Abstract
Organophosphorus (OP) and thiocarbamate (TC) agrochemicals are used worldwide as insecticides, herbicides, and fungicides, but their safety assessment in terms of potential off-targets remains incomplete. In this study, we used a chemoproteomic platform, termed activity-based protein profiling, to broadly define serine hydrolase targets in mouse brain of a panel of 29 OP and TC pesticides. Among the secondary targets identified, enzymes involved in degradation of endocannabinoid signaling lipids, monoacylglycerol lipase and fatty acid amide hydrolase, were inhibited by several OP and TC pesticides. Blockade of these two enzymes led to elevations in brain endocannabinoid levels and dysregulated brain arachidonate metabolism. Other secondary targets include enzymes thought to also play important roles in the nervous system and unannotated proteins. This study reveals a multitude of secondary targets for OP and TC pesticides and underscores the utility of chemoproteomic platforms in gaining insights into biochemical pathways that are perturbed by these toxicants.
Keywords: Activity-based protein profiling, cannabinoid, fatty acid amide hydrolase, monoacylglycerol lipase, organophosphorus, pesticides, serine hydrolase, thiocarbamate
INTRODUCTION
Most pesticides elicit their toxic action through blockade of a particular target enzyme in the pest species, whether it is in an insect, plant, fungus, or mammal. Safety of these pesticides relies in part on superior selectivity and/or in vivo efficacy of the pesticide towards the pest species compared to humans or off-target species. Nonetheless, pesticide poisoning is still a major concern worldwide due to accidental/suicidal, occupational, or by-stander exposures from off-target drift, or through environmental contamination (1-3). Multiple epidemiological studies have also shown health deficits associated with populations chronically exposed to elevated levels of certain pesticides (4-6). These effects often cannot be fully correlated with on-target toxicity, indicating there may be toxicologically-relevant secondary targets. Indeed, several secondary targets have been identified on an individual target-basis, which have allowed for further understanding of potential toxicities associated with these chemicals (7, 8).
There have been relatively few efforts to globally identify mammalian secondary targets on a proteome-wide scale for currently used pesticides. This is, in part, complicated by a large swath of the proteome that remains functionally uncharacterized and displays difficult physicochemical properties that complicate their analysis in biological samples (e.g. low abundance or difficulty in enrichment). This prevents direct assessment of enzyme activity (and inhibition thereof) by traditional substrate assays. Although conventional genomic and proteomic methods (9-13) that comparatively quantify the expression levels of transcripts and proteins have yielded many useful insights into pesticide mode of action, these platforms are still limited in their capacity to identify changes in protein activity or activities that may be regulated by post-translation modifications or processing. These methods also do not aid in identifying the direct molecular target of the pesticides that elicit transcriptional downstream changes.
Modern chemoproteomic platforms address many of these challenges by utilizing chemical probes to directly interrogate protein activity or assess post-translational modifications. One such powerful technology is termed activity-based protein profiling (ABPP) (Figure 1A) (14-16) that uses active-site directed chemical probes to read-out enzyme activities in complex proteomes. These activity-based probes only label active enzymes and not their inactive precursors (e.g. zymogens), allowing for discrimination of protein activity versus mere protein expression. There are currently ABPP probes for a multitude of enzyme classes, including many that play central roles in physiology, such as hydrolases, oxidoreductases, glycosidases, nitrilases, glutathione S-transferases, all major families of proteases (serine, cysteine, aspartyl, metallo, and proteosomal), kinases, and phosphatases (17). Importantly, because activity-based probes bind the active site of enzymes, inhibitors can directly compete with the probe to read out enzyme inhibition in native proteomes. The potency and selectivity of both reversible (18) and irreversible (19) inhibitors can be assessed concurrently since compounds are profiled against many mechanistically related enzymes in parallel (Figure 1B). Protein inhibition can be assayed even for uncharacterized enzymes that lack known substrates, since activity-based probes read-out protein activity regardless of functional annotation (20). Competitive ABPP has been used to comprehensively profile the off-targets of a bioactivated organophosphorus (OP) insecticide metabolite, chlorpyrifos oxon (CPO), and a sarin analog, isopropyl dodecylfluorophosphonate (IDFP) (21).
Figure 1. Activity-based protein profiling (ABPP).
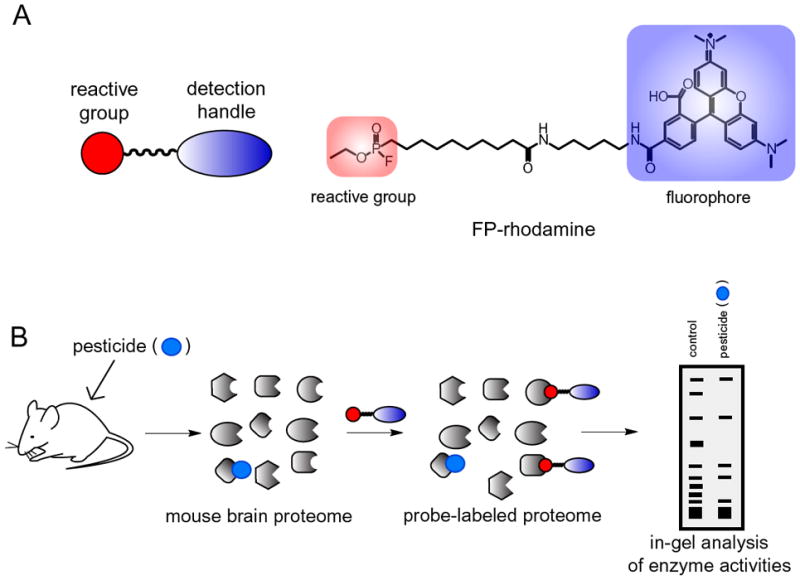
A. Activity-based probes consist of a reactive group that covalently reacts with a specific enzyme class, conjugated to a spacer arm and a detection handle such as a fluorophore or biotin. Shown on the right is the activity-based probe for the serine hydrolase superfamily with a fluorophosphonate (FP) reactive group and a rhodamine-detection handle (FP-rhodamine). B. For this study, we used ABPP to determine potential or putative secondary targets of pesticides. Mice were injected intraperitoneally with a pesticide and after 4 h they were euthanized and brain membranes were prepared, followed by incubation of the membrane proteome with FP-rhodamine. The reacted proteome was resolved by SDS-PAGE and visualized by in-gel fluorescence scanning. Pesticide-induced inhibition of serine hydrolase activity was recognized and quantitated by loss of a fluorescent band.
In this study, we broadly profiled 29 currently used OP and thiocarbamate (TC) insecticides, herbicides and fungicides to globally identify in vivo secondary targets of these pesticides in mouse brain. We focused our efforts on identifying targets within the serine hydrolase superfamily (i.e. active site serine) since the OP and carbamate chemotypes represent a privileged chemical scaffold for this enzyme class (22, 23). ABPP efforts showed multiple off-targets of OP and TC pesticides, including several enzymes with important roles in brain physiology as well as functionally uncharacterized enzymes. Of particular interest among these secondary targets were two serine hydrolases, monoacylglycerol lipase (MAGL) and fatty acid amide hydrolase (FAAH), that terminate the signaling of endogenous cannabinoid ligands (endocannabinoids) on the cannabinoid receptor (24, 25). Blockade of MAGL and FAAH caused robust elevations in brain endocannabinoid levels, and inhibition of MAGL also led to disruption in brain arachidonic acid metabolism. These studies underscore the importance of using chemoproteomic approaches to screen for off-targets of environmental toxicants that have population-wide exposures.
MATERIALS AND METHODS
Chemicals
OP and TC pesticides were purchased from ChemService and Sigma. FP-rhodamine was synthesized in Benjamin Cravatt’s laboratory at The Scripps Research Institute.
Mice
C57BL/6 mice were treated intraperitoneally with each pesticide in a vehicle of 18:1:1 saline:emulphor:ethanol (10 μL/g mouse) (preparations were sonicated until samples were in solution or homogenously suspended or emulsified). Doses were chosen as follows. Mice were treated with 100 mg/kg of the compound (or 30 or 10 mg/kg if LD50 values were previously reported to be <100 mg/kg). These doses did not cause overt toxic responses but, in some cases, elicited behavioral responses indicative of cannabinoid-related phenotypes. After 4 h, mice were sacrificed by cervical dislocation and brains were removed and flash frozen in liquid nitrogen. Animal experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of The Scripps Research Institute.
ABPP of Mouse Brain Proteomes
Membranes were isolated by homogenizing the brain in 50 mM Tris-HCl buffer, followed by a 100,000 × g centrifugation of the supernatant from a 1000 × g spin. The resulting membrane pellet was sonicated in 50 mM Tris-HCl buffer. For ABPP experiments, 50 μ g of brain membrane protein was incubated with 2 μM FP-rhodamine for 30 min in a 50 μL reaction volume. The reaction was quenched by addition of 30 μL of 4 × SDS-PAGE loading buffer and heated at 90°C for 10 min. Samples were run on an SDS-PAGE gel and scanned using the Hitachi FMBio IIe flatbed fluorescence scanner (MiraiBio). Activity of each enzyme was quantitated by measuring density of the fluorescent gel band using the Image J program (http://rsb.info.nih.gov/ij/).
Analysis of Brain Metabolite Levels
Brain metabolite levels were measured as described previously (2, 26) using an Agilent Triple Quadrupole LC/MS. One half brain was weighed and dounce homogenized in 2:1:1 v/v/v chloroform:methanol:Tris pH 8.0 (8 mL) containing internal standards for anandamide, 2-AG, and a fatty acid (2 pmol d4-anandamide, 0.5 nmol d5-2-AG, 10 nmol pentadecanoic acid). The mixture was vortexed and then centrifuged (1,400 × g, 10 min). The organic layer was removed, evaporated under a stream of nitrogen and resolubilized in 120 μL of chloroform. An aliquot of the extract (10 μL) was injected for analysis with an Agilent G6410B QQQ instrument. For LC separation, mobile phase A consisted of 95:5 water:methanol and mobile phase B consisted of 60:35:5 isopropanol:methanol:water. Formic acid (0.1 %) or ammonium hydroxide (0.1 %) was included to assist in ion formation in positive ionization and negative ionization modes, respectively. The flow rate for each run started at 0.1 mL/min with 0% B. At 5 min, the solvent was immediately changed to 60% B with a flow rate of 0.4 mL/min and increased linearly to 100% B over 10 min. This was followed by an isocratic gradient of 100% B for 5 min at 0.5 mL/min before equilibrating for 3 min at 0% B at 0.5 mL/min. The following MS parameters were used to measure the indicated metabolites (precursor ion, product ion, collision energy in n V): anandamide (348, 62, 11), d4-anandamide (352, 66, 11), 2-AG (379, 287, 8), d5-2-AG (384, 287, 8). Arachidonic acid was measured by targeting the parent mass and quantified based on targeted measurements of pentadecanoic acid. MS analysis was performed with an electrospray ionization source. The drying gas temperature was 350°C, the drying gas flow rate was 11 L/min, and the nebulizer pressure was 35 psi. Lipids were quantified by measuring the area under the peak in comparison to the internal standards.
Assessment of Catalepsy
Catalepsy was assessed as described previously (27) on a bar 0.7 cm in diameter placed 4.5 cm off of the surface. The mouse was placed with its front paws on the bar and a timer (Timer #1) was started. A second timer (Timer #2) was turned on only when the mouse was immobile on the bar, with the exception of respiratory movements. If the mouse moved off the bar, it was placed back on in the original position. The assay was stopped when either Timer #1 reached 60 s, or after the fourth time the mouse moved off the bar, and the cataleptic time was scored as the amount of time on Timer #2.
RESULTS AND DISCUSSION
ABPP reveals a multitude of OP and TC pesticide secondary targets in mouse brain
OP and carbamate compounds include potent inhibitors for the serine hydrolase superfamily (22, 23). Two examples of serine hydrolases inhibited by these compounds that also confer toxic effects are the primary target acetylcholinesterase (AChE) and the secondary target neuropathy target esterase (NTE) (8). Therefore, we used an activity-based probe to assess what other members of this class are targeted by OP and TC pesticides. The serine hydrolase activity-based probe consists of a fluorophosphonate (FP) reactive group, conjugated to a spacer arm and a detection handle [in this case, rhodamine (FP-rhodamine)] (Figure 1A) (14, 15). In a typical competitive ABPP experiment, inhibitor treatment can be performed in vivo, in situ, or in vitro, after which the proteome is incubated with the activity-based probe, separated by SDS-PAGE and monitored by in gel-fluorescent scanning, allowing for visualization of distinct protein activities (Figure 1B). For this study, we initially treated mice intraperitoneally with either 100 mg/kg or the maximal tolerated sublethal dose of 9 OP and TC pesticides for 4 h (Figure 2A, 2B). ABPP analysis revealed multiple secondary targets of the 9 OPs and TCs screened, including NTE, acyl peptide hydrolase (APEH), FAAH, KIAA1363, MAGL, alpha/beta hydrolase domain 6 (ABHD6) and acyl protein thioesterases (APT1 and APT2) (Figure 2B). These provocative results led us to expand our analysis to include an additional 20 pesticides to determine how common these off-targets were across a diverse chemical space of OP and TC pesticides (Figure 2C). These studies revealed that several of the off-targets were not only inhibited by specific individual agents, but also constituted a common inhibitory signature for a wide range of OP and TC pesticides, as shown in a heat-map of target inhibition across the 29 pesticide panel (Figure 2D). The most common off-targets inhibited by >30 % were FAAH by 16 pesticides, followed by APEH and ABHD6 by 9 pesticides each, KIAA1363 by 8 pesticides, MAGL by 6 pesticides, NTE and APT1 by 2 pesticides each, and APT2 by 1 pesticide.
Figure 2. Screening of a panel of 29 OP and TC pesticides reveals multiple secondary targets in mouse brain.
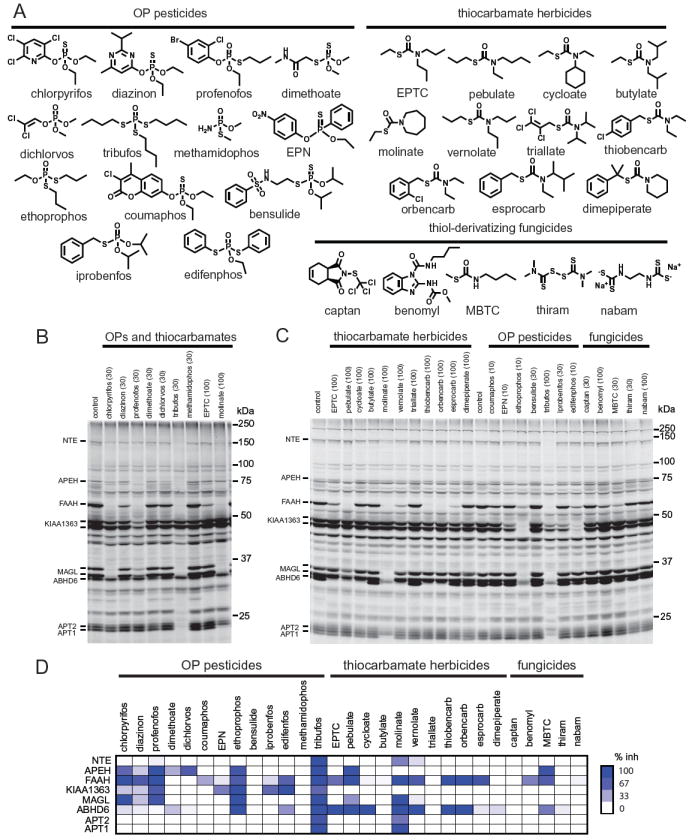
A. Structures of the OP and TC pesticides which include insecticides, herbicides, and fungicides screened by ABPP. B. Nine OP and TC pesticides were initially screened in vivo. The numbers in parentheses are the doses administered in mg/kg. A representative ABPP gel-based analysis is shown of brain membrane proteome labeled with FP-rhodamine and separated by SDS-PAGE. Each band represents a serine hydrolase activity and loss of a band compared to control indicates inhibition of that enzyme by the pesticide. C. A larger set of OP and TC pesticides were screened in vivo. D. Heat map showing average % inhibition of target by each pesticide. Protein identifications for NTE (21), FAAH (19, 21, 25, 28), KIAA1363 (29), MAGL (19, 21, 25), and ABHD6 (21, 22, 25) on the ABPP gels in B and C are based on mass spectrometry analysis or selective inhibitors used in previous studies. APEH, APT2, and APT1 identifications are based on previous mass spectrometry studies (21, 25) and selective inhibitor analyses that are forthcoming and not reported here. AChE is not observable on the gel due to its low abundance. Data represent n=3 mice/group. Gels are representative of n=3.
The endocannabinoid system is a major secondary target of several OP and TC pesticides
We were particularly interested in FAAH and MAGL, since these enzymes are involved in the breakdown of the endocannabinoid signaling lipids, anandamide and 2-arachidonoylglycerol (2-AG), respectively, to terminate their action on the cannabinoid receptor type 1 (CB1) (Figure 3A) (24, 25). The endocannabinoid system is involved in a vast array of physiological processes, including memory, neurogenesis, pain, mood, feeding, energy homeostasis, and metabolism (19, 30-35). We previously identified FAAH and MAGL as targets of CPO and IDFP (21). Dual blockade of FAAH and MAGL led to dramatic elevations in brain levels of both anandamide and 2-AG and robust cannabinoid-dependent behavioral phenotypes, including hypomotility, antinociception and catalepsy fully reversed with a CB1 receptor antagonist (21, 27). We have also previously shown that MAGL blockade decreases brain arachidonic acid levels. Since arachidonic acid is a precursor of prostaglandins, these results suggest that brain endocannabinoid and eicosanoid signaling pathways may be linked (21). In the present study, we show similar neurochemical changes with chlorpyrifos, profenofos, tribufos and molinate among the 9 OP and TC pesticides initially screened, causing elevations in brain levels of 2-AG and anandamide and reductions in brain arachidonic acid levels, with modest but selective elevation of anandamide by EPTC (Figure 3B). The pesticides which significantly blocked both MAGL and FAAH also elicited cataleptic behavior (Figure 3C), consistent with previous studies (21, 27).
Figure 3. Blockade of MAGL and FAAH by OP and TC pesticides alters brain endocannabinoid levels and elicits catalepsy.
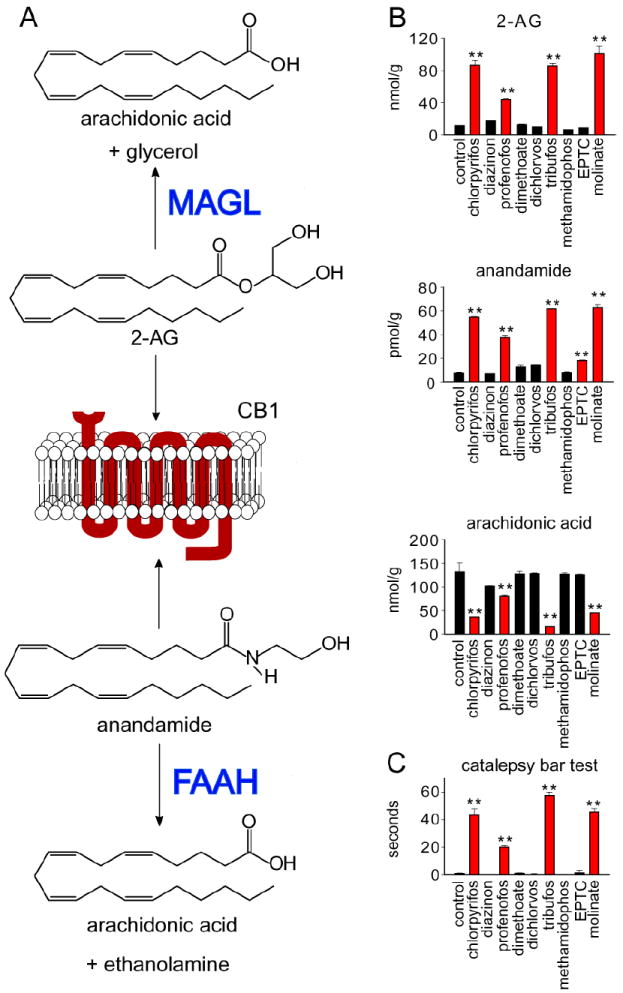
A.The endocannabinoid system has two endogenous ligands, 2-AG and anandamide, which are degraded by MAGL and FAAH, respectively, to terminate their action on the CB1 receptor. B. Brain 2-AG, anandamide and arachidonic acid levels of mice treated with 9 OP and TC pesticides. C. Degree of catalepsy, scored as seconds that a mouse is unresponsive on a bar during a 60 second period. Data represent n=3 mice group, with significance expressed as **p<0.01 compared to control. Data are given as mean ± standard error.
The recent advent of selective and efficacious MAGL inhibitors and dual MAGL/FAAH inhibitors has allowed further dissection of these two signaling pathways. Genetic and pharmacological ablation of FAAH leads to elevations in brain anandamide levels and a subset of cannabinoid behaviors such as antinociception, anxiolysis, and anti-inflammation without the full behavioral effects associated with direct CB1 agonists (i.e. antinociception, hypothermia, hypomotility and catalepsy) (30, 36-38). MAGL blockade by the selective carbamate MAGL inhibitor JZL184 leads to elevation in brain 2-AG and reduction in arachidonic acid levels to elicit a subset of the cannabinoid phenotypes, i.e. hypomotility and antinociception, but not catalepsy (19, 26). Blocking both endocannabinoid degrading enzymes by the selective carbamate MAGL/FAAH inhibitor JZL195 leads to hypomotility, additive antinociception, and catalepsy (27). Interestingly, MAGL and/or FAAH blockade also has ramifications for tetrahydrocannabinol (THC)-mediated drug abuse (27). Acute blockade of FAAH or MAGL significantly attenuated CB1-antagonist-precipitated withdrawal signs in THC-dependent mice (39). Dual FAAH/MAGL blockade, but not disruption of either FAAH or MAGL alone, also produced THC-like responses that were reversed by a CB1 antagonist in a drug-discrimination test (27). These studies indicate a provocative and important feature of the endocannabinoid system where ligand (rather than receptor) diversification is exploited to regulate specific mammalian behaviors. Electrophysiological studies have revealed that MAGL blockade also has profound effects on retrograde synaptic depression and synaptic plasticity. JZL184 prolongs depolarization-induced suppression of excitation in Purkinje neurons in cerebellar slices and depolarization-induced suppression of inhibition in CA1 pyramidal neurons in hippocampal slices (35, 40).
Dual blockade of MAGL and FAAH is also associated with peripheral metabolic phenotypes. The non-selective MAGL/FAAH inhibitor IDFP elicits hypertriglyceridemia in mice in a CB1-dependent manner, attributable to reduced plasma triglyceride clearance and an accumulation in plasma of apolipoprotein E-depleted triglyceride-rich lipoproteins (41). MAGL is also highly upregulated in aggressive human cancer cell lines and high-grade primary human tumors, where it controls a fatty acid network enriched in protumorigenic signaling lipids (42). Blocking MAGL impairs cancer cell pathogenicity and tumorigenicity (42).
Despite potential beneficial effects that could be yielded from pharmacological blockade of MAGL or FAAH in treating pain, inflammation, anxiety, and cancer, there may also be untoward effects from blocking MAGL or both MAGL and FAAH. These effects include alterations in synaptic plasticity, motility defects, potential ramifications for drug abuse, and metabolic abnormalities. Although the effect of chronically ablating MAGL remains unknown due to a lack of a MAGL knockout mouse model, it is also conceivable that CB1 may become desensitized to alter basal cannabinoid function in vivo in regulating pain, feeding, memory and metabolism. Chronic exposure to OP and TC pesticides also has the potential to elicit these toxic responses.
Other Secondary Targets of OP and TC Pesticides of Potential Toxicological Relevance
Several of the other secondary targets have been characterized as having roles in nervous system function. NTE, for example, is the target of OP-induced delayed neuropathy (8, 43). Brain-specific deletion of NTE in mice leads to defects in the hippocampus, thalamus and cerebellum, stemming from disruptions in the endoplasmic reticulum, vacuolation of nerve cell bodies, and abnormal reticular aggregates (44). Both pharmacological and genetic reduction in NTE activity lead to hyperactivity and global NTE deletion leads to embryonic lethality (45). The purported biochemical role of this enzyme is to hydrolyze lysophosphatidyl choline (46) or phosphatidylcholine (47), and there is evidence that the pathology stems from defects in phosphatidylcholine homeostasis in the endoplasmic reticulum (47, 48).
APTs have been implicated in depalmitoylating proteins localized on synaptic membranes, including α13 subunits of G proteins (49). To our knowledge, this is the first report that pesticides target APTs. Knockdown of APT1 has been shown to impair dendritic spine enlargement (49). The physiological functions of APEH are not fully understood, but it has been implicated in the hydrolysis of terminal acetylated amino acids from peptides to release an acetylated amino acid which is further processed by aminoacylase 1 (50). Terminal acetylation of proteins is widespread and thought to protect them from degradation. APEH has also been shown to degrade oligomeric forms of amyloid beta—a peptide that aggregates and is involved in the pathology of Alzheimer’s disease (51, 52). ABHD6 is an uncharacterized serine hydrolase that may contribute to the hydrolysis of 2-AG, perhaps in specific brain regions or specific cell-types (25).
Serine hydrolase KIAA1363 has been partially characterized and has garnered attention in multiple fields. It is a primary detoxifying enzyme for selected OP pesticides (27, 53, 54). Ablation of KIAA1363 results in hypersensitivity to OP toxicants and inhibition of other secondary targets (53, 54). KIAA1363 is also highly expressed in multiple types of aggressive human cancer cell lines and primary estrogen receptor negative human primary breast tumors where it regulates an ether lipid network to drive cancer cell migration and tumor growth (20, 55, 56). One possible substrate for this enzyme is acetyl monoalkylglycerol ether (acetyl MAGE) which is hydrolyzed by KIAA1363 to monoalkylglycerol ether (MAGE) and acetate (20, 53). Consistent with this premise, blocking this enzyme in cancer cells leads to depletion of MAGE and reduced levels of protumorigenic lipids such as alkyl lysophosphatidic acid (20). Almost all of the acetyl MAGE hydrolytic activity in the aggressive cancer cells is due to KIAA1363. In mice, KIAA1363 is the primary acetyl MAGE hydrolase in brain, lung, heart, and kidney (53). On incubating KIAA1363 +/+ and −/− brain membranes with acetyl MAGE cofactors for acetyl MAGE phosphocholine transferase, the absence of KIAA1363 activity dramatically increased de novo formation of platelet-activating factor, a highly potent signaling lipid in the brain (53). Recent studies have also provided evidence that KIAA1363 serves as a neutral cholesteryl ester hydrolase and genetic ablation of KIAA1363 promotes foam cell formation and the development of atherosclerosis in mice (57).
Bioactivation of OPs and TCs to Phosphorylating and Carbamoylating Agents
Many of the pesticides examined are bioactivated by cytochrome P450-catalyzed sulfoxidation to form phosphorylating or carbamoylating agents for hydrolases with serine or cysteine at their active sites. This includes at least three of the phosphorothiolates (P-S-R) (ethoprophos, profenofos, and tribufos) (58) and the six phosphorothionates (P=S) examined. Most or all of the TCs studied undergo sulfoxidation to TC sulfoxides which are more potent than the parent compounds as carbamoylating agents for glutathione and other tissue thiols (59, 60). In the case of MAGL which has a reactive cysteine within the active site, it is conceivable although not likely that the derivatization site of the activated TC pesticides may be cysteine (61) instead of the active site serine.
Insights Revealed from Secondary Targets of OPs and TCs
We show that several of the 29 OP and TC pesticides screened in this study target neurologically important enzymes such as those involved in regulating endocannabinoid function (MAGL and FAAH), endoplasmic reticulum integrity (NTE), and palmitoylation states of proteins and dendritic spine formation (APTs). We also find that several pesticides block MAGL and FAAH, resulting in neurochemical changes in brain endocannabinoid levels and arachidonic acid metabolism, resulting in cannabinoid-behaviors such as catalepsy. In addition, ABPP also reveals secondary targets that have poorly or completely uncharacterized physiological function such as ABHD6, APEH, and KIAA1363. Because several of the OPs and TCs tested have overlapping off-target profiles, cumulative exposure to these pesticides may potentially aggravate the risk of eliciting toxicity from these secondary sites. We caution that the doses used in this study are significantly higher than those that would be encountered in normal pest control practice, and that dose-response profiles and chronic low-dose exposure studies are warranted to address the physiological relevance (and the proteomic and metabolic correlates) of these studies. However, this investigation shows complete blockade of several secondary targets at sublethal doses (i.e. doses that do not fully inhibit AChE), implying higher in vivo sensitivity of these enzymes compared to AChE. Additionally, the pesticides used in this study may provide scaffolds for further inhibitor optimization to study the biology of these targets in vivo. Collectively, this study shows how ABPP can be used to study metabolic pathways that are dysregulated by pesticides to provide unique insights into toxicant mode of action.
Acknowledgments
We acknowledge Professor Benjamin F. Cravatt of The Scripps Research Institute for continued collaboration, support, and resources.
D.K.N. was supported by Award Number K99DA030908 from the National Institute on Drug Abuse. J.E.C. was funded by the University of California at Berkeley William Muriece Hoskins Chair in Chemical and Molecular Entomology.
ABBREVIATIONS USED
- ABHD6
alpha/beta hydrolase domain 6
- ABPP
activity-based protein profiling
- AChE
acetylcholinesterase
- APEH
acyl peptide hydrolase
- APT1
acyl protein thioesterase 1
- APT2
acyl protein thioesterase 2
- 2-AG
2-arachidonoylglycerol
- CB1
cannabinoid receptor type 1
- CPO
chlorpyrifos oxon
- FAAH
fatty acid amide hydrolase
- FP-rhodamine
fluorophosphonate-rhodamine
- IDFP
isopropyl dodecylfluorophosphonate
- MAGE
monoalkylglycerol ether
- MAGL
monoacylglycerol lipase
- MBTC
methyl butylthiocarbamate
- NTE
neuropathy target esterase
- OP
organophosphorus
- TC
thiocarbamate
- THC
tetrahydrocannabinol
LITERATURE CITED
- 1.Reeves M, Schafer KS. Greater risks, fewer rights: U.S. farmworkers and pesticides. Int J Occup Environ Health. 2003;9:30–9. doi: 10.1179/107735203800328858. [DOI] [PubMed] [Google Scholar]
- 2.Calvert GM, Plate DK, Das R, Rosales R, Shafey O, Thomsen C, Male D, Beckman J, Arvizu E, Lackovic M. Acute occupational pesticide-related illness in the US, 1998-1999: surveillance findings from the SENSOR-pesticides program. Am J Ind Med. 2004;45:14–23. doi: 10.1002/ajim.10309. [DOI] [PubMed] [Google Scholar]
- 3.Jeyaratnam J. Acute pesticide poisoning: a major global health problem. World Health Stat Q. 1990;43:139–44. [PubMed] [Google Scholar]
- 4.Eskenazi B, Bradman A, Castorina R. Exposures of children to organophosphate pesticides and their potential adverse health effects. Environ Health Perspect. 1999;107(Suppl 3):409–19. doi: 10.1289/ehp.99107s3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pilkington A, Buchanan D, Jamal GA, Gillham R, Hansen S, Kidd M, Hurley JF, Soutar CA. An epidemiological study of the relations between exposure to organophosphate pesticides and indices of chronic peripheral neuropathy and neuropsychological abnormalities in sheep farmers and dippers. Occup Environ Med. 2001;58:702–10. doi: 10.1136/oem.58.11.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoppin JA, Umbach DM, London SJ, Lynch CF, Alavanja MC, Sandler DP. Pesticides associated with wheeze among commercial pesticide applicators in the Agricultural Health Study. Am J Epidemiol. 2006;163:1129–37. doi: 10.1093/aje/kwj138. [DOI] [PubMed] [Google Scholar]
- 7.Casida JE, Quistad GB. Serine hydrolase targets of organophosphorus toxicants. Chem Biol Interact. 2005:157–158. 277–83. doi: 10.1016/j.cbi.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 8.Casida JE, Quistad GB. Organophosphate toxicology: safety aspects of nonacetylcholinesterase secondary targets. Chem Res Toxicol. 2004;17:983–98. doi: 10.1021/tx0499259. [DOI] [PubMed] [Google Scholar]
- 9.Patel S, Singh K, Singh S, Singh MP. Gene expression profiles of mouse striatum in control and maneb + paraquat-induced Parkinson’s disease phenotype: validation of differentially expressed energy metabolizing transcripts. Mol Biotechnol. 2008;40:59–68. doi: 10.1007/s12033-008-9060-9. [DOI] [PubMed] [Google Scholar]
- 10.Unver T, Bakar M, Shearman RC, Budak H. Genome-wide profiling and analysis of Festuca arundinacea miRNAs and transcriptomes in response to foliar glyphosate application. Mol Genet Genomics. 283:397–413. doi: 10.1007/s00438-010-0526-7. [DOI] [PubMed] [Google Scholar]
- 11.Pereira JL, Hill CJ, Sibly RM, Bolshakov VN, Goncalves F, Heckmann LH, Callaghan A. Gene transcription in Daphnia magna: effects of acute exposure to a carbamate insecticide and an acetanilide herbicide. Aquat Toxicol. 97:268–76. doi: 10.1016/j.aquatox.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 12.Harris W, Sachana M, Flaskos J, Hargreaves AJ. Proteomic analysis of differentiating neuroblastoma cells treated with sub-lethal neurite inhibitory concentrations of diazinon: identification of novel biomarkers of effect. Toxicol Appl Pharmacol. 2009;240:159–65. doi: 10.1016/j.taap.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 13.Jin J, Davis J, Zhu D, Kashima DT, Leroueil M, Pan C, Montine KS, Zhang J. Identification of novel proteins affected by rotenone in mitochondria of dopaminergic cells. BMC Neurosci. 2007;8:67. doi: 10.1186/1471-2202-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry. 2001;40:4005–15. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96:14694–9. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speers AE, Cravatt BF. Chemical strategies for activity-based proteomics. Chembiochem. 2004;5:41–7. doi: 10.1002/cbic.200300721. [DOI] [PubMed] [Google Scholar]
- 17.Saghatelian A, Cravatt BF. Assignment of protein function in the postgenomic era. Nat Chem Biol. 2005;1:130–42. doi: 10.1038/nchembio0805-130. [DOI] [PubMed] [Google Scholar]
- 18.Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat Biotechnol. 2003;21:687–91. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 19.Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavon FJ, Serrano AM, Selley DE, Parsons LH, Lichtman AH, Cravatt BF. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13:1041–50. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Nomura DK, Blankman JL, Simon GM, Fujioka K, Issa RS, Ward AM, Cravatt BF, Casida JE. Activation of the endocannabinoid system by organophosphorus nerve agents. Nat Chem Biol. 2008;4:373–8. doi: 10.1038/nchembio.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Blankman JL, Cravatt BF. A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. J Am Chem Soc. 2007;129:9594–5. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- 23.Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chem Rev. 2006;106:3279–301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 24.Ahn K, McKinney MK, Cravatt BF. Enzymatic pathways that regulate endocannabinoid signaling in the nervous system. Chem Rev. 2008;108:1687–707. doi: 10.1021/cr0782067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blankman JL, Simon GM, Cravatt BF. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem Biol. 2007;14:1347–56. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long JZ, Nomura DK, Cravatt BF. Characterization of monoacylglycerol lipase inhibition reveals differences in central and peripheral endocannabinoid metabolism. Chem Biol. 2009;16:744–53. doi: 10.1016/j.chembiol.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long JZ, Nomura DK, Vann RE, Walentiny DM, Booker L, Jin X, Burston JJ, Sim-Selley LJ, Lichtman AH, Wiley JL, Cravatt BF. Dual blockade of FAAH and MAGL identifies behavioral processes regulated by endocannabinoid crosstalk in vivo. Proc Natl Acad Sci U S A. 2009;106:20270–5. doi: 10.1073/pnas.0909411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahn K, Johnson DS, Mileni M, Beidler D, Long JZ, McKinney MK, Weerapana E, Sadagopan N, Liimatta M, Smith SE, Lazerwith S, Stiff C, Kamtekar S, Bhattacharya K, Zhang Y, Swaney S, Van Becelaere K, Stevens RC, Cravatt BF. Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem Biol. 2009;16:411–20. doi: 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nomura DK, Leung D, Chiang KP, Quistad GB, Cravatt BF, Casida JE. A brain detoxifying enzyme for organophosphorus nerve poisons. Proc Natl Acad Sci U S A. 2005;102:6195–200. doi: 10.1073/pnas.0501915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piomelli D. The molecular logic of endocannabinoid signalling. Nat Rev Neurosci. 2003;4:873–84. doi: 10.1038/nrn1247. [DOI] [PubMed] [Google Scholar]
- 31.Gao Y, Vasilyev DV, Goncalves MB, Howell FV, Hobbs C, Reisenberg M, Shen R, Zhang MY, Strassle BW, Lu P, Mark L, Piesla MJ, Deng K, Kouranova EV, Ring RH, Whiteside GT, Bates B, Walsh FS, Williams G, Pangalos MN, Samad TA, Doherty P. Loss of retrograde endocannabinoid signaling and reduced adult neurogenesis in diacylglycerol lipase knock-out mice. J Neurosci. 2010;30:2017–24. doi: 10.1523/JNEUROSCI.5693-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osei-Hyiaman D, Harvey-White J, Batkai S, Kunos G. The role of the endocannabinoid system in the control of energy homeostasis. Int J Obes (Lond) 2006;30(Suppl 1):S33–8. doi: 10.1038/sj.ijo.0803276. [DOI] [PubMed] [Google Scholar]
- 33.Osei-Hyiaman D, DePetrillo M, Pacher P, Liu J, Radaeva S, Batkai S, Harvey-White J, Mackie K, Offertaler L, Wang L, Kunos G. Endocannabinoid activation at hepatic CB1 receptors stimulates fatty acid synthesis and contributes to diet-induced obesity. J Clin Invest. 2005;115:1298–305. doi: 10.1172/JCI23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Marzo V, Goparaju SK, Wang L, Liu J, Batkai S, Jarai Z, Fezza F, Miura GI, Palmiter RD, Sugiura T, Kunos G. Leptin-regulated endocannabinoids are involved in maintaining food intake. Nature. 2001;410:822–5. doi: 10.1038/35071088. [DOI] [PubMed] [Google Scholar]
- 35.Straiker A, Hu SS, Long JZ, Arnold A, Wager-Miller J, Cravatt BF, Mackie K. Monoacylglycerol lipase limits the duration of endocannabinoid-mediated depolarization-induced suppression of excitation in autaptic hippocampal neurons. Mol Pharmacol. 2009;76:1220–7. doi: 10.1124/mol.109.059030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cravatt BF, Demarest K, Patricelli MP, Bracey MH, Giang DK, Martin BR, Lichtman AH. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc Natl Acad Sci U S A. 2001;98:9371–6. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pattipati SN, Kinsey SG, Guo T, Cravatt BF, Lichtman AH. Regulation of Inflammatory Pain by Inhibition of Fatty Acid Amide Hydrolase (FAAH) J Pharmacol Exp Ther. 2010 doi: 10.1124/jpet.109.164806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kathuria S, Gaetani S, Fegley D, Valino F, Duranti A, Tontini A, Mor M, Tarzia G, La Rana G, Calignano A, Giustino A, Tattoli M, Palmery M, Cuomo V, Piomelli D. Modulation of anxiety through blockade of anandamide hydrolysis. Nat Med. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- 39.Schlosburg JE, Carlson BL, Ramesh D, Abdullah RA, Long JZ, Cravatt BF, Lichtman AH. Inhibitors of endocannabinoid-metabolizing enzymes reduce precipitated withdrawal responses in THC-dependent mice. AAPS J. 2009;11:342–52. doi: 10.1208/s12248-009-9110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan B, Wang W, Long JZ, Sun D, Hillard CJ, Cravatt BF, Liu QS. Blockade of 2-arachidonoylglycerol hydrolysis by selective monoacylglycerol lipase inhibitor 4-nitrophenyl 4- (dibenzo[d][1,3]dioxol-5-yl(hydroxy)methyl)piperidine-1-carboxylate (JZL184) Enhances retrograde endocannabinoid signaling. J Pharmacol Exp Ther. 2009;331:591–7. doi: 10.1124/jpet.109.158162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruby MA, Nomura DK, Hudak CS, Mangravite LM, Chiu S, Casida JE, Krauss RM. Overactive endocannabinoid signaling impairs apolipoprotein E-mediated clearance of triglyceride-rich lipoproteins. Proc Natl Acad Sci U S A. 2008;105:14561–6. doi: 10.1073/pnas.0807232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casida JE, Nomura DK, Vose SC, Fujioka K. Organophosphate-sensitive lipases modulate brain lysophospholipids, ether lipids and endocannabinoids. Chem Biol Interact. 2008;175:355–64. doi: 10.1016/j.cbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akassoglou K, Malester B, Xu J, Tessarollo L, Rosenbluth J, Chao MV. Brain-specific deletion of neuropathy target esterase/swisscheese results in neurodegeneration. Proc Natl Acad Sci U S A. 2004;101:5075–80. doi: 10.1073/pnas.0401030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winrow CJ, Hemming ML, Allen DM, Quistad GB, Casida JE, Barlow C. Loss of neuropathy target esterase in mice links organophosphate exposure to hyperactivity. Nat Genet. 2003;33:477–85. doi: 10.1038/ng1131. [DOI] [PubMed] [Google Scholar]
- 46.Quistad GB, Barlow C, Winrow CJ, Sparks SE, Casida JE. Evidence that mouse brain neuropathy target esterase is a lysophospholipase. Proc Natl Acad Sci U S A. 2003;100:7983–7. doi: 10.1073/pnas.1232473100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muhlig-Versen M, da Cruz AB, Tschape JA, Moser M, Buttner R, Athenstaedt K, Glynn P, Kretzschmar D. Loss of Swiss cheese/neuropathy target esterase activity causes disruption of phosphatidylcholine homeostasis and neuronal and glial death in adult Drosophila. J Neurosci. 2005;25:2865–73. doi: 10.1523/JNEUROSCI.5097-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vose SC, Fujioka K, Gulevich AG, Lin AY, Holland NT, Casida JE. Cellular function of neuropathy target esterase in lysophosphatidylcholine action. Toxicol Appl Pharmacol. 2008;232:376–83. doi: 10.1016/j.taap.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 49.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hubel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, Martinez J, Schratt GM. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–16. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrier J, Durand A, Giardina T, Puigserver A. Catabolism of intracellular N-terminal acetylated proteins: involvement of acylpeptide hydrolase and acylase. Biochimie. 2005;87:673–85. doi: 10.1016/j.biochi.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 51.Yamin R, Zhao C, O’Connor PB, McKee AC, Abraham CR. Acyl peptide hydrolase degrades monomeric and oligomeric amyloid-beta peptide. Mol Neurodegener. 2009;4:33. doi: 10.1186/1750-1326-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamin R, Bagchi S, Hildebrant R, Scaloni A, Widom RL, Abraham CR. Acyl peptide hydrolase, a serine proteinase isolated from conditioned medium of neuroblastoma cells, degrades the amyloid-beta peptide. J Neurochem. 2007;100:458–67. doi: 10.1111/j.1471-4159.2006.04251.x. [DOI] [PubMed] [Google Scholar]
- 53.Nomura DK, Fujioka K, Issa RS, Ward AM, Cravatt BF, Casida JE. Dual roles of brain serine hydrolase KIAA1363 in ether lipid metabolism and organophosphate detoxification. Toxicol Appl Pharmacol. 2008;228:42–8. doi: 10.1016/j.taap.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nomura DK, Durkin KA, Chiang KP, Quistad GB, Cravatt BF, Casida JE. Serine hydrolase KIAA1363: toxicological and structural features with emphasis on organophosphate interactions. Chem Res Toxicol. 2006;19:1142–50. doi: 10.1021/tx060117m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3, Jeffrey SS, Cravatt BF. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–7. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 56.Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci U S A. 2002;99:10335–40. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sekiya M, Osuga J, Nagashima S, Ohshiro T, Igarashi M, Okazaki H, Takahashi M, Tazoe F, Wada T, Ohta K, Takanashi M, Kumagai M, Nishi M, Takase S, Yahagi N, Yagyu H, Ohashi K, Nagai R, Kadowaki T, Furukawa Y, Ishibashi S. Ablation of neutral cholesterol ester hydrolase 1 accelerates atherosclerosis. Cell Metab. 2009;10:219–28. doi: 10.1016/j.cmet.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 58.Glickman AH, Wing KD, Casida JE. Profenofos insecticide bioactivation in relation to antidote action and the stereospecificity of acetylcholinesterase inhibition, reactivation, and aging. Toxicol Appl Pharmacol. 1984;73:16–22. doi: 10.1016/0041-008x(84)90047-4. [DOI] [PubMed] [Google Scholar]
- 59.Casida JE, Kimmel EC, Lay M, Ohkawa H, Rodebush JE, Gray RA, Tseng CK, Tilles H. Thiocarbamate sulfoxide herbicides. Environ Qual Saf Suppl. 1975;3:675–9. [PubMed] [Google Scholar]
- 60.Staub RE, Quistad GB, Casida JE. Mechanism for benomyl action as a mitochondrial aldehyde dehydrogenase inhibitor in mice. Chem Res Toxicol. 1998;11:535–43. doi: 10.1021/tx980002l. [DOI] [PubMed] [Google Scholar]
- 61.Saario SM, Salo OM, Nevalainen T, Poso A, Laitinen JT, Jarvinen T, Niemi R. Characterization of the sulfhydryl-sensitive site in the enzyme responsible for hydrolysis of 2-arachidonoyl-glycerol in rat cerebellar membranes. Chem Biol. 2005;12:649–56. doi: 10.1016/j.chembiol.2005.04.013. [DOI] [PubMed] [Google Scholar]


