Abstract
Most human somatic cells do not express telomerase. Consequently, with each cell division their telomeres progressively shorten until replicative senescence is induced. Approximately 15% of human cancers maintain their telomeres using telomerase-independent, recombination-based mechanisms collectively termed Alternative Lengthening of Telomeres (ALT). In the yeast Saccharomyces cerevisiae, ALT cells are referred to as “survivors”. One type of survivor (type II) resembles human ALT cells in that both are defined by the amplification of telomeric repeats. We analyzed recombination-mediated telomere extension events at individual telomeres in telomerase-negative yeast during type II survivor formation and find that long telomeres are preferentially extended. Furthermore, we find that senescent cells with long telomeres are more efficient at bypassing senescence via the type II pathway. We speculate that telomere length may be important in determining whether cancer cells utilize telomerase or ALT to bypass replicative senescence.
Telomeres are regions of guanine-rich, repetitive DNA at the ends of chromosomes bound by proteins that protect chromosome ends from being recognized as DNA double-stranded breaks1. The telomeres of most human somatic cells undergo progressive shortening due to incomplete DNA replication and nucleolytic degradation. This process ultimately leads to replicative senescence and is thought to be a barrier to tumorigenesis2. Most cancer cells bypass this senescence via the activation of telomerase2,3, the specialized reverse transcriptase that elongates telomeres4. However, 15% of human cancers bypass senescence by using telomerase-independent length maintenance mechanisms collectively referred to as Alternative Lengthening of Telomeres (ALT), which are thought to involve DNA recombination3,5.
Telomerase-independent telomere maintenance was first discovered in the budding yeast Saccharomyces cerevisiae6. Yeast cells lacking telomerase senesce after 60–80 generations, but a small subset of cells can bypass this senescence to become “survivors”6. Subsequent studies revealed that two main classes of survivors exist (type I and type II), both of which require Rad52-dependent recombination and Pol32-dependent break-induced replication6-10. Type I formation involves the amplification of subtelomeric Y’ sequences and depends on Rad51, Rad52, Rad54, Rad55, and Rad576-8. Type II survivors exhibit amplification of the TG1-3 telomeric repeats and requires Rad52, the MRX complex (consisting of Mre11, Rad50, and Xrs2), Sgs1, and Rad597,9,11-14.
Type I survivors occur more frequently but grow very poorly while growth of type II survivors is comparable to telomerase-positive cells6,8. Live cell imaging has shown that senescent cells are arrested at the G2/M transition, with telomeres moving back and forth between the mother and the bud15. Telomeres of type I survivors exhibit the same oscillating motion, but telomeres of type II survivors behave like telomeres in telomerase-positive cells15. This observation correlates with the slow growth of type I survivors and wild type growth of type II survivors, and furthermore, suggests that type I telomeres do not return to a properly capped state.
Type II survivors resemble human ALT cells in that both are defined by the presence of long, heterogeneous telomeric repeats8,16,17. Both type II survivors and human ALT cells are thought to amplify their telomeric DNA through recombination-dependent DNA replication18, likely involving extrachromosomal circular DNA containing telomeric repeat sequences19,20. The similarities between type II survivors and human ALT cells make yeast an attractive model for studying recombination-based telomere maintenance.
In this study, we analyzed recombination-mediated telomere extension events of S. cerevisiae type II survivors immediately following their emergence from a senescent culture. Surprisingly, we find that long telomeres are preferentially extended during type II survivor formation, which is in striking contrast to telomerase-mediated telomere maintenance, where short telomeres are preferentially elongated21. Furthermore, we find that increasing the length of telomeres at the point of senescence also increases the efficiency by which cells bypass senescence via type II recombination. Our results suggest that telomere length at senescence may be an important factor in determining whether neoplastic cells reactivate telomerase or use ALT to maintain their telomeres.
Results
Assaying telomere extension events during survivor formation
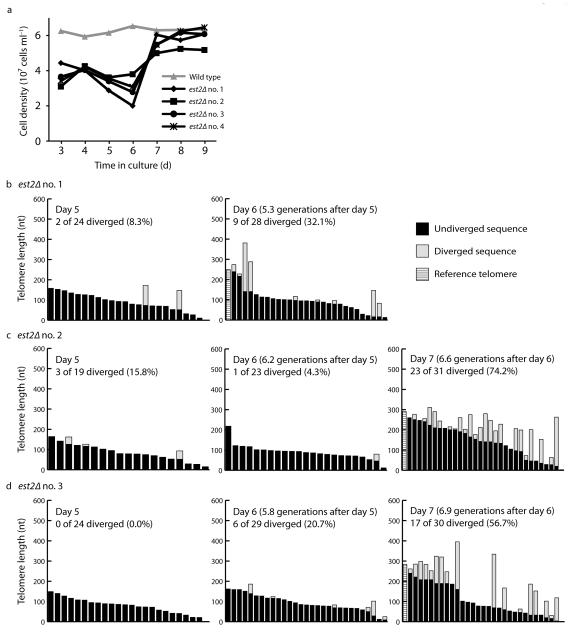
The Single Telomere EXtension (STEX) assay was previously developed to analyze telomerase-mediated telomere extension events at individual telomeres at nucleotide resolution21,22. Here, we modified the assay to allow us to study recombination-mediated telomere extension events as cells senesce and as survivors emerge. For this assay, one copy of EST2, which encodes the catalytic subunit of telomerase23, was deleted in a diploid strain. The resulting est2Δ/+ heterozygous diploid was sporulated and the haploid meiotic progeny were isolated on agar plates. After ~25 generations of growth, wild type and telomerase-negative est2Δ strains were transferred to liquid culture. After 24 h, cell density was measured and the cultures were diluted to a cell density of 5 × 105 cells ml−1 in fresh media. The measurements and dilutions were repeated 7 times at 24 h intervals. Growth in liquid culture allowed us to look specifically at type II survivors because type II survivors have a substantial growth advantage compared to type I survivors8. Although type I survivors arise more frequently, they grow poorly, and in liquid culture, the less frequently-arising type II survivors quickly outgrow the type I survivors and dominate the culture8. As expected, the wild type strain grew consistently throughout the course of the experiment while growth of the est2Δ strains progressively declined (Fig. 1a). Senescence occurred around days 5 and 6 for four independent est2Δ isolates. By day 7, all four isolates had bypassed senescence.
Figure 1.
STEX analysis of type II survivor formation. (a) Senescence was monitored in liquid culture by serially passaging four est2Δ spore products derived from an est2Δ/+ heterozygous diploid. (b–d) Analysis of sequenced VI-R telomeres from three est2Δ strains at days 5, 6, and 7 – (b)est2Δ no. 1, (c) est2Δ no. 2, (d) est2Δ no. 3. Analysis of telomeres from est2Δ no. 4 is shown in Supplementary Figure 1. Each bar represents an individual VI-R telomere, which are sorted by the length of the undiverged sequence. The black portion of each bar represents the undiverged, or unextended, region of the telomere. The light grey portion of each bar represents the diverged, or extended, region of the telomere. For each est2Δ strain, the longest telomere without exhibiting divergent sequence, represented by a hashed bar, is used as a reference telomere to which all other telomeres are compared to determine if divergence has occurred.
DNA was isolated from the est2Δ isolates from days 5, 6, and 7. Telomere VI-R was amplified by telomere PCR24,25. The amplified telomeres were then cloned and sequenced. Divergence of telomeric sequences is due to the recombination of the imperfect 5′-(TG)0-6TGGGTGTG(G)0-1-3′ repeats of S. cerevisiae telomeres21. Prior to the emergence of survivors in day 7, telomeric recombination events are rare (Fig. 1b–d, Supplementary Fig. 1, and data not shown), consistent with previous observations21. In contrast, most telomeres from day 7 samples (less than 7 population doublings after day 6) showed sequence divergence indicative of telomere extension events (Fig. 1c, d and Supplementary Fig. 1). We also analyzed telomeres I-L and XIV-R from day 6 and day 7 samples from est2Δ isolate no. 4 (Supplementary Fig. 1 and data not shown). The data from these telomeres are similar to those from telomere VI-R, suggesting that our results are applicable to all telomeres.
Preferred extension of long telomeres in type II survivors
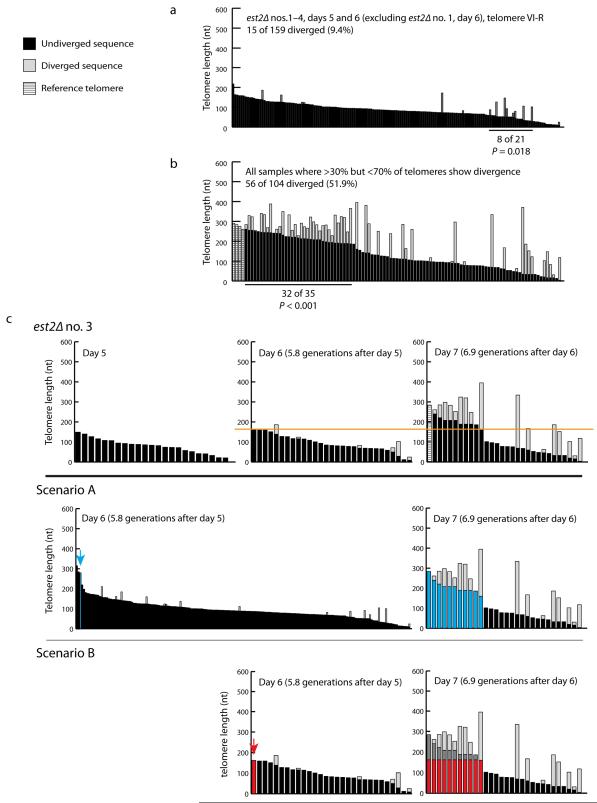
Pooling the sequenced telomeres from pre-survivor samples (Fig. 1b–d and Supplementary Fig. 1: est2Δ no. 1, day 5; est2Δ no. 2, days 5 and 6; est2Δ no. 3, days 5 and 6; est2Δ no. 4, days 5 and 6), we find that there is a modest preference for the extension of short telomeres (Fig. 2a), which is consistent with previous observations21,22,26,27.
Figure 2.
Long telomeres are preferentially extended during type II survivor formation. (a) The sequenced VI-R telomeres from samples from pre-survivor cells (est2Δ no. 1, day 5; est2Δ no. 2, days 5 and 6; est2Δ no. 3, days 5 and 6; est2Δ no. 4, days 5 and 6) were pooled and analyzed together. A run of telomeres with a significant increase in telomeres that show divergence is indicated. The number of extended telomeres and total number of telomeres in the run is shown, as well as the p-value (determined by scan statistics; see Methods). (b) The sequenced telomeres from all samples where more than 30% but less than 70% of the telomeres show sequence divergence (est2Δ no. 1, day 6, telomere VI-R; est2Δ no. 3, day 7, telomere VI-R; est2Δ no. 4, day 7, telomeres VI-R and XIV-R) were pooled and analyzed together. A run of telomeres with a significant increase in telomeres that show divergence is indicated. The number of extended telomeres and total number of telomeres in the run is shown, as well as the p-value (determined by scan statistics; see Methods). (c) Long unextended telomeres from day 7 samples can be explained by one of two models, both involving the preferential, recombination-mediated extension of long telomeres. The top panel shows the est2Δ no. 3 data from Figure 1d. An orange line highlights the observation that a significant number of telomeres from the day 7 sample contains undiverged regions that are longer than the longest day 6 telomere. In Scenario A, the long telomeres from day 7 originated from a long telomere from day 6 (blue bar indicated by blue arrow) that was not sequenced because such long telomeres were too rare in the day 6 sample to be detected in our assay. In Scenario B, a long telomere from day 6 (red bar indicated by red arrow) was extended between days 6 and 7. This telomere was then duplicated several times, giving rise to the long telomeres in day 7. See text for more detail. Importantly, in both models, it is the long telomeres that are preferentially extended.
We next analyzed the recombination events in samples from emerging survivors. In pre-survivor cultures after 30 generations of clonal expansion, approximately 7% of telomeres show sequence divergence21. Therefore we analyzed samples where more than 30% of the telomeres show sequence divergence, ensuring that these samples were from emerging survivors. We did not examine samples where more than 70% of the telomeres show sequence divergence because the process of survivor formation had progressed too far, rendering the data uninformative. In these samples, it was no longer possible to determine how many extension events occurred on a given telomere and whether there was a telomere length preference for extension events. In samples where the frequency of telomeres showing sequence divergence was between 30% and 70%, long telomeres show preferential extension, whether the samples are analyzed individually (Fig. 1b–d and Supplementary Fig. 1: est2Δ no. 1, day 6, telomere VI-R; est2Δ no. 3, day 7, telomere VI-R; est2Δ no. 4, day 7, telomeres VI-R and XIV-R) or grouped together (Fig. 2b).
The preferential extension of long telomeres can also be seen by comparing day 6 and day 7 telomeres from the same est2Δ isolate. The undiverged regions of the day 7 telomeres can actually be longer than the longest day 6 telomeres (Fig. 2c, top panel, black bars above orange line), which initially appears counterintuitive. However, this observation can be explained in one of two ways. First, a very rare long telomere from day 6 (Fig. 2c, Scenario A, blue bar indicated by blue arrow), too infrequent in the population to be detected among the 20–30 telomeres sequenced, was replicated several times (with daughter telomeres eroding with each round of DNA replication), giving rise to the long telomeres in day 7 (blue bars), most of which were then extended (light grey portion of blue bars). Alternatively, the longest detectable telomere from day 6 (Fig. 2c, Scenario B, red bar indicated by red arrow) was extended and, after being replicated several times (with daughter telomeres eroding with each round of DNA replication), gave rise to the set of longer telomeres in day 7 (red/dark grey bars; dark grey portion indicating the extended region), most of which were extended yet again (light grey portion of red/dark grey bars). Cells harboring this set of long telomeres in day 7 (blue bars in Scenario A, red bars in Scenario B) outcompete other senescing cells with shorter (more extensively eroded) telomeres, resulting in the enrichment of these longer telomeres. Importantly, for both explanations long telomeres are preferentially extended, which is consistent with the statistical analysis (Fig. 2b).
Telomere length affects type II survivor emergence
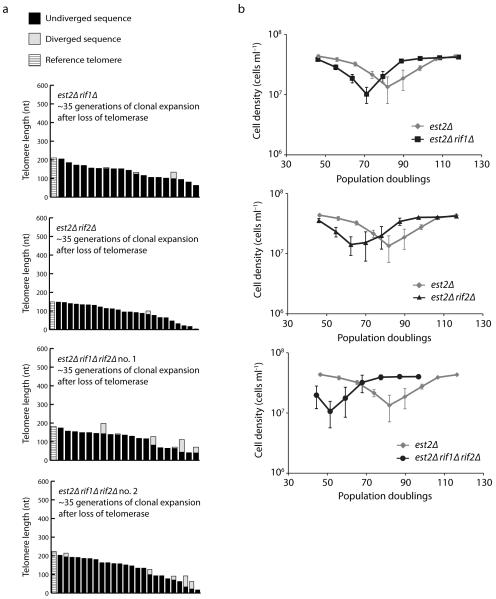
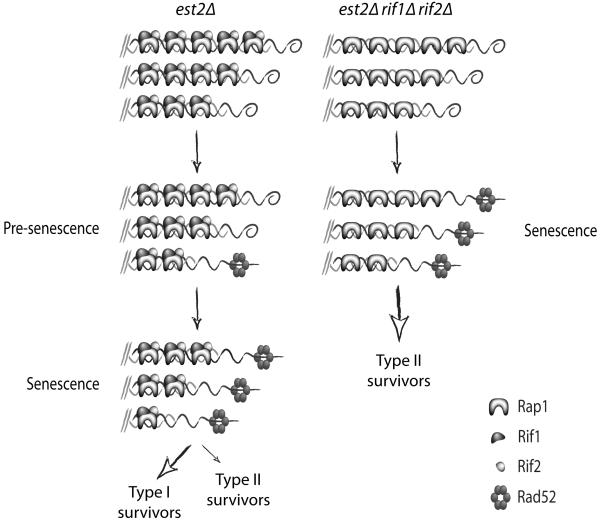
Given that long telomeres are preferentially extended during the emergence of type II survivors, we reasoned that senescent cells with long telomeres would favor the bypass of senescence via the type II pathway as opposed to the type I pathway. Therefore, we next examined telomere length in mutant backgrounds where type II survivors are increased. Rif1 and Rif2, recruited to telomeres via the C-terminal domain of Rap128,29, inhibit exonucleolytic processing at telomeres by the MRX complex30. In a telomerase-negative background, removing Rif1 or Rif2, especially Rif2, greatly increases the fraction of type II survivors11. In fact, all survivors generated from a telomerase-negative rif1Δ rif2Δ strain are type II11. Since it was proposed that the increase in type II survivors was due to a role for Rif1 and Rif2 in inhibiting type II recombination11, telomeric recombination should be increased even in rifΔ pre-survivor cells. However, when we sequenced telomeres from pre-survivor est2Δ rif1Δ, est2Δ rif2Δ, and est2Δ rif1Δ rif2Δ mutants after ~35 generations of clonal expansion following loss of telomerase, we found no marked increase in type II recombination events (Fig. 3a). Although there appears to be a slight increase of recombination events in est2Δ rif1Δ rif2Δ pre-survivor mutants, the levels are nowhere near as high as seen in survivors. These observations indicate that the greater proportion of type II survivors in est2Δ rif1Δ rif2Δ mutant strains is not due to inhibition of recombination by the Rif proteins. Therefore, we speculated that the increase in type II survivors might actually be due to telomerase-negative rifΔ mutants having longer telomeres at senescence than telomerase-negative RIF+ mutants.
Figure 3.
Telomerase-negative rifΔ strains exhibit accelerated senescence. (a) Telomere VI-R was sequenced from est2Δ rif1Δ, est2Δ rif2Δ, and two different isolates of est2Δ rif1Δ rif2Δ after ~35 generations of clonal expansion following sporulation of an est2Δ/+ rif1Δ/+ rif2Δ/+ diploid. Sequenced telomeres were analyzed as in Figure 1. (b) Senescence rates were measured in liquid culture by serial passaging of est2Δ, est2Δ rif1Δ, est2Δ rif2Δ, and est2Δ rif1Δ rif2Δ strains derived from the same diploid used in (a). The means and standard errors for at least four independent spore isolates for each genotype are shown.
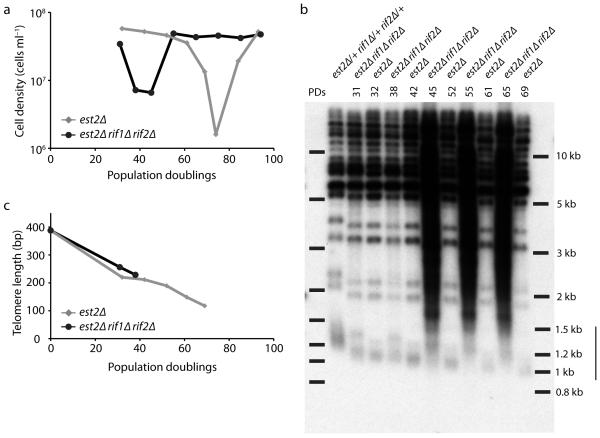
To test this hypothesis, we sporulated an est2Δ/+ rif1Δ/+ rif2Δ/+ diploid and monitored the growth of the telomerase-negative haploid progeny in liquid culture by serial dilution of cells to 5 × 104 cells ml−1 every 24 h (Fig. 3b). Interestingly, we find that est2Δ rifΔ mutants senesce after fewer population doublings than est2Δ strains. We then monitored the telomere lengths of est2Δ and est2Δ rif1Δ rif2Δ strains as they senesced and find that telomeres shorten at the same rate in both strains (Fig. 4). However, since the est2Δ rif1Δ rif2Δ strain senesced earlier, its telomeres were more than 100 bp longer at the point of maximum senescence (Fig. 4c). These observations support our model that longer telomeres recombine more efficiently, which explains the increase in type II survivors in telomerase-negative rifΔ mutants.
Figure 4.
Telomere shortening rate is unchanged when RIF1 and RIF2 are deleted. (a) One isolate each of est2Δ and est2Δ rif1Δ rif2Δ was passaged as in Figure 3b. (b) At each passage, telomere lengths were determined by denaturing in-gel hybridization (see Methods). The vertical bar to the right of the gel indicates the position of the terminal restriction fragments of Y’ telomeres, which are present in over half of yeast telomeres. The larger bands represent non-Y’-containing telomeres. A smear of telomeric signal appears in the est2Δ rif1Δ rif2Δ strain after 45 population doublings (PDs), which arise from type II survivors that have undergone telomere repeat amplification. (c) The telomere lengths from (b) were quantified and plotted.
Finally, we find that deleting both RIF1 and RIF2 dramatically accelerates senescence and that rif2Δ has a bigger effect than rif1Δ (Fig. 3b). This finding correlates with the increase in type II survivors in telomerase-negative rifΔ mutants where rif2Δ has more of an effect than rif1Δ on the percentage of type II survivors11. Interestingly, Rif2 has recently been shown to have a more prominent role than Rif1 in inhibiting telomeric 5′ end-resection by the MRX complex30. Taken together, these observations suggest that the function of the Rif proteins, in particular Rif2, during senescence is to prevent premature uncapping of telomeres.
Discussion
In this study, we analyzed type II recombination-mediated telomere extension events as survivors emerge from a senescent culture. We find that long telomeres are preferentially extended (Fig. 2b and 2c), which is a surprising result considering previous studies showing that the shortest telomeres trigger senescence in both mouse and yeast studies31-33 and that the shortest telomeres are preferentially elongated via telomerase in yeast and human fibroblasts expressing telomerase21,34. However, it is well known from studies in prokaryotes, yeast, and mammalian cells that recombination efficiency is directly proportional to the length of the substrate DNA35-40. Therefore, we propose that once a senescent cell becomes a survivor, the long telomeres are better substrates for recombination and will recombine first. Importantly, any short telomeres do eventually get extended as well – otherwise the cell would not be able to escape senescence.
Although Rad52 is important for virtually all recombination events in budding yeast, Rad52-independent recombination can occur if the substrate DNA is sufficiently long41. Recently, two reports show that type II-like survivors, exhibiting amplification of the telomeric repeats, can form in the absence of Rad52 if longer telomeres are present during senescence42,43. These results are consistent with our findings that long telomeres recombine more efficiently and that these long telomeres are preferentially extended during type II survivor formation.
Interestingly, long telomeres are not preferentially extended in pre-survivor cells (Fig. 2a). In fact, in the pre-survivor samples we analyzed, there is a modest preference for the extension of short telomeres (Fig. 2a), consistent with previous observations21,22,26,27. Such short telomeres are more likely to be uncapped and recruit DNA damage checkpoint proteins including Rad5232, which may explain why short telomeres are more likely to be extended by recombination in pre-survivor cells. A recent study has shown that the recruitment of these proteins at eroded telomeres can start many generations before senescence32. In our model, these senescing cells continue to divide until most or all telomeres have been sufficiently eroded and Rad52 is present at all of the telomeres (Fig. 5). Consequently, when a senesced cell transforms into a survivor, all of the Rad52-coated telomeres are competent for recombination, with the longer ones being more efficiently extended via recombination.
Figure 5.
Model for the senescence of est2Δ and est2Δ rif1Δ rif2Δ strains. In an est2Δ strain (on the left), telomeres progressively shorten with each cell division. Short telomeres are uncapped and recruit DNA damage factors like Rad52. The recruitment of DNA damage factors at one or a small number of shortened telomeres is insufficient to cause senescence until most or all telomeres have been sufficiently eroded. Bypass of senescence occurs either by the type I pathway (involving subtelomeric Y’ elements), or less frequently, the type II pathway (involving the telomeric repeats). The type II pathway involves the preferential extension of long telomeres by recombination. In an est2Δ rif1Δ rif2Δ strain (on the right), telomeres become uncapped earlier leading to accelerated senescence. However, since the telomere shortening rate is unaffected by deletion of the RIF genes, telomeres are relatively long at senescence as compared to telomeres in an est2Δ strain. The longer telomeres recombine more efficiently, resulting in a dramatic increase in the fraction of est2Δ rif1Δ rif2Δ survivors that are type II. The Rad52 protein complex is depicted as a hexamer for simplicity.
Alternatively, the modest increase in short telomeres that have been extended in pre-survivor samples (Fig. 2a) may not be due to preferential extension of short telomeres, but may be due to the senescence of cells with short telomeres. Cells that extend such telomeres would be selected to survive, giving the illusion that short telomeres are preferentially extended. This explanation may also account for the slight increase of short telomeres exhibiting divergence even in emerging survivors (Fig. 2b).
We also find that telomere length in senescent cells determines the probability that those cells will bypass senescence via the type II pathway. Long telomeres recombine more efficiently and thus favor the utilization of the type II pathway over the type I pathway. This preference for long telomeres is clearly seen in telomerase-negative rifΔ mutants (Fig. 4). Rif1 and Rif2 inhibit MRX-mediated telomere end-resection30. In telomerase-negative RIF+ strains, as telomeres get progressively shorter with each round of cell division, the amount of telomere-associated Rif1 and Rif2 declines44. Eventually, critically short telomeres lack sufficient Rif1 and Rif2 to inhibit the MRX complex, allowing end-resection, telomere uncapping, and induction of senescence. We find that, in telomerase-negative strains lacking Rif1 and Rif2, senescence is accelerated (Fig. 3b and Fig. 4a) since MRX-mediated telomere uncapping likely occurs earlier after loss of telomerase in this background. In addition, the rate of telomere shortening is not accelerated in these strains, therefore telomerase-negative rifΔ mutants senesce with longer telomeres than telomerase-negative RIF+ strains (Fig. 4c), resulting in an increase in the fraction of type II survivors (Fig. 5).
It is unclear why some cancer cells maintain telomeres by reactivating telomerase and why some cancer cells do so by using ALT. We suggest that the length of the telomeres in neoplastic cells just before activation of a telomere maintenance mechanism may play a key role in determining which mechanism is used. Our work predicts that mutations in genes that cause cells to senesce with long telomeres would favor utilization of ALT so it will be of great interest to determine whether these genes are indeed mutated in ALT cells.
Supplementary Material
Acknowledgments
We thank Kara Bernstein, Brian Luke, and Peter Thorpe for constructive comments on the manuscript. MC was supported by a Long-term Fellowship Award from the International Human Frontier Science Program (HFSP) Organization and a Terry Fox Foundation (TFF) Fellowship Award. This work was supported by funds from the National Institutes of Health (CA009503 and GM008798 to JCD; GM50237 to RR).
Appendix
Methods
Yeast media and strains
Standard yeast media and growth conditions were used45. Yeast strains used in this study are listed in Supplementary Table 1.
Telomere PCR and sequencing
Telomeres VI-R and I-L were amplified by PCR as previously described21,25. The telomere I-L PCR also amplified telomere XIV-R, which could be differentiated by analysis of the subtelomeric sequence just upstream of the telomeric repeats. Sequencing was performed by GENEWIZ, Inc. To catch the est2Δ cultures immediately after the emergence of survivors, STEX samples where greater than 30%, but less than 70%, of the telomeres exhibited sequence divergence were pooled and analyzed (Fig. 2b). These threshold values were determined empirically. Most samples from survivors (as determined by the senescence assays) contain telomeres where more than 30% exhibit sequence divergence. And from our experience, samples where more than 70% of telomeres exhibit sequence divergence do not yield very meaningful data.
Scan statistics
To determine which telomeres were preferentially elongated, a scanning statistic was performed. The lengths of the undiverged regions of the sequenced telomeres were recorded, as well as if an extension event occurred. The results were then ordered based on telomere length (longest to shortest) and a scanning statistic was performed using this list on the basis of whether or not an extension event occurred. The scanning statistic was scored by calculating a lambda using a generalized likelihood ratio test (GLRT) with window sizes ranging from 5 to half the number of telomeres considered. For this GLRT, the null hypothesis was that the probability of having an extension event within any given window of data was the same as an event occurring outside the window. Following this calculation, the window with the smallest lambda value was considered to be the most significantly enriched for extension events. To calculate a p-value for this lambda value, a permutation test was performed by randomizing the data 20,000 times, each time calculating the most significant (smallest lambda) value. These random and ordered lambdas were then ranked and the ordered lambda’s p-value was assigned by: p-value = rank / 20,00046,47.
Senescence assays
Each senescence assay started with est2Δ/+ diploids that were propagated for at least 50 generations before sporulation to ensure that telomeres were at a stable equilibrium length. Freshly dissected spores were allowed to form colonies on YPD agar plates after 2 days of growth at 30°C, or approximately 25 population doublings. Cells from these colonies were serially passaged in liquid YPD media at 24 h intervals. For each passage, the cell density in each culture was determined and the cultures were diluted back into fresh YPD media at a cell density of 5 × 104 cells ml−1. For the STEX experiments (Fig. 1), the cultures were diluted to a cell density of 5 × 105 cells ml−1 to reduce the number of population doublings between time points and maximize the chances of catching the cultures soon after survivors had emerged.
DNA isolation and denaturing in-gel hybridization
Yeast genomic DNA was isolated using a Yeast DNA Extraction Kit (Thermo Scientific). The DNA was digested with XhoI restriction endonuclease before running on a 0.8% agarose gel. Denaturing in-gel hybridization using a telomeric CA oligonucleotide radiolabeled probe was performed as described48.
References
- 1.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–52. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–5. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 3.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–91. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 4.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–13. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 5.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–30. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 6.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–60. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 7.Le S, Moore JK, Haber JE, Greider CW. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–52. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–93. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, Ijpma A, Greider CW. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol. 2001;21:1819–27. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–3. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 11.Teng SC, Chang J, McCowan B, Zakian VA. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell. 2000;6:947–52. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- 12.Huang P, et al. SGS1 is required for telomere elongation in the absence of telomerase. Curr Biol. 2001;11:125–9. doi: 10.1016/s0960-9822(01)00021-5. [DOI] [PubMed] [Google Scholar]
- 13.Johnson FB, et al. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001;20:905–13. doi: 10.1093/emboj/20.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsukamoto Y, Taggart AK, Zakian VA. The role of the Mre11-Rad50-Xrs2 complex in telomerase- mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol. 2001;11:1328–35. doi: 10.1016/s0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- 15.Straatman KR, Louis EJ. Localization of telomeres and telomere-associated proteins in telomerase-negative Saccharomyces cerevisiae. Chromosome Res. 2007;15:1033–50. doi: 10.1007/s10577-007-1178-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–8. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–4. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 18.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–35. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 19.Henson JD, et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. 2009;27:1181–5. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 20.Larrivee M, Wellinger RJ. Telomerase- and capping-independent yeast survivors with alternate telomere states. Nat Cell Biol. 2006;8:741–7. doi: 10.1038/ncb1429. [DOI] [PubMed] [Google Scholar]
- 21.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase-extendible and - nonextendible states. Cell. 2004;117:323–35. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 22.Arneric M, Lingner J. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 2007;8:1080–5. doi: 10.1038/sj.embor.7401082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingner J, et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–7. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 24.Forstemann K, Hoss M, Lingner J. Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res. 2000;28:2690–4. doi: 10.1093/nar/28.14.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–94. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JY, Kozak M, Martin JD, Pennock E, Johnson FB. Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol. 2007;5:e160. doi: 10.1371/journal.pbio.0050160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak ML, et al. Inactivation of the Sas2 histone acetyltransferase delays senescence driven by telomere dysfunction. EMBO J. 2009;29:158–70. doi: 10.1038/emboj.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–14. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 29.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–60. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 30.Bonetti D, et al. Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet. 2010;6:e1000966. doi: 10.1371/journal.pgen.1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 32.Khadaroo B, et al. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat Cell Biol. 2009;11:980–7. doi: 10.1038/ncb1910. [DOI] [PubMed] [Google Scholar]
- 33.Abdallah P, et al. A two-step model for senescence triggered by a single critically short telomere. Nat Cell Biol. 2009;11:988–93. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Britt-Compton B, Capper R, Rowson J, Baird DM. Short telomeres are preferentially elongated by telomerase in human cells. FEBS Lett. 2009;583:3076–80. doi: 10.1016/j.febslet.2009.08.029. [DOI] [PubMed] [Google Scholar]
- 35.Singer BS, Gold L, Gauss P, Doherty DH. Determination of the amount of homology required for recombination in bacteriophage T4. Cell. 1982;31:25–33. doi: 10.1016/0092-8674(82)90401-9. [DOI] [PubMed] [Google Scholar]
- 36.Watt VM, Ingles CJ, Urdea MS, Rutter WJ. Homology requirements for recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1985;82:4768–72. doi: 10.1073/pnas.82.14.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen P, Huang HV. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986;112:441–57. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jinks-Robertson S, Michelitch M, Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3937–50. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubnitz J, Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984;4:2253–8. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liskay RM, Letsou A, Stachelek JL. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics. 1987;115:161–7. doi: 10.1093/genetics/115.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grandin N, Charbonneau M. Telomerase-and Rad52-independent immortalization of budding yeast by an inherited-long-telomere pathway of telomeric repeat amplification. Mol Cell Biol. 2009;29:965–85. doi: 10.1128/MCB.00817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lebel C, et al. Telomere maintenance and survival in saccharomyces cerevisiae in the absence of telomerase and RAD52. Genetics. 2009;182:671–84. doi: 10.1534/genetics.109.102939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levy DL, Blackburn EH. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol. 2004;24:10857–67. doi: 10.1128/MCB.24.24.10857-10867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 46.Janeja VP, Atluri V. LS3: A Linear Semantic Scan Statistic Technique for Detecting Anomalous Windows; ACM Symposium on Applied Computing; 2005.pp. 493–497. [Google Scholar]
- 47.Shi L, Janeja VP. Anomalous Window Discovery through Scan Statistics for Linear Intersecting Paths (SSLIP); Proceedings of the 15th ACM SIGKDD international conference on Knowledge discovery and data mining; 2009.pp. 767–775. [Google Scholar]
- 48.Dionne I, Wellinger RJ. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc Natl Acad Sci U S A. 1996;93:13902–7. doi: 10.1073/pnas.93.24.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.