Abstract
A catalytic beacon sensor for Pb2+ has been developed based on the first DNAzyme discovered in the field, and such a sensor has shown a much higher metal ion selectivity (40 000 times) than the previously reported Pb2+ sensor based on 8–17 DNAzyme and thus is suitable for a wider range of practical applications.
Wide-spread Pb2+ contamination has posed adverse effects on human health, especially on children.1 To reverse the negative effects, accurate, on-site and real-time detection and quantification of Pb2+ in the environment or in vivo is an important first step. Toward this goal, a number of Pb2+ sensors have been developed.2–5 Among them, DNAzymes have emerged recently as a promising class of molecules to build sensors. DNAzymes that are selective for a number of metal ions have been obtained from a large DNA library through in vitro selection.7,8 More importantly, these DNAzymes exhibit high catalytic turnovers that allow signal amplification and employ rate-based measurement that is much less vulnerable to the sample background interferences that have hampered other intensity-based measurement methods. Because of these features, DNAzymes have been converted into fluorescent, colorimetric and electrochemical sensors for a wide range of metal ions, including Pb2+.3–6
Currently, all DNAzyme-based Pb2+ sensors are built on the 8–17 DNAzyme, which is capable of catalyzing a phosphodiester bond cleavage reaction in the presence of Pb2+. This DNAzyme has been obtained through in vitro selection by several groups under different conditions.9 Even though the same 8–17 DNAzyme was selected against different metal ions such as Mg2+, Zn2+ and Ca2+, a survey of the metal-ion-dependent activities showed that the 8–17 DNAzyme displays substantially higher activity in the presence of Pb2+ than any other metal ion,4,10 and thus this DNAzyme has been converted into sensors for Pb2+.3–5 Although being selective for Pb2+, the 8–17 DNAzyme is still active in the presence of other metal ions, such as Mg2+, Zn2+, Mn2+, Co2+ and Ca2+. For example, the activity of the 8–17 DNAzyme at millimolar concentrations of Zn2+ has been shown to be equivalent to nanomolar concentrations of Pb2+.9,10 As a result, the 8–17 DNAzyme would be a good Pb2+ sensor in the presence of equal concentrations of Pb2+ and Zn2+, but it would become ineffective if Zn2+ concentration is much higher. Therefore, a more selective DNAzyme sensor is required in such a situation.
The first DNAzyme was selected using Pb2+ as the cofactor more than a decade ago.8 Similar to the 8–17 DNAzyme, this classic DNAzyme (to avoid confusion, we name this DNAzyme GR-5 DNAzyme) can also catalyze the cleavage of an RNA base embedded in the DNA substrate in the presence of Pb2+. To find out if this DNAzyme has a higher selectivity for Pb2+ over other competing metal ions, we carried out metal-ion dependent activity assays with this DNAzyme and found that it has excellent selectivity for Pb2+ (Fig. S1†). For example, under single turnover conditions, the kobs (observed rate constant was obtained by fitting the equation y = y0 + Ae−x/t) was measured to be 0.64 min−1 for 100 μM Pb2+ in HEPES buffer (pH 7.0 plus 500 mM NaCl, 500 mM KCl, 50 mM MgCl2). In contrast, under the same conditions, the kobs was measured to be 0.014 min−1 for 20 mM Zn2+, and no other metal ion surveyed has a kobs higher than 0.0034 min−1 (Table S1†). Encouraged by this high selectivity, we report here a new catalytic beacon sensor for Pb2+ based on the GR-5 DNAzyme and compare its performance with that of the 8–17 DNAzyme.
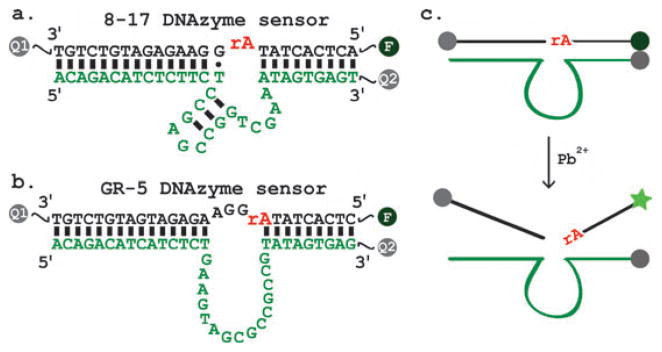
The new sensor is designed similarly to a temperature independent 8–17 DNAzyme sensor variant called +5_17E reported previously (Fig. 1a).4d It consists of a substrate strand (in black) labelled with a fluorophore (F) at the 5′ end and a quencher (Q1) at the 3′ end, and an enzyme strand (in green) labelled with a second quencher (Q2) at the 3′ end. Previous studies have shown that while the intermolecular quenching between Q2 in the enzyme strand and F in the substrate accounts for the majority of the quenching because of their close proximity to one another,4a a small percentage of dehybridization of the substrate from the enzyme at room temperature resulted in F being away from Q2 and thus high fluorescent background.4b To improve the quenching efficiency, Q1 was added to the 3′ end of the substrate to promote intramolecular quenching when the substrate strand was dehybridized from the enzyme strand, lowering the fluorescent background by ~75%.4b Fluorescein (FAM) was used as the fluorophore in both sensors in Fig. 1. For the 8–17 DNAzyme sensor, a black hole quencher (BHQ-1 ®) was used at 3′ end of the substrate strand while Dabcyl was used at the 3′ end of the enzyme strand, as reported previously.4a,b,d For the new sensor design, we chose to use BHQ-1® at the 3′ ends of both the substrate and the enzyme strands to simplify the design even further (Fig. 1b). Previous studies indicate that both Dabcyl and BHQ-1 quench FAM efficiently at such short a distance.12 In the absence of Pb2+, the arms of the enzymesubstrate complex maintain a double helical structure, placing the FAM molecule close to a BHQ-1® quencher, resulting in the fluorescent signal being quenched. When Pb2+ is present in the solution, it facilitates the cleavage of the phosphodiester bond of the internal RNA base (rA) by the enzyme strand (Fig. 1c). After cleavage, the base pairing between the enzyme and the substrate is destabilized and the cleaved substrate disassociates from the complex. As a result, the fluorophore is no longer quenched by BHQ-1®, resulting in an increased fluorescent signal.
Fig. 1.
(a) +5_17E Lead sensor design based on a temperature independent 8–17 DNAzyme.4d (b) Design of current lead sensor based on the GR-5 DNAzyme. Substrate (black) is 5′ functionalized with a FAM molecule and 3′ functionalized with a BHQ-1® quencher. The enzyme strand (green) is 3′ functionalized with a BHQ-1® quencher. In both sensors in (a) and (b), all sequences represent DNA, except the “rA” in the middle of the substrate which represents adenosine ribonucleotide. The secondary structures are predicted by mfold.11 (c) Schematic representation of the proposed catalytic beacon sensing mechanism.
Prior to testing the sensor, the buffer conditions were optimized for the best sensor performance (see Fig. S2†). Under these optimized conditions (50 mM NaHEPES, 50mMNaCl and 5mMMgCl2, pH 7.26), the GR-5 DNAzyme’s observed rate was higher than the observed rate obtained under the selection conditions.
In consistent with the original study that showed Mg2+ played no role in the catalysis,8 Mg2+ alone did not generate an enhanced fluorescent signal in the study here. Since a low concentration of Mg2+ (<5 mM) has no significant effect on the rate of the sensor, while maintaining a more stable enzyme–substrate complex over the course of the experiment, we chose the buffer conditions for the GR-5 DNAzyme sensor in the subsequent experiments to be 50 mM NaHEPES, 50 mM NaCl and 5 mM MgCl2 at pH 7.26. Interestingly, pH ~ 7.2 has been used in previous investigations of the 8–17 DNAzyme catalytic beacon sensors.4 Therefore the performance of the two generations of the sensors can be fairly compared at the same pH.
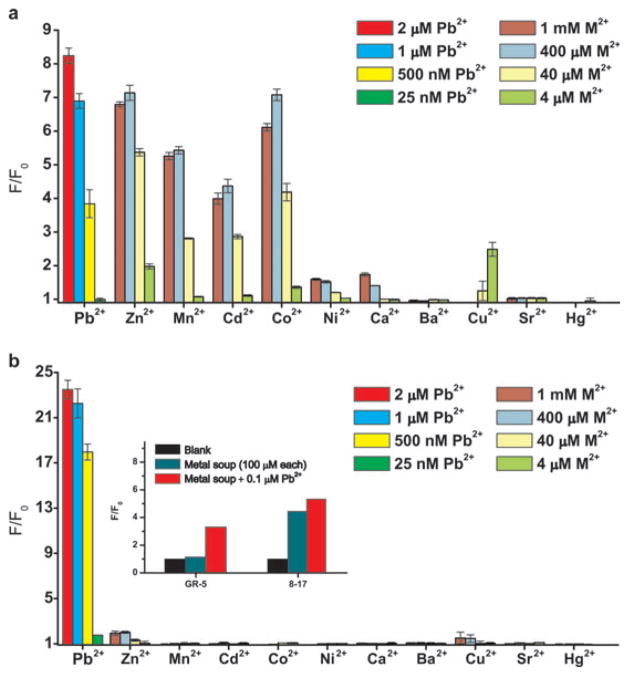
Although the 8–17 DNAzyme sensor was shown to be selective for Pb2+, the interference from other metal ions at very higher concentration has not been thoroughly investigated. In 50 mM NaHEPES, 50 mM NaCl, at pH 7.2, the 8–17 DNAzyme catalytic beacon exhibited a high fluorescent enhancement over the background (F/F0) in the presence of 25 nM–2 μM of Pb2+ (Fig. 2a), similar to those reported previously.4 When the competing metal concentration was ≥ 400 μM, both Co2+ and Zn2+ gave a fluorescent enhancement that was comparable to 2 μM Pb2+. Although to a lesser degree, Mn2+ and Cd2+ also gave a strong signal with concentration ≥40 μM and their signal saturated at ~ 400 μM. The 8–17 DNAzyme sensor was less active with Ni2+, Cu2+ and Ca2+ (note that Cu2+ had a quenching effect on the fluorophore used, Fig. S3†).6c Ba2+, Sr2+ and Hg2+ were the only metal ions that the 8–17 DNAzyme sensor did not respond to. Based on these measurements, the sensor offers no more than 160-fold selectivity against the most active interfering ion, Zn2+.
Fig. 2.
Selectivity comparison of both DNAzyme catalytic beacons. The fluorescent enhancement (F/F0) was measured at 6 min after the addition of each metal ion to the 8–17 DNAzyme sensor (a) and the GR-5 DNAzyme sensor (b). All experiments have been carried out in 50 mM NaHEPES, 50 mM NaCl at pH 7.2 for the 8–17 sensor and 50 mM NaHEPES, 50 mM NaCl and 5 mM MgCl2 at pH 7.26 for the GR-5 sensor. Inset: fluorescent response of both Pb2+ sensors in the presence of other eight metal ions.
In contrast, the fluorescent enhancement (F/F0) for the GR-5 Pb2+ DNAzyme sensor, shown in Fig. 1b, was much more selective for Pb2+ under the same conditions. First the F/F0 for the GR-5 sensor in the presence of 25 nM–2 μM Pb2+ were higher than those of the 8–17 DNAzyme sensor, reaching ~24 (Fig. 2b) instead of ~8 (Fig. 2a). Remarkably, other competing metal ions displayed little F/F0 for the new sensor based on the GR-5 Pb2+ DNAzyme. Even in the presence of 1 mM of the competing metal ions, only a very small F/F0 (~2) was observed with Zn2+ and Cu2+. Other metal ions did not give any fluorescent signal increase with concentrations up to 1 mM. This GR-5 DNAzyme sensor is about 40 000 times more selective for Pb2+ against Zn2+, the most active interfering metal ions.
To investigate the selectivity further, the fluorescent response to Pb2+ in the presence of a “metal soup” containing 100 μM of Zn2+, Mn2+, Cd2+, Co2+, Ni2+, Ca2+, Sr2+ and Ba2+ was tested for both sensors. For the 8–17 DNAzyme sensor (Fig. 2b inset), a high fluorescent enhancement was observed with the metal soup in the absence of Pb2+. Addition of 0.1 μM Pb2+ to the metal soup resulted in a similar fluorescent response, suggesting poor selectivity. Remarkably, for the GR-5 DNAzyme sensor, no fluorescent enhancement was observed with the metal soup without Pb2+, and ~3-fold fluorescent enhancement was observed with 0.1 μM of Pb2+. Therefore the new sensor based on the GR-5 DNAzyme has demonstrated an excellent selectivity for Pb2+ when tested against the metal ions individually or as a mixture of many metal ions.
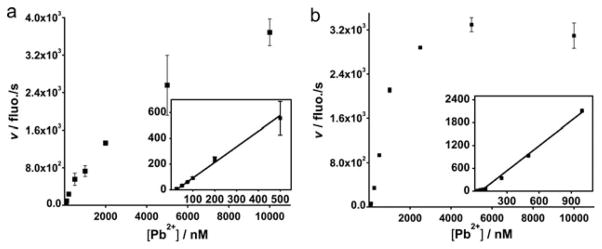
In addition to selectivity, the sensitivity of the two sensors was also investigated and compared under the same condition. The kinetics of the 8–17 DNAzyme sensor shown in Fig. S4 is similar to those reported previously.4 In contrast, the GR-5 sensor displayed faster kinetics, as its signal reached saturation in about 100 s with 2 μM Pb2+ (Fig. S4b) while the 8–17 sensor took almost 200 s (Fig. S4a). The faster kinetics is advantageous for practical applications because it allows rapid detection. The initial rates were clearly faster for the GR-5 Pb2+ DNAzyme sensor (Fig. 3b) than those of the 8–17 DNAzyme sensor (Fig. 3a) at almost all concentrations investigated (Fig.3). The linear range for the GR-5 Pb2+ DNAzyme sensor was wider than the 8–17 sensor while the dynamic range was narrower because its signal plateaued at a lower Pb2+ concentration. From the linear range of each sensor’s initial rate, the detection limit for the two sensors has been estimated based on 3σ/slope. The 8–17 DNAzyme sensor has a detection limit of 7.8 nM and the GR-5 DNAzyme sensor has a detection limit of 3.7 nM. While both detection limits are considerably lower than 72 nM, the EPA-defined maximal contamination level for Pb2+ in drinking water, the new sensor offers a slightly lower detection limit with a narrower dynamic range at lower Pb2+ concentrations.
Fig. 3.
Initial rate profile of the 8–17 DNAzyme sensor (a) and the GR-5 DNAzyme sensor (b). The insets display rates at lower Pb2+ concentrations.
In summary, we have constructed a new fluorescent sensor for the detection of Pb2+ based on the first DNAzyme (GR-5) selected through in vitro selection. When compared with the previously reported 8–17 DNAzyme based sensors, this new sensor offers a much higher selectivity and even a slightly lower detection limit. This GR-5 Pb2+ DNAzyme can not only be applied for sensing based on the fluorescent catalytic beacon, it can also be used in place of the other 8–17 DNAzyme based sensors.3,5 The higher selectivity of the GR-5 DNAzyme can be attributed mostly to the DNAzyme being selected in the presence of Pb2+,8 while the 8–17 DNAzyme was selected in the presence of other metal ions.9 During the selection process, Mg2+ was included in the buffer during the wash before the addition of Pb2+, and thus any DNA that is active with Mg2+, or other divalent metal ions with similar properties, would have probably been removed. In addition, the inclusion of Mg2+ may play a role in lowering the detection limit, since Mg2+ can stabilize the enzymesubstrate complex, therefore leading to a lower fluorescent background. Interestingly, the scissile “rA” in the GR-5 DNAzyme may be in a different position from that in the 8–17 DNAzyme, as predicted by mfold. While the “rA” is base-paired to a T in the GR-5 DNAzyme, it is in a bulge region without any base pairing partner in the 8–17 DNAzyme. This difference may have contributed to its superior selectivity and sensitivity. Further investigations are under way to elucidate the origin of such high selectivity.
Supplementary Material
Acknowledgments
We thank Hannah Ihms and Nandini Nagraj for proofreading the manuscript, and the US Department of Energy (DE-FG02-08ER64568), National Institutes of Health (Grant No. ES016865), and the National Science Foundation (Grant No. CTS-0120978 and DMI-0328162) for financial support.
Footnotes
Electronic supplementary information (ESI) available: Initial biochemical assays (Fig. S2 and S3), Pb2+ concentration dependence of the two sensors (Fig. S4), optimization (Fig. S1 and Table S1) and the DNAzyme sensor preparations, details of fluorescence measurements. See DOI: 10.1039/b926910j
Notes and references
- 1.Godwin HA. Curr Opin Chem Biol. 2001;5:223–227. doi: 10.1016/s1367-5931(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 2.(a) Chen P, Greenberg B, Taghavi S, Romano C, van der Lelie D, He C. Angew Chem, Int Ed. 2005;44:2715–2719. doi: 10.1002/anie.200462443. [DOI] [PubMed] [Google Scholar]; (b) Deo S, Godwin HA. J Am Chem Soc. 2000;122:174–175. [Google Scholar]; (c) Blake DA, Jones RM, Blake RC, Pavlov AR, Darwish IA, Yu H. Biosens Bioelectron. 2001;16:799–809. doi: 10.1016/s0956-5663(01)00223-8. [DOI] [PubMed] [Google Scholar]; (d) Telting-Diaz M, Bakker E. Anal Chem. 2002;74:5251–5256. doi: 10.1021/ac025596i. [DOI] [PubMed] [Google Scholar]; (e) Chen C, Huang W. J Am Chem Soc. 2002;124:6246–6247. doi: 10.1021/ja025710e. [DOI] [PubMed] [Google Scholar]; (f) Li J, Guo S, Zhai Y, Wang E. Anal Chim Acta. 2009;649:196–201. doi: 10.1016/j.aca.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 3.(a) Xiao Y, Rowe AA, Plaxco KW. J Am Chem Soc. 2007;129:262–263. doi: 10.1021/ja067278x. [DOI] [PubMed] [Google Scholar]; (b) Shen L, Chen Z, Li Y, He S, Xie S, Xu X, Liang Z, Meng X, Li Q, Zhu Z, Li M, Le XC, Shao Y. Anal Chem. 2008;80:6323–6328. doi: 10.1021/ac800601y. [DOI] [PubMed] [Google Scholar]; (c) Li T, Wang E, Dong S. J Am Chem Soc. 2009;131:15082–15083. doi: 10.1021/ja9051075. [DOI] [PubMed] [Google Scholar]
- 4.(a) Li J, Lu Y. J Am Chem Soc. 2000;122:10466–10467. [Google Scholar]; (b) Liu J, Lu Y. Anal Chem. 2003;75:6666–6672. doi: 10.1021/ac034924r. [DOI] [PubMed] [Google Scholar]; (c) Wang H, Kim Y, Liu H, Zhu Z, Bamrungsap S, Tan W. J Am Chem Soc. 2009;131:8221–8226. doi: 10.1021/ja901132y. [DOI] [PubMed] [Google Scholar]; (d) Nagraj N, Liu J, Sterling S, Wu J, Lu Y. Chem Commun. 2009:4103–4105. doi: 10.1039/b903059j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Liu J, Lu Y. J Am Chem Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]; (b) Liu J, Lu Y. J Am Chem Soc. 2004;126:12298–12305. doi: 10.1021/ja046628h. [DOI] [PubMed] [Google Scholar]; (c) Liu J, Lu Y. J Fluoresc. 2004;14:343–354. doi: 10.1023/b:jofl.0000031816.06134.d3. [DOI] [PubMed] [Google Scholar]; (d) Wei H, Li B, Li J, Dong S, Wang E. Nanotechnology. 2008;19:095501–095505. doi: 10.1088/0957-4484/19/9/095501. [DOI] [PubMed] [Google Scholar]; (e) Li J, Zhang J, Wei H, Wang E. Analyst. 2009;134:273–277. doi: 10.1039/b804670k. [DOI] [PubMed] [Google Scholar]; (f) Wang Z, Lee JH, Lu Y. Adv Mater. 2008;20:3263–3267. [Google Scholar]; (g) Liu J, Lu Y. Chem Mater. 2004;16:3231–3238. [Google Scholar]; (h) Dalavoy TS, Wernette DP, Gong M, Sweedler JV, Lu Y, Flachsbart BR, Shannon MA, Bohn PW, Cropek DM. Lab Chip. 2008;8:786–793. doi: 10.1039/b718624j. [DOI] [PubMed] [Google Scholar]; (i) Chang I, Tulock JJ, Liu J, Kim W, Cannon DM, Jr, Lu Y, Bohn PW, Sweedler JV, Cropek DM. Environ Sci Technol. 2005;39:3756–3761. doi: 10.1021/es040505f. [DOI] [PubMed] [Google Scholar]; (j) Mazumdar D, Liu J, Lu G, Zhou J, Lu Y. Chem Commun. 2010;46:1416–1418. doi: 10.1039/b917772h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. Proc Natl Acad Sci U S A. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu J, Lu Y. J Am Chem Soc. 2007;129:9838–9839. doi: 10.1021/ja0717358. [DOI] [PubMed] [Google Scholar]; (c) Liu J, Lu Y. Chem Commun. 2007:4872–4874. doi: 10.1039/b712421j. [DOI] [PubMed] [Google Scholar]; (d) Lee JH, Wang Z, Liu J, Lu Y. J Am Chem Soc. 2008;130:14217–14226. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]; (b) Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 8.Breaker RR, Joyce GF. Chem Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 9.(a) Faulhammer D, Famulok M. Angew Chem, Int Ed Engl. 1996;35:2837–2841. [Google Scholar]; (b) Faulhammer D, Famulok M. J Mol Biol. 1997;269:188–202. doi: 10.1006/jmbi.1997.1036. [DOI] [PubMed] [Google Scholar]; (c) Santoro SW, Joyce GF. Proc Natl Acad Sci U S A. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Li J, Zheng W, Kwon AH, Lu Y. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Peracchi A. J Biol Chem. 2000;275:11693–11697. doi: 10.1074/jbc.275.16.11693. [DOI] [PubMed] [Google Scholar]; (f) Cruz RPG, Withers JB, Li Y. Chem Biol. 2004;11:57–67. doi: 10.1016/j.chembiol.2003.12.012. [DOI] [PubMed] [Google Scholar]; (g) Schlosser K, Li Y. Biochemistry. 2004;43:9695–9707. doi: 10.1021/bi049757j. [DOI] [PubMed] [Google Scholar]; (h) Schlosser K, Gu J, Lam JC, Li Y. Nucleic Acids Res. 2008;36:4768–4777. doi: 10.1093/nar/gkn396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Brown AK, Li J, Pavot CMB, Lu Y. Biochemistry. 2003;42:7152–7161. doi: 10.1021/bi027332w. [DOI] [PubMed] [Google Scholar]; (b) Peracchi A, Bonaccio M, Clerici M. J Mol Biol. 2005;352:783–794. doi: 10.1016/j.jmb.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 11.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marras SAE, Kramer FR, Tyagi S. Nucleic Acids Res. 2002;30:122e. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.