibiting gene expression in leaves.
Keywords: ADP-glucose pyrophosphorylase, cis-element, fruit-specific promoter, transcriptional factors, transgenic tomato, watermelon
Introduction
In genetic engineering, genetic transformation is an important tool for crop improvement and gene function analysis. Many transgenic plants with genes of industrial or agronomic interest have been generated through Agrobacterium-mediated or biolistic transformation. Some constitutive promoters such as the cauliflower mosaic virus (CaMV) 35S promoter and maize ubiquitin are widely used in this regard, and usually result in high and constitutive expression of the genes in host plants. However, constitutive expression of the foreign gene(s) could be harmful to the growth and development of plants because overexpression can lead to metabolic stress. Therefore, it is important to achieve temporal and tissue-specific expression of the foreign gene(s) through promoter control.
Many ripening-related genes have been shown to exhibit organ- or tissue-specific expression patterns. However, few promoters have been well characterized compared with the number of genes studied, partly due to complicated interactions between a large number of cis-elements in promoters and their associated nuclear transcription factors. Some fruit-specific genes have been well characterized and their promoters, such as PG (Montgomery et al., 1993), E4 (Xu et al., 1996), E8 (Deikman et al., 1998; Coupe and Deikman, 1997), 2A11 (van Haaren and Houck, 1993), and ACO1 (Moon and Callahan, 2004), have been demonstrated to direct the ripening-specific expression of the reporter genes in fruits. However, the mechanisms of the specific promoting activities and related specific cis-elements remain illusive.
Starch is an important storage carbohydrate in plants, and its accumulation in roots, tubers, and fruits positively affects the yield and quality of a number of crops. ADP-glucose pyrophosphorylase (AGPase, EC 2.7.7.27), a key enzyme in starch biosynthesis, catalyses the conversion of glucose-1-phosphate to ADP-glucose, which serves as a direct substrate for starch synthesis (Preiss, 1998). AGPase in plants is a heterotetramer, consisting of two small and two large subunits (Morell et al., 1987). The large subunit has multiple isoforms with tissue-specific expression patterns. Northern blot analysis indicated that one of them, AGPL1, is specifically expressed in the fruit of watermelon, but not in the leaves. However, the mechanism underlying this fruit-specific expression of AGPL1 remains unknown. To understand the mechanism(s) of fruit-specific gene expression and explore possible application(s) of fruit-specific gene promoters, a detailed analysis of the AGPL1 promoter was conducted. The isolation and characterization of a 1573 bp 5′-flanking region of AGPL1 is also reported and the cis-acting elements involved in fruit-specific directing activity are defined.
Materials and methods
Cloning of the AGPL1 promoter
Genomic DNA was isolated from leaf tissue of young watermelon (Citrullus vulgaris S. cv. Jingxin NO.1) growing under greenhouse conditions using the CTAB (cetyltrimethyl ammonium bromide) method.
A pair of nested PCR primers was synthesized based on the sequence of watermelon AGPL1 (GenBank accession no. AF032472) (Kim et al., 1998): P1, 5′-AAACTTCCTCTAACTTTCTCACC-3′; and P2, 5′-TCCAGACCACCCCAATTACCCTT-3′. Uneven PCRs were conducted according to the methods of Chen et al. (1997). Primary PCRs were performed in 40 μl volumes containing 10 mmol l−1 TRIS-HCl (pH 8.3), 50 mmol l– KCl, 1.5 mmol l−1 MgCl2, 20 ng of watermelon genomic DNA, 0.25 μmol l−1 primer P1, 0.05 μmol l−1 non-specific primer (5′-TGCGAAGGCT-3′), 200 μmol l−1 each of dNTPs, and 2.0 U of polymerase. The cycle parameters were as follows: 94 °C for 30 s, 55 °C for 1 min, 72 °C for 1.5 min, 94 °C for 30 s, 42 °C for 1 min, and 72 °C for 1.5 min, except for an initial denaturation step of 94 °C for 2 min, for 20 cycles. A secondary PCR was performed in 0.5 μl of primary PCR product, using 0.25 μmol l−1 primer P2, 0.05 μmol l−1 non-specific primer, and the same reaction composition as the primary PCR. The cycles were as follows: 94 °C for 15 s, 57 °C for 30 s, 72 °C for 1 min, 94 °C for 15 s, 45 °C for 30 s, and 72 °C for 1 min, for 20 cycles, with an initial denaturation step of 2 min at 94 °C. Based on the sequence of AGPL1, an identification primer, P3 (5′-TGAGATATGGTTGCGATGGA-3′), upstream of primer P2 was designed. The amplification products were screened by specific PCRs using primer pair P2 and P3 with 0.25 μmol l−1 each in a 20 μl reaction volume. The reaction parameters were as follows: 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, except for an initial denaturation step of 94 °C for 5 min. A total of 30 cycles were performed. PCR products were examined and fragments of interest were isolated and sequenced.
The tomato (Solanum lycopersicum) E8 promoter was isolated from genomic DNA of young leaves. The PCR primers were as follows: sense primer, 5′-CGAAGCTTGAGGAAAATGTACGATGA-3′; and antisense primer, 5′-CGTCTAGACACTGTGAATGATTAGAA-3′.
Determination of the TSS of the AGPL1 promoter
A 1 g aliquot of fresh watermelon fruit, 10 d after anthesis, was used for total RNA extraction using Trizol reagent, and mRNA was separated using the PolyA Tract mRNA isolation Kit (Promega). The transcriptional start site (TSS) was determined via primer extension analysis as described previously (Dey and Maiti, 1999). The sequence of the specific extension primer was 5′-GTGGAGAAAGGGGATTAATG-3′.
Construction of expression vectors with deleted AGPL1 promoter fragments
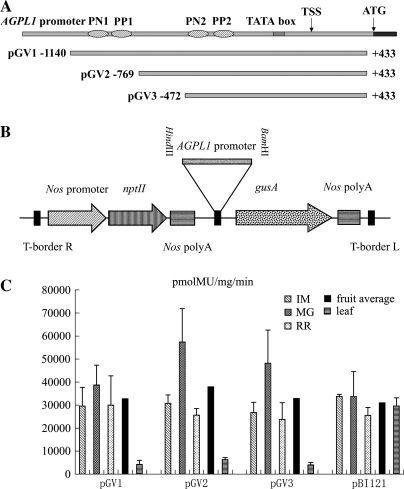
Three AGPL1 promoter fragments with progressive 5′ deletions were obtained through digestion with restriction enzymes, and each was inserted into a HindIII+BamHI site in pBI121 by replacing the CaMV 35S promoter to generate pGV1 (containing the AGPL1 promoter, –1140 to +433), pGV2 (containing the AGPL1 promoter, –769 to +433), and pGV3 (containing the AGPL1 promoter, –472 to +433). The vector pBI121 with the AGPL1 promoter–GUS constructs were introduced into Agrobacterium tumefaciens strain GV3101 via tri-parental mating. The vector pBI121 harboring the CaMV 35S promoter was used as a control.
Plant transformation
Tomato (S. lycopersicum cv. UC204C) cotyledon transformation was performed essentially as described by McBride et al. (1990). Tomato seeds were germinated in MS medium in darkness. After germination, the seedlings were placed under light for 2 d, and the cotyledons were cut transversally and pre-cultured in co-cultivation medium [MS agar medium supplemented with 2 mg l−1 zeatin+0.05 mg l−1 indole acetic acid (IAA)] for 48 h and transferred to a diluted culture of Agrobacterium for 15 min. Following 2 d of co-cultivation, the cotyledons were placed upside down on screening medium containing 100 mg l−1 kanamycin+250 mg l−1 carbenicillin, and subcultured every 10 d. After 6 weeks, the green shoots were regenerated and screened on kanamycin-containing medium. The shoots resistant to kanamycin were rooted on one-half strength MS medium supplemented with 0.1 mg l−1 IAA and antibiotics (kanamycin 100 mg l−1). Positive transgenic plantlets were confirmed by PCR analysis and GUS staining.
Construction of vectors for fruit transient expression assay
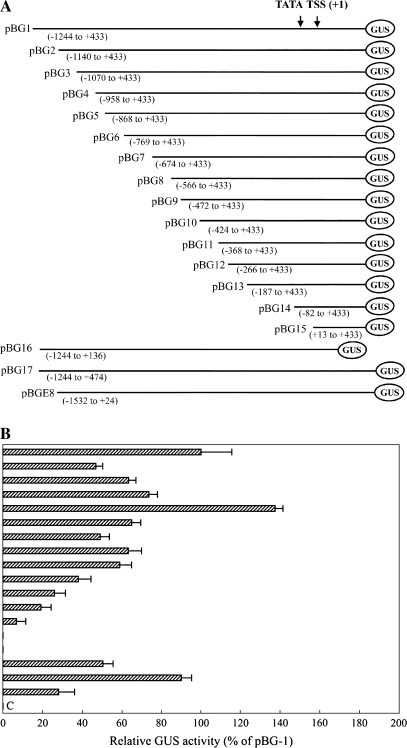
A series of AGPL1 promoter fragments (Fig. 4) were amplified by PCR performed for 35 cycles under the following standard conditions: denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min using LA Taq DNA polymerase (Takara, Ohtsu, Japan). Each PCR-amplified fragment was gel purified and digested with HindIII and XbaI, and the resulting fragments were cloned and sequenced. The following deletion constructs were generated: pBG1 (–1244 to +433), pBG2 (–1140 to +433), pBG3 (–1070 to +433), pBG4 (–958 to +433), pBG5 (–868 to +433), pBG6 (–769 to +433), pBG7 (–674 to +433), pBG8 (–566 to +433), pBG9 (–472 to +433), pBG10 (–424 to +433), pBG11 (–368 to +433), pBG12 (–266 to +433), pBG13 (–187 to +433), pBG14 (–82 to +433), pBG15 (+13 to +433), pBG16 (–1244 to +136), and pBG17 (–1244 to +474).
Fig. 4.
(A) A schematic map of the GUS constructs (number 1–18) developed for deletion analysis of the AGPL1 promoter. The 5′ and 3′ end coordinates of the relative deletion fragments are given in parentheses. At the top is shown the relative position of the TATA box, the TSS (+1), and the AGPL1 promoter coordinates. (B) AGPL1 promoter expression analysis in the fruit transient expression assay using the gusA reporter gene. Soluble protein extracts (∼60 mg) from fruit were used for the GUS assay. Each construct was assayed at least three times in independent experiments. The average GUS activity (as a percentage of pBG) with standard division is presented in the histogram. Error bars show the 95% confidence intervals of the means. (C) Untransformed control, extract from fruit.
The E8 promoter spanning 1556 bp was digested by HindIII and XbaI, and the obtained fragment was cloned into pBI426 (coordinates –1532 to +24 from the TSS), resulting in pBGE8 as a control promoter in watermelon fruit.
Fusion of the promoter deletions with GFP/RFP
The CaMV 35S promoter driving the red fluorescent protein (RFP) gene in an expression vector containing two cascades, including 35S::GFP::Nos and 35S::RFP::Nos, was substituted by the promoter fragments released from vectors pBG1–pBG17 to generate pGR1–pGR17, respectively.
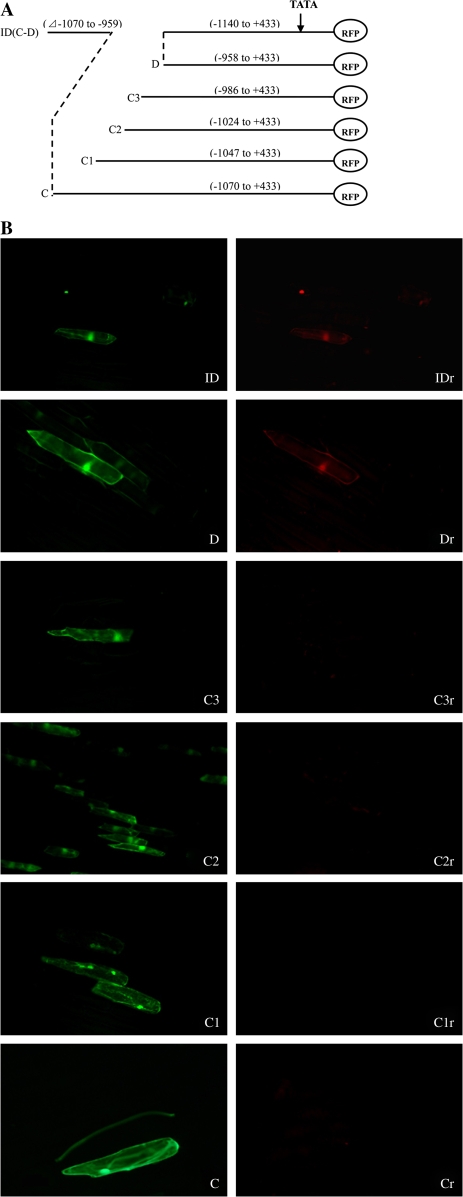
The region specified for internal deletion was excluded from the fragment by separately amplifying the upstream (first half) and downstream (second half) regions of the desired fragment from the full-length promoter with appropriately designed primers to generate a fragment with the following structure: 5′-HindIII-first half-GTAGGGTTGGTTTAG-3′ and 5′-CATCCCAACCAAATC-second half-XbaI-3′ (Fig. 6a). Both fragments were annealed for deletion amplification, and fine deletions were amplified by PCR with specific primers. All DNA sequences were checked before use. The following deletion constructs were generated: pGRID (–1244 to +433, internal deletion from –1068 to –959), pGR31 (–1047 to +433), pGR32 (–1024 to +433), and pGR33 (–986 to +433).
Fig. 6.
(A) A schematic map of RFP/GFP double marker gene transient expression vectors harbouring different fine truncated deletions of the AGPL1 promoter. ID (C and D), pGRID (–1140 to +433, –1070 to –959 deleted); C, pGR3 (–1070 to +433); C1, pGR31 (–1047 to +433); C2, pGR32 (–1024 to +433); C3, pGR33 (–986 to +433); D, pGR4 (–958 to +433). (B) C–ID are fluorescent images under an exciter filter (450–490 nm). Cr–IDr are fluorescent images of (C–ID) under an exciter filter (510–560 nm). All images were observed under 40× magnification. (This figure is available in colour at JXB online.)
Constructs for gain-of-function experiments
For gain-of-function experiments, the following cis-regions (–989 to –959 and –464 to –424) were individually inserted upstream of the minimal CaMV 35S promoter (coordinates –89 to +19 relative to the TSS) to develop the transient expression vectors pmGR1 and pmGR2. The cis-regions from the AGPL1 promoter were separately amplified by PCR and inserted into the site before the CaMV 35S minimal promoter. The following general structures were obtained and used for transient expression analysis to confirm the function of the putative regulation regions: 5′-putative cis region::35S mini::RFP::Nos and 35S::RFP::Nos-3′.
Transient expression assay
Immature watermelon fruits (∼10 days after anthesis) growing in the greenhouse were cut horizontally into slices (2 mm thickness) and placed in dishes. Each sample was bombarded three times with gold particles coated with plasmids. In all experiments, an equimolar amount of each construct's DNA (5 μg) was mixed with 1 mg of gold particles and suspended in 60 μl of ethanol. Each plant's material was bombarded with a 12 μl aliquot of the suspension per shot using a helium-driven Biolistic PDS-1000/He system (Bio-Rad) with a 28 mmHg vacuum. The distances between the rupture disc (1100 psi) and macrocarrier and the macrocarrier and samples were 3.0 cm and 7.0 cm, respectively. After bombardment, the tissues were incubated in the dark for 20 h at 25 °C. All constructs used in the transient assay were repeated three times.
Fresh onion epidermis was prepared for fluorescent transient expression assays. The procedures were performed as above.
GUS activity assay and histochemical staining
Histochemical staining for GUS enzyme activity was performed as described by Rech et al. (2003). Fragments of fruits, stems, leaves, and roots were taken from plants growing in a greenhouse for 2 months. These were vacuum infiltrated for 15–20 min in a reaction mixture containing 50 mM Na2HPO4 pH 7.0, 0.5% Triton X-100, 10 mM EDTA sodium salt, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, and 2 mM X-Gluc. Infiltrated tissues were incubated in GUS assay mixture for 16–18 h at 37 °C or until blue indigo dye precipitate was observed. Tissues were then post-fixed in 60% ethanol, 5% acetic acid, and 3.7% formaldehyde for a minimum of 2 h, and dehydrated in a series of aqueous ethanol solutions (v/v) of 70, 80, 90, and 100%, each for a minimum of 60 min.
Transgenic tomato fruits growing in a greenhouse were randomly selected for GUS activity analysis. GUS activity in tomato fruits, leaves, and transiently transformed watermelon fruit discs was analysed using a fluorometric assay as described in Jefferson et al. (1987) using 1 mM 4-methylumbelliferyl-β-D-glucuronide (4-MUG) as substrate. The reaction was performed for 1 h at 37 °C, and the released fluorescent product, 4-methylumbelliferone (4-MU), was read with a F-4500 fluorometer. Total soluble proteins of leaves and fruits were isolated and the protein concentrations were determined as described by Bradford (1976).
Visualization of GFP and RFP in transient expression
Fresh onion epidermis was placed onto MS medium for transient transformation. After transient transformation as described above, the epidermal cells were cultured for 20 h. Green fluorescent protein (GFP) fluorescence was visualized with a GFP filter set comprising an excitation filter (450–490 nm), a dichromatic beam splitter (505 nm), and a barrier filter (500 nm). RFP fluorescence was visualized with an RFP filter set comprising an excitation filter (510–560 nm), a dichromatic beam splitter (575 nm), and a barrier filter (590 nm). The light source was an HBO 100 W mercury lamp. Photos were taken with a Nikon digital colour camera coupled to the microscope. The fluorescence status of different epidermal cell was analysed in 10 independent transiently transformed tissue samples and the representative results were presented.
Isolation of nuclei and nuclear proteins
Nuclear isolation procedures were essentially the same as those described by Smirnova et al. (2000). All procedures were performed at 4 °C. The leaves (50 mg) were cut into small pieces and homogenized in 500 μl of cell lysis buffer [10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol (DTT), and 0.2 mM phenylmethylsulphonyl fluoride (PMSF)]. The mixture was incubated on ice for 10 min and 27.5 μl of 20× cytoplasmic extraction buffer (1× cytoplasmic extraction buffer: 30 mM HEPES (pH 7.9) at 4 °C, 140 mM KCl, 3 mM MgCl2) were added and mixed thoroughly. After centrifugation of the mixture at 16 000 g for 5 min, the pellet was resuspended in 250 μl of nuclear extraction buffer (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 400 mM KCl, 0.5 mM DTT, 0.2 mM PMSF, and 25% glycerol). The nuclear suspension was stirred on ice for 40 min and mixed thoroughly every 10 min. The supernatant was collected after centrifugation at 16 000 g for 10 min at 4 °C and stored at –80 °C.
Gel mobility shift assay and competition experiment
The electrophoretic mobility shift assay (EMSA) was performed as described by the supplier (Pierce product number 20148). The following oligonucleotides were synthesized and used as probes: PN1, 5′-AGGGGAGAACCAATCCAAAATTTTTAAATTTC-3′; PP1, 5′-TGGATTCTCAAGTTATTTGTGCATCTTAGACGTTAATATGT-3′; PN2, 5′-CTTAATTTTATTTAGGAATTTAATAATTGGGTGTGGTTTTG-3′; and PP2, 5′-CATAAAAATAAAGAAAGCTATAAACAGAGGTCCTAGGATGCAGATTGGTTACA-3′. Single-stranded oligonucleotides (5 pmol) were labelled at the 3′ end with biotin using terminal deoxynucleotidyl transferase (TdT) in labelled reaction buffer [100 mM sodium cacodylate, 2 mM CoCl2, 0.2 mM TRIS (2-carboxyethyl)phosphine (TCEP), 0.5 μmol biotin-11-UTP; pH 7.2]. Then, the end-labelled complementary oligos were mixed together in equal amounts and denatured at 90 °C for 1 min, cooled down slowly, and incubated at the melting temperature for 30 min to generate labelled, double-stranded probes.
Gel mobility shift reactions were performed in a 20 μl mixture containing 20 fmol of biotin-labelled oligonucleotide probes and 3 μl of nuclear protein in binding buffer [10 mM TRIS-HCl, 100 mM KCl, 1 mM DTT, 2.5% (v/v) glycerol, 0.05% NP-40, 5 mM MgCl2, 1 mM EDTA; pH 7.5]. A total of 50 ng μl−1 poly(dI–dC) was added as a non-specific competitive binding inhibitor. The unlabelled probes were used for competition assays. After incubation for 30 min at 25 °C, the reaction mixture was loaded onto a 6% native polyacrylamide gel containing 45 mM TRIS borate and 1 mM EDTA (0.5× TBE buffer) and subjected to electrophoresis at 100 V for 3 h. The binding reactions were then electrophoretically transferred to a nylon membrane and cross-linked. Detection of biotin-labelled nucleic acids was completed using the chemiluminescent nucleic acid detection module of Pierce (product no. 89880).
Results
Isolation and sequence analysis of AGPL1 5′-flanking regions
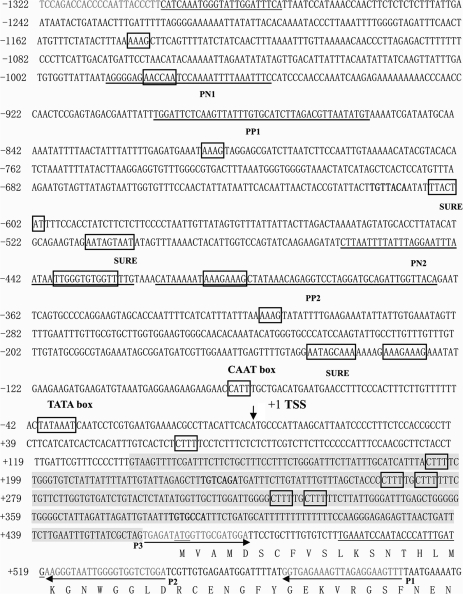
The nucleotide sequence of the clone is presented in Fig. 1. The length of the insert is 1864 bp, with 129 bp at the 3′ end that are identical to the 5′ end of the AGPL1 gene. The sequence is a non-specific amplification by primer P2 in the second step of uneven PCR. A search of GenBank revealed no significantly similar sequence. There is also a 46 bp inverted repeat at the two ends of the clone. There is no translation start (ATG) in the upstream inverted sequence and it therefore appears to be a pseudo gene. The AGPL1 promoter sequence contains several consensus eukaryotic regulatory domains such as TATA box- and CAAT box-like sequences. The TATA box is present 34 bp upstream of the transcriptional start site. Deletion analysis (presented in the following section) indicated that the TATA box sequence of the AGPL1 promoter is essential for its activity. A CAAT box sequence, CCATT, is present 82 bp upstream of the transcriptional start site.
Fig. 1.
The DNA sequence of the promoter region of ADP-glucose pyrophosphorylase large subunit (AGPase). A 1920 bp fragment (–1322 to +598 with respect to the TSS) including the intron of the 5′-untranslated region. Nucleotides are numbered on the left side, with the position of the TSS designated as +1. The translation start codon at position +466 is in bold. The dark sequence is the intron. (Deposited in the Nucleotide Sequence Database under the accession number AY262035.)
A search of the AGPL1 promoter for documented cis-regulatory elements discovered in other angiosperms revealed a number of elements that are potential binding sites for transcription factors (Table 1). Thirty-one AAAG boxes, recognized by Dof proteins, a class of plant-specific transcription factor, were found in the promoter region, among which 23 had inverse orientation (Yanagisawa, 2000). Two gibberellin (GA)-responsive elements (i.e. TAACAAA-like boxes) with inverse orientation were identified, which is a conserved GA response element in the promoters of GA-regulated genes expressed in cereal aleurone cells (Gubler et al., 1999). Four TGTCACA-like boxes were found in the promoter and two of them had inverse orientation and are known enhancer elements necessary for fruit-specific expression of the cucumisin gene in melon (Yamagata et al., 2002).
Table 1.
The putative cis-regulatory elements in the AGPL1 promoter
| Trans-factor | Core sequencea | Sequenceb | Position | Reference |
| SURE | AATAGAAAA | AATAGtAAt (+) | −512 to –504 | Grierson et al. (1994) |
| AATAGtAAA (−) | −608 to −600 | |||
| AATAGcAAA (+) | −150 to −142 | |||
| SURE-like fragments | AATAAAA | aAATAAAAt (−) | −459 to −452 | Kwak et al. (2006) |
| cAATAAAAt (−) | +212 to +218 | |||
| Dof/PBF | nnnwAAAGnnn | ttaAAAAGctt (+) | −1149 to −1139 | Yanagisawa and Schmidt (1999) |
| aaaTAAAGtag (+) | −814 to −804 | |||
| aaaTAAAGaaa (+) | −414 to −404 | |||
| ttaAAAAGtat (+) | −323 to −313 | |||
| aaaAAAAGaaa (+) | −144 to −134 | |||
| caaTAAAGaga (−) | −1254 to −1244 | |||
| tttTAAAGtat (−) | −1154 to −1144 | |||
| tttTAAAGttg (−) | −1125 to −1115 | |||
| aaaAAAAGtct (−) | −1093 to −1083 | |||
| attTAAAGtca (−) | −729 to −719 | |||
| Fruit-specific motif | TGTCACA | ctTGTTACA(+) | −618 to −612 | Yamagata et al. (2002) |
| TtTGTCAGA (+) | +231 to +237 | |||
| TaTTTCACA (−) | −294 to −288 | |||
| E-box | CANNTG | AtCAAATG (+) | −1291 to −1298 | |
| TcCAATTG (+) | −781 to −788 | |||
| GtCAGATG (+) | +232 to +239 | |||
| GARE | TAACAAA | TAACAAA (−) | −1114 to −1108 | Gubler et al. (1999) |
N=A+T+C+G, W=A+T.
(−), DNA sequence of the antisense strand; (+), DNA sequence of the sense strand.
Mapping the TSS of the AGPL1 promoter
The TSS of the AGPL1 promoter was determined by primer extension analysis with mRNA isolated from 10-d-old watermelon fruit after anthesis. A major extension product was detected and mapped to an A residue located 35 nucleotides downstream of the TATA element. It most probably represents the 5′ end of the AGPL1 transcript in this context. Sequence comparisons of the TSS with other fruit-specific promoters revealed low sequence homology.
Fruit-specific expression of the AGPL1 promoter::GUS in transgenic tomato plants
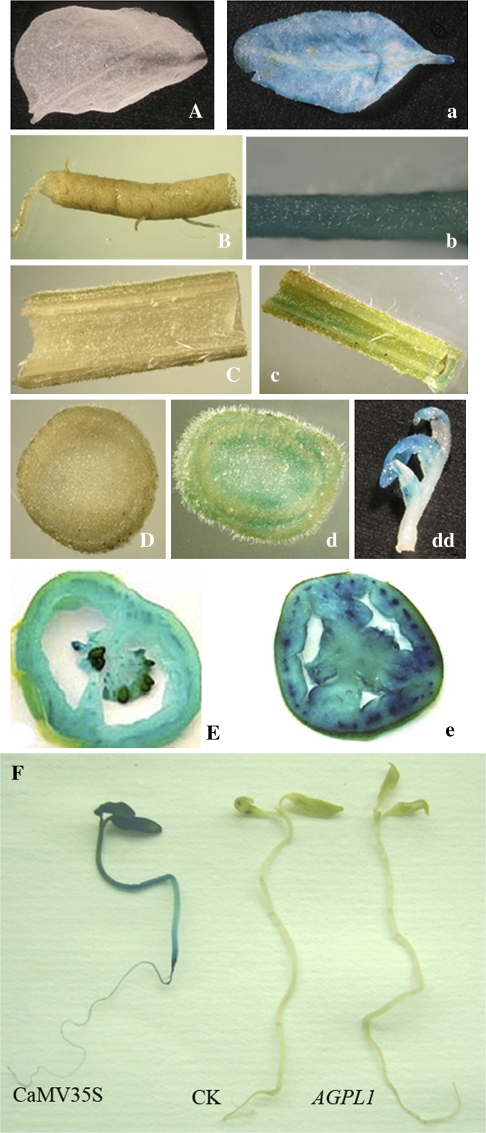
The promoting patterns of the AGPL1 promoter were investigated in stable transgenic plants. The 1.5 kb promoter of AGPL1 was used to replace the CaMV 35S promoter in the plant binary vector pBI121, and was transferred into tomato plants. Intense GUS staining was observed in all tissues including roots, stems, leaves, and fruits from transgenic plants transformed with pBI121 (35S::GUS). However, GUS staining was only detected in fruits from transgenic plants transformed with pBGV1 containing the AGPL1 promoter (AGPL1::GUS), confirming the fruit-specific activity of the AGPL1 promoter (Fig. 2A–E, a–e). During seed germination and seedling development, no GUS staining was visualized in AGPL1::GUS transgenic lines, whereas intense GUS staining was observed in 35S::GUS transgenic lines (Fig. 2F). During fruit development, the AGPL1 promoter showed high activity similar to the CaMV 35S promoter in young fruits, but with different spatial expression patterns. The CaMV 35S promoter drove gusA expression in all fruit tissues, including pericarp, placental tissue, and columella, with the highest expression in the vascular bundle, whereas the AGPL1 promoter drove gusA expression most abundantly in the pericarp and placental tissues around the seeds. Total GUS activity analysis revealed that GUS activity was ∼10-fold higher in fruits than in leaves in AGPL1::GUS transgenic lines, whereas it was the same in both organs in 35S::GUS transgenic lines (Fig. 3).
Fig. 2.
Histochemical localization of GUS activity in transgenic tomato plants under control of the AGPL1 and CaMV 35S promoters. Transgenic tomato carrying the AGPL1 promoter fused to GUS (A–E): (A) leaf; (B) root; (C) longitudinal section of stem; (D) transverse section of stem; (E) fruit. Transgenic tomato carrying the CaMV 35S promoter fused to GUS (a–e): (a) leaf; (b) root; (c) longitudinal section of stem; (d) transverse section of stem; (dd) bud; (e) fruit. (E and e) Note that the driving patterns for the AGPL1 promoter and CaMV 35S promoter plants are different. (F) Histochemical staining of 10-d-old transgenic tomato seedlings. (This figure is available in colour at JXB online.)
Fig. 3.
(A) Schematic map of the T-DNA region of binary vectors. The rectangular box represents the AGPL1 genome and the horizontal lines below it represent the different 5′ deletions (pGV1, pGV2, and pGV3). The T-DNA regions are not drawn to scale. (B) The T-DNA region of the binary vector, pBI121. The promoter fragments were cloned in the HindIII–BamHI sites. The gusA gene was driven by the AGPL1 promoter deletions (coordinates −1140 to +433, −769 to +433, −472 to +433 from the TSS). The relative positions of the gusA gene and the NPTII gene are shown with respect to the left border (LB) and right border (RB) of the T-DNA. (C) Reporter gene expression in fruit and leaf of stably transformed tomato plants containing the AGPL1 promoter and CaMV 35S promoter, respectively. Fruits and leaves from transgenic tomato plants grown in a growth chamber were pooled and assayed in duplicate. The fruits were growing: immature (IM), mature green (MG), and red ripe (RR). Error bars represent the standard error. Dark bars represent the average for each construct containing the relative promoter.
Another two constructs were made with the promoter fragment −769 to +433 (pBGV2) and −472 to +433 (pBGV3), and introduced into tomato plants by Agrobacterium-mediated transformation. pBI121 was used as a control. The promoter with these deletions did not alter the gene expression pattern. Even the shortest promoter fragment of −472 to +433 in pBGV3 led to the specific expression of gusA in fruits, with little decrease in activity compared with pBGV1 and pBGV2, suggesting that this 905 bp promoter with cis-elements is long enough for a fruit-specific promoter to function fully. All three constructs had the same GUS activity as pBI121, indicating high AGPL1 promoter activity in fruits.
5′ and 3′ end deletion analysis of the AGPL1 promoter in fruit
To determine the cis-acting elements, a 1677 bp AGPL1 promoter fragment (from –1244 to +433) was subjected to 5′ and 3′ end deletion analysis. Seventeen promoter deletions fused with the gusA gene were introduced into watermelon fruit discs and subjected to a transient expression assay. A schematic map of the deletions is presented in Fig. 4a, and expression analyses are shown in Fig. 4b. The expression level of construct 1 (pBG1, −1244 to +433 from the TSS) demonstrated high promoter activity, which was considered full promoter activity (100%). Construct 5 (pBG5, −868 to +433 from the TSS) gave maximum promoter activity that was ∼1.5-fold higher than that of construct 1. These data suggest that the upstream promoter sequence from −1244 to −868 from the TSS may not be essential for maximum promoter activity. In contrast, successive deletion of 99 bp and 95 bp from the 5′ end of construct 5 (as in constructs 6 and 7, respectively) dramatically reduced promoter activity compared with construct 5 (coordinates −868 to +433 from the TSS), indicating that a positive regulatory element is present in the region between −868 and −769, which up-regulates the AGPase large subunit gene promoter in the 1.3 kb promoter constructs in fruit. Further successive deletions from the 5′ end of constructs 8, 9, 10, 11, 12, and 13 decreased promoter activity accordingly. Deletion of 105 bp and 95 bp in constructs 14 and 15 (pG14 and pG15, coordinates −82 and +13 to +433 from the TSS, respectively) eliminated promoter activity, suggesting that deletion of the trans-acting elements results in undetectable levels of promoter activity. The construct E8 containing the promoter of the tomato E8 gene (−1532 to +24 from TSS) showed 28.1% less activity than that of construct 1, pBG1.
An intron spanning 323 bp located downstream of the promoter (+136 to +459) was found in the 5′ untranslated region (UTR) of AGPL1. The intron is rich in A/T (65%) and has GT–AG located at the two boundaries, an essential characteristic for splicing. To study its effect on promoter activity, two more constructs were made, one with the full-length intron (pG16, from −1244 to +474) and one without the intron from −1244 to +136 (pG17). The results of transient expression experiments in fruits showed that deletion of the intron led to a reduction in promoter activity, indicating that this intron has a positive effect on promoter activity (Fig. 4).
Deletion analysis on promoter activity in epidermal cells
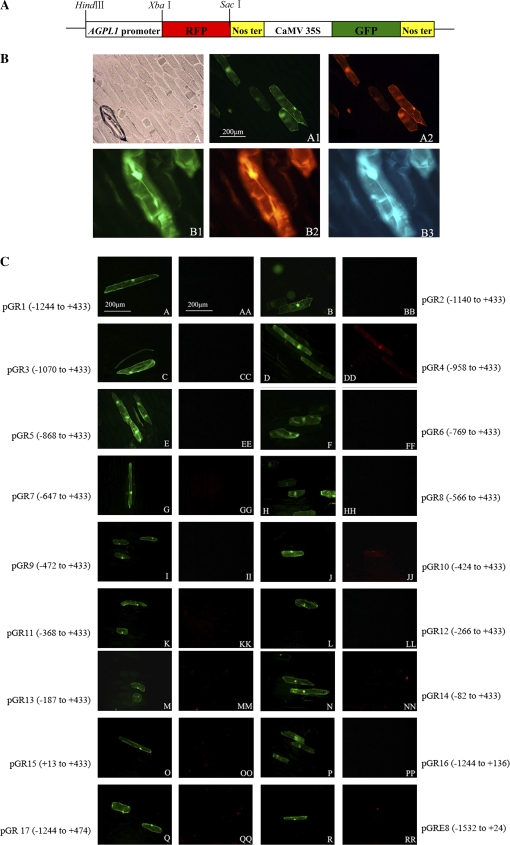
Although β-glucuronidase (gusA) has been widely used as a reporter gene for expression profiles of promoters or histochemical localization (Jefferson et al., 1987; Sunilkumar et al., 2002), it always suffers from a high background in plants, especially in leaves or other chlorophyll-rich tissues. In this study, a series of transient expression vectors were constructed using RFP and GFP as marker or internal standard genes to determine the specific expression profile of the promoter (Fig. 5a). Figure 5b shows the fluorescence images of cells transformed with the pGR vector harbouring CaMV 35S promoters for both GFP and RFP. GFP and RFP fluorescence with the same fluorescence intensities was observed in the same transformed cells. This system was further validated using the fruit-specific promoter E8. No red fluorescence was observed in green epidermal cells transformed with pGRE8, a construct containing the tomato fruit-specific E8 promoter upstream of the RFP gene.
Fig. 5.
(A) The schematic map of RFP/GFP double marker genes in the transient expression vector pBI221 with CaMV 35S promoter-driven GFP and RFP. (B) CaMV 35S promoter-driven GFP and RFP transient expression in onion epidermal cells. The results shown are from transient transformed epidermal cells using the DNA delivery particle bombardment system. (A) Light images of transiently transformed epidermal cells. (A1) Fluorescent images of (A) under an exciter filter (450–490 nm) that expressed GFP. (A2). Fluorescent images of (A1) under an exciter filter (510–560 nm) that expressed RFP. (B1) Fluorescent images that expressed GFP. (B2) Fluorescent images of (B1) that expressed RFP. (B3) Fluorescent images of (B1) under an exciter filter (380–420 nm). (C) Variable-length truncated AGPL1 promoter-driven RFP transient expression in epidermal cells. Single letters: fluorescent images expressed GFP driven by the CaMV 35S promoter. Double letters: fluorescent images expressed RFP driven by the AGPL1 promoter deletions. (This figure is available in colour at JXB online.)
Next, promoter fragments from constructs 1–17 (pBG1–17) were obtained and inserted into pGR to substitute for the CaMV 35S promoter before RFP. The resulting constructs were introduced into epidermal cells for transient expression assay. The results are shown in Fig. 5c. In GFP-positive cells, no RFP fluorescence was visualized when the cells were transformed with the three constructs pGR1 (from −1244 to +422), pGR2 (from −1140 to +422), and pGR3 (from −1070 to +422) (Fig. 5cAA, BB, CC), which were able to drive gusA gene expression at a level high enough to be detected in fruits. In contrast, no constitutive activity was detected in leaves. The data indicate that these promoter fragments have fruit-specific activity. Further deletion of the next 112 bp from −1070 to −958 resulted in the truncated promoter (pGR4, from −958 to +433) driving RFP expression in epidermal cells (Fig. 5cDD), suggesting that this region from −1070 to −958 contains a negative element that inhibits RFP expression only in the leaf.
Further encroachment into the promoter revealed another negative element for expression in the leaf. When a 48 bp fragment from −472 to −424 was deleted, the resulting promoter of –424 to +433 showed constitutive activity in epidermal cells (Fig. 5cJJ), although it was not as high as the fragment −958 to +433. Interestingly, in both cases, the constitutive activity in epidermal cells was eliminated when the fragments next to the fruit-specific elements were deleted, indicating a complex mechanism involved in the regulation of promoter specificity and activity (Fig. 5cEE, KK).
Fine-deletion analysis of the cis-domain (coordinates −1070 to −959 from TSS)
To confirm the function of the 112 bp putative cis-domain, an internal deletion construct pGRID (coordinates −1140 to +433, with deleted regions −1070 to −959 from the TSS) was generated. Transient expression analysis showed that the internal deleted promoter was able to drive RFP expression in epidermal cells, suggesting that the region containing negative element(s) only inhibits RFP expression in the leaf (Fig. 6IDr).
To dissect further the regulatory sequence present in the domain (coordinates −1070 to −959), a detailed deletion analysis was conducted. A schematic deletion map and the results are shown in Fig. 6a and b, respectively. In GFP-positive cells, no RFP fluorescence was visualized when the cells were transformed with three constructs, pGR31 (from −1047 to +433), pGR32 (from −1024 to +433), and pGR33 (from −986 to +433), indicating that these promoter fragments have no constitutive-like activity in leaves (Fig. 6C1r, C2r, C3r). Further deletion of the next 28 bp from −986 to −959 resulted in the truncated promoter (pGR4, from −958 to +433) driving RFP expression in epidermal cells, indicating that this region contains a negative element that only inhibits RFP expression in leaves (Fig. 6Dr). The next 41 bp region deletion (from −464 to 424) had the same result as described above, indicating that there are two negative regulatory regions responsible for fruit-specific promoting activity.
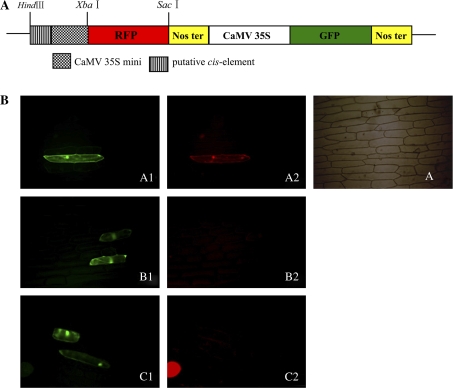
Gain-of-function experiment using putative regulatory domains
To confirm the functions of the two fruit-specific regions described above, the two fragments of −989 to −959 and −466 to −425 were fused with the CaMV 35S minimal promoter to generate two constructs, pmGR1 and pmGR2.
As shown in Fig. 7, in all green epidermal cells transformed with the control construct, red fluorescence could be visualized with the same expression pattern as found for green fluorescence, but with a weaker intensity, indicating that the CaMV 35S minimal promoter had lower expression activity (Fig. 7bA2). Fusion of the 30 bp region from −986 to −959 and the 42 bp region from −466 to −425 to the CaMV 35S minimal promoter led to the loss of the basal red fluorescence, confirming that these elements can inhibit the constitutive expression activity of the promoter in epidermal cells (Fig. 7bB2, C2). Several other regions flanking the two elements were analysed for expression activity in leaf epidermal cells (coordinates −1070 to −1047, −1047 to −1024, –1024 to −986, and −424 to −368, respectively, from the TSS). None of these regions was found to inhibit CaMV 35S minimal promoter activity completely in this organ (data not shown).
Fig. 7.
Functional analysis of putative cis-elements. (A) RFP/GFP double marker gene vector harbouring the CaMV 35S minimal promoter. (B) Functional analysis of transient epidermal cells. A, A1, and A2, CaMV 35S minimal promoter as the positive control in the fluorescent image assays. B1 and B2, fluorescent images harbouring a negative regulatory element (−464 to −424 from the TSS) of 41 bp that expressed GFP and RFP, respectively. C1 and C2, images harbouring a negative regulatory element (−989 to −959 from the TSS) of 30 bp that expressed GFP and RFP respectively. (This figure is available in colour at JXB online.)
Detection of nuclear factors that interact with the putative regulatory regions
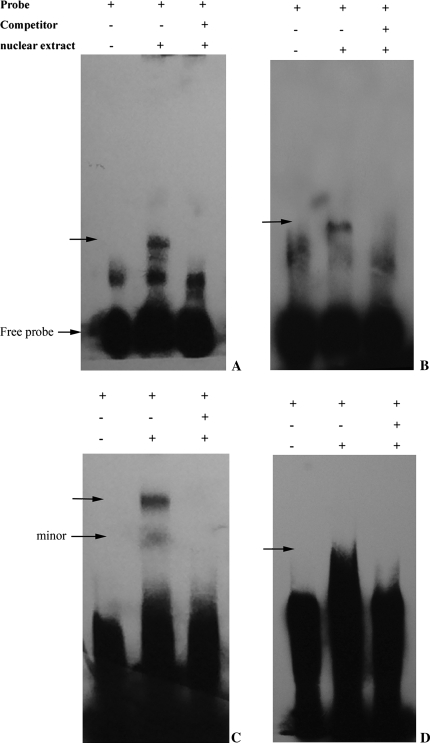
Transient expression assays suggested that the 30 bp sequences from −989 to −959 designated as PN1 and the 41 bp sequence from −464 to −424 designated as PN2 contain cis-element(s) responsible for fruit-specific expression, while both regions from −899 to −859 designated as PP1 and from −419 to −367 designated as PP2 coupled with PN1 and PN2, respectively, have a positive influence on gene expression in leaves. To evaluate whether these sequences interact specifically with proteins in nuclei from watermelon leaves, EMSAs were conducted. Nuclear protein extracts were prepared from watermelon leaves. The putative regions with both positive and negative regulatory element(s) were divided as described above. When these sequences were used as probes in the EMSAs, binding of nuclear proteins to PN1 was detected as a broad retarded band (Fig. 8a). The signal of the leaf protein–probe complex was very strong. To test further the specificity of the interaction, the probe was labelled and allowed to react with the protein extract in the presence or absence of an unlabelled competitor. Binding to the biotin-labelled fragment was efficiently inhibited by a 200-fold excess of the unlabelled fragment. When using PN2, one major band and some minor bands (Fig. 8c) were observed, indicating an interaction between the probe and the nuclear extract. Both DNA–protein complexes could be efficiently challenged using unlabelled PN2 (200-fold molar excess), demonstrating the specific nature of the above interaction.
Fig. 8.
EMSAs of nuclear proteins from watermelon leaves with the synthetic oligonucleotides. Binding of nuclear proteins from watermelon leaves to the labelled probes. (A) Identification of nuclear proteins that bind specifically to the 30 bp fragment from −989 to −959 (PN1) of the AGPL1 promoter. (B) Gel shifts using a 41 bp fragment from −899 to −859 (PP1) representing the positive element and leaf nuclear extracts. (C) Gel shifts using a 41 bp fragment from −464 to −424 (PN2). (D) Gel shifts using a 53 bp fragment from −419 to −367 (PP2). The nuclear extract was prepared from watermelon leaves as described in Materials and methods. A gel retardation assay was performed using a DNA fragment as a probe, as described in Materials and methods. The DNA−protein complexes in (A), (B), (C), and (D) were formed in the presence of 1 μg of non-specific competitor, poly dI:dC–dI:dC. All of the probes were incubated in the presence (lane 2 and lane 3) or absence (lane1) of nuclear extract. Competitors were added in 200-fold molar excess (lane 3). The arrow indicates the position of DNA–protein complexes. The gel retardation assay was performed as described in Materialsand methods. All were electrophoresed in a 6% polyacrylamide gel.
Similarly, for the positive element fragments PP1 and PP2, specific DNA–protein complexes (Fig. 8b, d) could also be obtained.
Critical nucleotides responsible for the binding of nuclear proteins in the motif
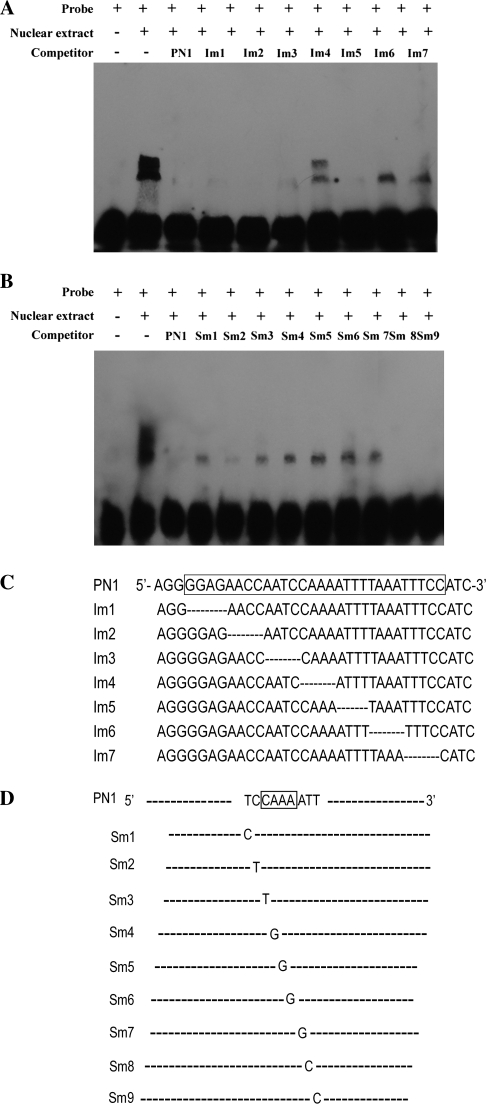
To pinpoint the cis-element responsible for binding with leaf nuclear proteins, a series of mutated PN1 oligonucleotides were synthesized in which every four nucleotides was successively deleted. These oligonucleotides were used as competitors in the EMSAs with PN1 fragments as labelled probes (Fig. 9a). The addition of a 200-fold excess of seven competitors to the binding mixture significantly diminished the retarded bands. Surprisingly, each of two mutated competitors, Im4 and Im6, also showed weak competition (Fig. 9b). Im6 diminished one retarded band, while Im4 diminished both bands, suggesting that Im6 competed more strongly with the PN1 probe than Im4. These results confirmed that there are at least two binding regions for the nuclear proteins within the PN1 probe, or two forms of one binding factor, and that the CAAA deleted in Im4 binds more strongly to the nuclear proteins than the TAAA in Im6.
Fig. 9.
Binding of leaf nuclear proteins to the cis-element in the PN1 region. (A and B) Determination of the binding site of the nuclear protein in gel mobility shift experiments. PN1 was used as a DNA probe. Competitors, the sequences of which are listed in C and D, were added in 200-fold molar excess to the corresponding probe. (C) Sequences of mutated derivatives of PN1. Only the mutated nucleotides in the upper strands are shown below the sequences of the wild type.
To confirm further the binding of nuclear protein to the motif, detailed analysis of sequence-specific binding was performed using mutated derivatives of these sequences in EMSAs. Successive single base pair mutations were introduced into these fragments (Fig. 9), and the mutated fragments were used as competitors in competition experiments with the wild-type fragment. As shown in Fig. 9, the specificity of binding of nuclear protein to the CAAA region was demonstrated by the ability to compete this complex effectively with a 200-fold molar excess of the sequence. However, competitors containing a point mutation located at positions 1, 3, 4, 5, 6, or 7 in the sequence failed to bind the nuclear protein fully to the wild-type fragment. In particular, oligonucleotides with a point mutation at positions 4, 5, 6, or 7 in the sequence showed little competition. In contrast, oligonucleotides containing mutations at the middle position of this sequence efficiently competed with the binding of nuclear protein. This indicates that one of the nuclear protein-binding sites in the CAAA region is located in a region covering the sequence, and that the nucleotides important for binding are T1, C3, A4, A5, A6, A7.
An analogous experiment was conducted for the motif in Im6. A 200-fold molar excess of the PN1 fragment efficiently interfered with protein binding to the labelled PN1 fragment. However, competitors containing point mutations could not abolish the binding of nuclear protein to the wild-type fragment. These observations indicate that the AATTTC motif is not responsible for the binding of nuclear proteins, and that perhaps the role of this motif is only as a cis-acting enhancer element in the AGPL1 promoter.
Discussion
AGPase is a heterotetrameric enzyme containing two small and two large subunits, between which there are differences in the number of isoforms and expression patterns in higher plants. The large subunit is present as multiple isoforms and shows tissue-specific expression. There are at least three isoforms of the large subunit encoded by independent nuclear genes in maize (Giroux et al., 1995), sweet potato (Harn et al., 2000), tomato (Chen et al., 1998; Park and Chung, 1998), barley (Eimert et al., 1997), oriental melon (Park et al., 1998), sugar beet (Müller-Röber et al., 1995), chickpea (Singh et al., 2002), and Arabidopsis (Kleczkowski et al., 1999). The different isoforms of the large subunit are expressed in different organs in tomato. For example, AgpL1 of tomato is highly expressed in stems and poorly expressed in roots; AgpL2 is expressed in roots and fruits; and AgpL3 is exclusively expressed in leaves (Park and Chung, 1998). Kim et al. (1998) reported that the large subunit of watermelon was strongly expressed in fruits and hardly expressed in other tissues, which is consistent with the fact that AGPase large subunits exhibit fruit-specific expression. However, the 5′-flanking regions of the gene and the mechanism underlying the tissue-specific expression pattern have not been studied.
A 1.8 kb 5′-flanking region of one AGPase large subunit gene was isolated from watermelon genomic DNA that has universal characteristics of plant promoters with a TATA box (TATAAAT) and CAAT box (CATT). A remarkable feature of the AGPL1 promoter structure is that it contains many Dof cis-elements, which are involved in the expression of a variety of signal response and/or tissue-specific genes in plants (Baumann et al., 1999; Yanagisava and Schmidt, 1999). In this study, the 5′-UTR did not match the previous sequence (AF032472) reported by Kim et al. (1998). Specific primers were designed for the 5′-UTR of the sequence shown in Fig. 1 (AY262035) and the 5′-UTR of the previously reported cDNA sequences (AF032472), and were used to amplify RNA from 10-d-old watermelon fruit. Unexpectedly, only the primer from AY262035 had the correct ampification product, not AF032472, suggesting that the gene reported here is truly expressed in watermelon fruit. The 1.8 kb promoter region of the AGPL1 promoter augmented gene expression in a fruit-specific manner in transgenic tomato plants. The transient expression analysis in watermelon fruits revealed that the core promoter might be the sequence from −187 to +136 from the TSS for basic gene expression, which contains a TATA box element at −34, two Dof cis-elements, plus the 5′-UTRs without the intron.
To determine the specific expression profile of the promoter, a series of 5′ deletions were isolated, fused with gusA, and subjected to a transient expression assay in watermelon fruits. Unexpectedly, the truncated promoters still exhibited high activity in fruit even when all the sequences upstream of the TATA box were deleted. Therefore, it is proposed that the specific expression was not contributed by the binding of cis-elements with trans-factors in fruits, but in leaves. Then, GFP and RFP take the place of conventional gusA reporter genes to avoid the influence of a high background resulting in localization artefacts (Mascarenhas and Hamilton, 1992). In a series of transient expression assays, two regions (PN1, from −986 to −958; and PN2, from −472 to −424) responsible for the switch from fruit-specific expression to constitutive-like expression in epidermal cells were determined. This assumption is supported by the fact that the two regions inhibited CaMV 35S minimal promoter activity in leaves. Nuclear factors that specifically bind to the regions have been identified by gel shift assay with nuclear extracts prepared from leaves of watermelon plants (Fig. 8a, c). It is therefore hypothesized that there are two potential sites responsible for trans-factors in leaves, in which binding leads to inhibition of promoter activity in leaves but not in fruits. Furthermore, competition experiments in the EMSA clearly showed that CAAA was the most important site in the region. In addition, when the regulatory regions were searched for homology with other cis-elements, the PN1 region revealed an AACCAA motif, and the PN2 region had an inverted AACCAA motif (5′-TTGGGTGTGGTT-3′), which is a necessary cis-element in the Lhcb21 promoter for phytochrome regulation. The binding activity of the transcription factors that bind to the Lhcb21 element is much greater in etiolated plants than in green plants (Degenhardt and Tobin, 1996). However, in the present experiments, deletion of the PN1 and PN2 elements, which include the Lhcb21 motif, had no influence on binding activity with nuclear proteins in leaves (Im2 in Fig. 9). Single base pair mutation analysis of sequences containing CAAA was performed, and the results confirmed that TCCAAAA is the regulatory cis-element responsible for specific expression. Interestingly, a reverse TCCAAAA-like (5′-TCCTAAA-3′) motif was found in the PN2 region. This observation is consistent with the above results of a negative regulatory region for specific promoting activity. To our knowledge, the TCCAAAA motif is a novel cis-element in the promoter of plant genes.
Another interesting aspect of the AGPL1 promoter is that the truncated promoter with a 41 bp deletion from −899 to −859 or a 53 bp deletion from −419 to −367 showed no promoting activity in leaves, suggesting that the remaining portions of the promoter harbour positive cis-elements for leaf expression. A DNA-binding factor was also detected in the nuclear fraction prepared from leaves, which formed complexes with both promoters (Fig. 8b, d). These observations suggest that transcriptional factors are constantly present in the nucleus and bind to PP1 and PP2. There are several possible roles for the PP1- and PP2-binding factors that interact with PN1- and PN2-binding factors, respectively, in leaves or other non-fruit tissues. Because the two regions are thought not to be involved in specific inhibitory expression in leaves, these elements were not examined further in regard to regulated gene expression. The function of the two regions containing the specific related cis-element was also confirmed in stable transgenic plants. When one or two cis-elements were harboured, the promoter fragments showed fruit-specific promoting patterns. There are also cis-regulatory enhancer elements in the positively regulated activators. The binding of specific factors to the coupled factor led to expression inhibition in leaves. The flanking factor functions when its inhibitor is deleted. Therefore, the presence and interaction of these putative cis-elements may be a significant factor in gene expression assays and in fruits (Fig. 10).
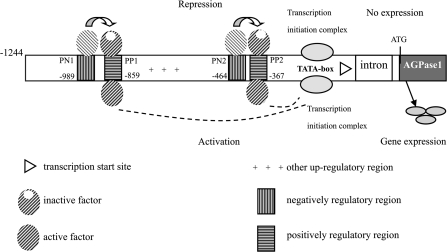
Fig. 10.
Organization of cis-elements in the 5′-flanking region of the AGPL1 gene. The numbers refer to positions relative to the transcriptional start site, which is indicated by a triangle. +, up-regulated elements; in leaves, trans-factors binding to the PN1 and/or PN2 elements repressed transcriptional activity of the promoter. Deletion of PN-binding sites relieves this repression of PP-binding factors. When the PN binding activity to the trans-factors is reduced or less able to bind, this renders trans-factors unable to repress gene activity. Instead, the remaining complex, which may involve different PP binding activity, activates transcription.
Introns are usually located in the coding regions of a gene in most organisms, with a few found in the 5′-UTR, an important region of the genome (Ng et al., 2004; Salvador et al., 2004). Tissue-preferential expression of the OSTubA1 gene may be mediated by intron 1 (Jeon et al., 2000). The intron found in the AGPL1 5′-UTR in this study contains two inverted TGTCACA motifs that were enhancer elements necessary for fruit-specific expression of the cucumisin gene in melon (Yamagata et al., 2002). However, deletion of the intron had no significant effect on the cell-specific expression pattern. In the present study, the intron increased the gene expression level by 1.5-fold in transient expression assays. In other studies, introns were also found to enhance gene expression by 30- to 800-fold in protoplast transient expression systems or in stably transformed plants (Bhattacharyya et al., 2003; Kim et al., 2006). Introns can enhance the maturation of mRNA after transcription (Rose and Beliakoff, 2000), and this enhancing effect is position and orientation dependent. An inverted cis-acting sequence element (G/C)CGA(C/T)N(A/G)N15(T/C/A)(A/T/G) was found in the intron that acted as an enhancer in Chlamydomonas reinhardtii (von Gromoff et al., 2006). The present studies further support a role for introns in regulating gene expression.
Tissue-specific expression is presumably due to interactions between a set of regulatory proteins and a special combined set of cis-regulatory elements in the promoter region (Singh, 1998). Therefore, expression of the designed gene is determined by trans-factors present in differentiated tissues. In this study, it was demonstrated that a TCCAAAA motif functions as a negative regulatory element in the AGPL1 promoter causing a loss of promoting activity in leaves but not in fruits. DNA-binding factors that specifically bind to the four putative regions in vitro were identified. All of these factors were detected in nuclear extracts from watermelon leaves.
In maize, the C4-type PEPC promoter was a target for Dof1 binding (Yanagisawa, 2000). Dof1 differentially elevated promoter activity in etiolated and greening protoplasts, suggesting the involvement of Dof1 in light-regulated gene expression (Yanagisawa and Sheen, 1998; Yanagisawa, 2000). Dof2 blocked transactivation by Dof1 and repressed the activity of the C4-type PEPC promoter. Based on the constitutive expression of Dof1 in leaves, stems, and roots, and predominant expression of Dof2 in stems and roots, a regulatory mechanism for tissue-specific gene expression was proposed (for a review, see Yanagisawa, 2004). Hence, this study has illustrated a mechanism for fruit-specific gene expression that is different from other well-characterized positive regulatory mechanisms, which operates through interactions between cis-elements in the 5′-flanking regions, such as ethylene-induced ripening-related genes and the ethylene-independent genes E4, E8, 2A11, PG, and ACO1 (Montgomery et al., 1993; van Haaren and Houck, 1993; Xu et al., 1996; Coupe and Deikman, 1997; Deikman et al., 1998; Moon and Callahan, 2004).
A model of the mechanism for gene regulation that is supported by data from the current study is shown in Fig. 10. In leaves, the binding of a protein complex to the TCCAAAA motif represses the initiation of gene transcription. Deletion of any one of the regions that harbour a cis-element would somehow relieve the transcription repression of the PP regions, so that gene expression could be activated. When the first coupled positive and negative trans-factor-binding sites are deleted, the second coupled complex of trans-factors is sufficient to mediate inactivation in leaves involved in the fruit-specific expression pattern. This pattern is most probably established via the action of fruit-specific transcriptional regulators interacting with cis-acting DNA sequences and/or trans-acting regulators within the analysed promoter fragment, where both positive and negative regulators were found. Further studies of their potential interactions with transcriptional factors might provide new insights into the mechanisms of organ-specific transcription in plants. It remains to be determined which types of trans-acting factors are responsible for the repression mechanism of the fruit-specific promoter.
Acknowledgments
This research was funded by the National Natural Science Grant (No. 30671430). We specially thank Dr Xiping Wang for advice and supporting.
Glossary
Abbreviations
- AGPase
ADP-glucose pyrophosphorylase
- CaMV
cauliflower mosaic virus
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- MS
Murashige and Skoog
- PCR
polymerase chain reaction
- RFP
red fluorescent protein
- TSS
transcription start site
- UTR
untranslated region
- X-Gluc
5-bromo-4-chloro-3-indolyl-β-d-glucuronide
References
- Baumann K, Paolis AD, Costantino P, Gualberti G. The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. The Plant Cell. 1999;11:323–333. doi: 10.1105/tpc.11.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Pattanaik S, Maiti IB. Intron-mediated enhancement of gene expression in transgenic plants using chimeric constructs composed of the Peanut chlorotic streak virus (PCISV) promoter-leader and the antisense orientation of PCISV ORF VII (p7R) Planta. 2003;218:115–124. doi: 10.1007/s00425-003-1078-4. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;7:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chen BY, Wang Y, Janes HW. ADP-glucose pyrophosphorylase is located to both the cytoplasm and plastids in developing pericarp of tomato fruit. Plant Physiology. 1998;116:101–106. doi: 10.1104/pp.116.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XF, Wu R. Direct amplification of unknown genes and fragments by uneven polymerase chain reaction. Gene. 1997;185:195–199. doi: 10.1016/s0378-1119(96)00637-3. [DOI] [PubMed] [Google Scholar]
- Coupe SA, Deikman J. Characterization of a DNA-binding protein that interacts with 5′ flanking regions of two fruit-ripening genes. The Plant Journal. 1997;11:1207–1218. doi: 10.1046/j.1365-313x.1997.11061207.x. [DOI] [PubMed] [Google Scholar]
- Degenhardt J, Tobin EM. A DNA binding activity for one of two closely defined phytochrome regulatory elements in an Lhcb promoter is more abundant in etiolated than in green plants. The Plant Cell. 1996;8:31–41. doi: 10.1105/tpc.8.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deikman J, Xu R, Kneissl ML, Ciardi JA, Kim KN, Pelah D. Separation of cis elements responsive to ethylene, fruit development, and ripening in the 5′-flanking region of the ripening-related E8 gene. Plant Molecular Biology. 1998;37:1001–1011. doi: 10.1023/a:1006091928367. [DOI] [PubMed] [Google Scholar]
- Dey N, Maiti IB. Structure and promoter/leader deletion analysis of mirabilis mosaic virus (MMV) full-length transcript promoter in transgenic plants. Plant Molecular Biology. 1999;40:771–782. doi: 10.1023/a:1006285426523. [DOI] [PubMed] [Google Scholar]
- Eimert K, Luo C, Déjardin A, Villand P, Thorbjørnsen T, Kleczkowski LA. Molecular cloning and expression of the large subunit of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves. Gene. 1997;189:79–82. doi: 10.1016/s0378-1119(96)00837-2. [DOI] [PubMed] [Google Scholar]
- Giroux M, Smith-White B, Gilmore V, Hannah LC, Preiss J. The large subunit of the embryo isoform of ADP glucose pyrophosphorylase from maize. Plant Physiology. 1995;108:1333–1334. doi: 10.1104/pp.108.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C, Du JS, de Torres Zabala M, Beggs K, Smith C, Holdsworth M, Bevan M. Separate cis sequences and trans factors direct metabolic and developmental regulation of a potato tuber storage protein gene. The Plant Journal. 1994;5:815–826. doi: 10.1046/j.1365-313x.1994.5060815.x. [DOI] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. The Plant Journal. 1999;17:1–9. doi: 10.1046/j.1365-313x.1999.00346.x. [DOI] [PubMed] [Google Scholar]
- Harn CH, Bae JM, Lee SS, Min SR, Liu JR. Presence of multiple cDNAs encoding an isoform of ADP-glucose pyrophosphorylase large subunit from sweet potato and characterization of expression levels. Plant and Cell Physiology. 2000;41:1235–1242. doi: 10.1093/pcp/pcd049. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Kim C, An G. Tissue preferential expression of a rice α-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiology. 2000;123:1005–1014. doi: 10.1104/pp.123.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IJ, Kahng HY, Chung WI. Characterization of cDNAs encoding small and large subunits of ADP-glucose pyrophosphorylases from watermelon (Citrullus vulgaris S.) Bioscience Biotechnology and Biochemistry. 1998;62:550–555. doi: 10.1271/bbb.62.550. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Kim H, Shin JS, Chung CH, Ohlrogge JB, Suh MC. Seed-specific expression of sesame microsomal oleic acid desaturase is controlled by combinatorial properties between negative cis-regulatory elements in the SeFAD2 promoter and enhancers in the 5′-UTR intron. Molecular Genetics and Genomics. 2006;276:351–368. doi: 10.1007/s00438-006-0148-2. [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA, Sokolov LN, Luo C, Villand P. Molecular cloning and spatial expression of an ApL1 cDNA for the large subunit of ADP-glucose pyrophosphorylase from Arabidopsis thaliana. Zeitscrift für Naturforschung, C. 1999;54:353–358. doi: 10.1515/znc-1999-5-610. [DOI] [PubMed] [Google Scholar]
- Kwak MS, Noh SA, Oh MJ, Huh GH, Kim KN, Lee SW, Shin JS, Bae JM. Two sweet potato ADP-glucose pyrophosphorylase isoforms are regulated antagonistically in response to sucrose content in storage roots. Gene. 2006;366:87–96. doi: 10.1016/j.gene.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Mascarenhas JP, Hamilton DA. Artifacts in the localization of GUS activity in anthers of petunia transformed with a CaMV 35S–GUS construct. The Plant Journal. 1992;2:405–408. [Google Scholar]
- McBride KE, Summerfelt KR. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Molecular Biology. 1990;14:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- Montgomery J, Pollard V, Deikman J, Fischer RL. Positive and negative regulatory regions control the spatial distribution of polygalacturonase transcription in tomato fruit pericarp. The Plant Cell. 1993;5:1049–1062. doi: 10.1105/tpc.5.9.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H, Callahan AM. Developmental regulation of peach ACC oxidase promoter–GUS fusions in transgenic tomato fruits. Journal of Experimental Botany. 2004;55:1519–1528. doi: 10.1093/jxb/erh162. [DOI] [PubMed] [Google Scholar]
- Morell MK, Bloom M, Knowles V, Preiss J. Subunits structure of spinach leaf ADP-glucose pyrophosphorylase. Plant Physiology. 1987;85:182–187. doi: 10.1104/pp.85.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Röber B, Nast G, Willmitzer L. Isolation and expression analysis of cDNA clones encoding a small and a large subunit of ADP-glucose pyrophosphorylase from sugar beet. Plant Molecular Biology. 1995;27:191–197. doi: 10.1007/BF00019190. [DOI] [PubMed] [Google Scholar]
- Ng DW, Chandrasekharan MB, Hall TC. The 5′ UTR negatively regulates quantitative and spatial expression from the ABI3 promoter. Plant Molecular Biology. 2004;54:25–38. doi: 10.1023/B:PLAN.0000028767.06820.34. [DOI] [PubMed] [Google Scholar]
- Park SW, Chung WI. Molecular cloning and organ-specific expression of three isoforms of tomato ADP-glucose pyrophosphorylase gene. Gene. 1998;206:215–221. doi: 10.1016/s0378-1119(97)00588-x. [DOI] [PubMed] [Google Scholar]
- Park SW, Kahng HY, Kim IJ, Park JO, Chung WI. Molecular cloning and characterization of small and large subunits of ADP-glucose pyrophosphorylase from oriental melon. Journal of Plant Research. 1998;111:59–63. [Google Scholar]
- Preiss J. Biosynthesis of starch and its regulation. In: Stumpf PK, Conn EE, editors. The Biochemistry of Plants. Vol 14. New York: Academic Press; 1998. pp. 181–254. [Google Scholar]
- Rech P, Grima-Pettenati J, Jauneau A. Fluorescence microscopy: a powerful technique to detect low GUS activity in vascular tissues. The Plant Journal. 2003;33:205–209. doi: 10.1046/j.1365-313x.2003.016017.x. [DOI] [PubMed] [Google Scholar]
- Rose AB, Beliakoff JA. Intron-mediated enhancement of gene expression independent of unique intron sequences and splicing. Plant Physiology. 2000;122:535–542. doi: 10.1104/pp.122.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador ML, Suay L, Anthonisen IL, Klein U. Changes in the 5′-untranslated region of the rbcL gene accelerate transcript degradation more than 50-fold in the chloroplast of Chlamydomonas reinhardtii. Current Genetics. 2004;45:176–182. doi: 10.1007/s00294-003-0470-8. [DOI] [PubMed] [Google Scholar]
- Singh KB. Transcriptional regulation in plants: the importance of combinatorial control. Plant Physiology. 1998;118:1111–1120. doi: 10.1104/pp.118.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Choi SB, Modi MK, Okita TW. Isolation and characterization of cDNA clones encoding ADP-glucose pyrophosphorylase (AGPase) large and small subunits from chickpea (Cicer arietinum L.) Phytochemistry. 2002;59:261–268. doi: 10.1016/s0031-9422(01)00457-5. [DOI] [PubMed] [Google Scholar]
- Smirnova IV, Bittel DC, Ravindra R, Jiang H, Andrews GK. Zinc and cadmium can promote rapid nuclear translocation of metal response element-binding transcription factor-1. Journal of Biological Chemistry. 2000;275:9377–9384. doi: 10.1074/jbc.275.13.9377. [DOI] [PubMed] [Google Scholar]
- Sunilkumar G, Mohr L, Lopata-Finch E, Emani C, Rathore KS. Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Molecular Biology. 2002;50:463–474. doi: 10.1023/a:1019832123444. [DOI] [PubMed] [Google Scholar]
- van Haaren MJJ, Houck CM. A functional map of the fruit-specific promoter of the tomato 2A11 gene. Plant Molecular Biology. 1993;21:626–640. doi: 10.1007/BF00014546. [DOI] [PubMed] [Google Scholar]
- von Gromoff ED, Schroda M, Oster U, Beck CF. Identification of a plastid response element that acts as an enhancer within the Chlamydomonas HSP70A promoter. Nucleic Acids Research. 2006;34:4767–4779. doi: 10.1093/nar/gkl602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Goldman S, Coupe S, Deikman J. Ethylene control of E4 transcription during tomato fruit ripening involves two cooperative cis elements. Plant Molecular Biology. 1996;31:1117–1127. doi: 10.1007/BF00040829. [DOI] [PubMed] [Google Scholar]
- Yamagata H, Yonesu K, Hirata A, Aizono Y. TGTCACA motif is a novel cis-regulatory enhancer element involved in fruit-specific expression of the cucumisin gene. Journal of Biological Chemistry. 2002;277:11582–11590. doi: 10.1074/jbc.M109946200. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. The Plant Journal. 2000;21:281–288. doi: 10.1046/j.1365-313x.2000.00685.x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant and Cell Physiology. 2004;45:386–391. doi: 10.1093/pcp/pch055. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Schmidt RJ. Diversity and similarity among recognition sequences of Dof transcription factors. The Plant Journal. 1999;17:209–214. doi: 10.1046/j.1365-313x.1999.00363.x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Sheen J. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. The Plant Cell. 1998;10:75–89. doi: 10.1105/tpc.10.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]