Abstract
The first component of the bacterial phosphotransferase system, enzyme I (EI), is a multidomain 128 kDa dimer that undergoes large rigid body conformational transitions during the course of its catalytic cycle. Here we investigate the solution structure of a non-phosphorylatable active site mutant in which the active site histidine is substituted by glutamine. We show that perturbations in the relative orientations and positions of the domains and subdomains can be rapidly and reliably determined by conjoined rigid body/torsion angle/Cartesian simulated annealing calculations driven by orientational restraints from residual dipolar couplings and shape and translation information afforded by small and wide angle X-ray scattering. Although histidine and glutamine are isosteric, the conformational space available to a Gln side chain is larger than that for the imidazole ring of His. An additional hydrogen bond between the side chain of Gln189 located on the EINα/β subdomain and an aspartate (Asp129) on the EINα subdomain results in a small (~9°) reorientation of the EINα and EINα/β subdomains that is in turn propagated to a larger reorientation (~26°) of the EIN domain relative to the EIC dimerization domain, illustrating the positional sensitivity of the EIN domain and its constituent subdomains to small structural perturbations.
Enzyme I (EI), the first component of the bacterial phosphotransferase system (PTS),1 is a 128 kDa multidomain dimer:2 the C-terminal dimerization domain (EIC) contains the phosphoenolpyruvate binding site; the N-terminal domain (EIN) is subdivided into two structural subdomains,3,4 the EINα/β subdomain that bears the active site histidine, His189, and the EINα subdomain that binds the next component of the pathway, the histidine phosphocarrier protein HPr.5,6 The EIC domain binds phosphoenolpyruvate and autophosphorylates the active site His189 on the EIN domain.2,7 Following concerted rigid body conformational transitions within the EIN domain involving reorientation of the EINα/β subdomain relative to the EIC domain and of the EINα subdomain relative to the EINα/β subdomain,8,9 the phosphoryl group is transferred to HPr. The structure of the EIC domain, on the other hand, remains unchanged in different states of the enzyme as well as across different species.8,10–12 Using residual dipolar couplings (RDCs)13 to determine the orientation of the symmetry-related EIN domains relative to the EIC dimerization domain, and small and wide angle X-ray scattering (SAXS/WAXS) to provide shape and translational information,14 we recently succeeded in determining the solution structures of free E. coli EI and the ~146 kDa EI-HPr complex based on the known structures of the individual domains in conjunction with conjoined rigid body/torsion angle/Cartesian simulated annealing.9 We showed that the relative orientation of the EINα and EINα/β subdomains in both free EI and the EI-HPr complex is identical to that in the structure of the isolated EIN domain,3,4,6 and differs by a ~70° reorientation from that observed in the trapped phosphorylated intermediate state of EI captured by crystallography.8 Further, the transition from the trapped phosphorylated state8 to the free enzyme9 also involves a ~95° reorientation of the EINα/β subdomain relative to the EIC domain. In this paper we show that RDCs and SAXS/WAXS can be used to rapidly determine the orientation of the EIN domain and its two sub-domains in the non-phosphorylatable active site mutant, H189Q, whose biochemical properties have been previously characterized.7 Comparison with the structure of wild-type EI9 reveals a small but significant change in the relative orientation of the two subdomains of EIN that is propagated to a much larger change in orientation of the EIN domain relative to the EIC domain. Although His and Gln are isosteric, the conformational space available to the side chains of His and Gln are different, and the structural perturbations can be directly attributed to changes in hydrogen-bonding pattern involving residue 189.
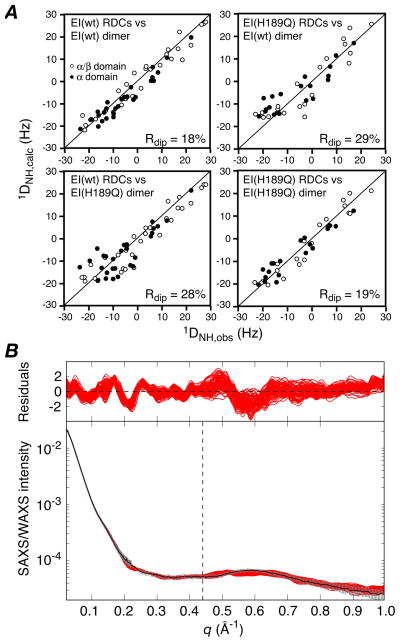
The EI(H189Q) mutant in the presence of 4 mM Mg2+ and 100 mM NaCl at pH 7.4 is dimeric under the conditions of the NMR (0.3 mM in subunits) and SAXS/WAXS (80 μM in subunits) experiments with a dimerization constant of ~0.7 μM that is unchanged from that of wild-type EI (~0.8 μM),9 as determined by sedimentation velocity. The 1H-15N TROSY correlation spectrum of EI(H189Q) is also essentially unchanged from that of wild-type EI, permitting the facile transfer of assignments for well-resolved resonances of the EIN domain from the spectrum of isolated EIN as described previously9 (see Supplementary Fig. S1). Backbone amide RDCs measured on well-resolved 1HN/15N cross-peaks of the EIN domain in intact U-[15N/2H]-EI(H189Q) using the TROSY-based ARTSY technique15 display excellent agreement with the NMR internal coordinates of the EINα and EINα/β subdomains6 with RDC R-factors of 16.3 and 17.3%, respectively (see Supplementary, Fig. S2). However, singular value decomposition (SVD) fitting to the structure of the whole EIN domain yields an R-factor of 20.2%, 3% higher than the weighted average of the R-factors for the two subdomains, suggesting a small change in relative orientation of the EINα and EINα/β subdomains in the EI(H189Q) mutant relative to wild-type. (The R-factor for the EIN domain with the EINα and EINα/β subdomains oriented as in the trapped phosphorylated state of EI8 is 38%). In contrast, the agreement of the wild-type RDCs with the structure of the wild-type EIN domain is only minimally larger (≤1%) than the weighted average for the two subdomains.9 More significantly, when the RDCs for the EIN domain of EI(H189Q) are fitted to the structure of the wild-type EI dimer, the R-factor is increased to 29% (Fig. 1A, top right) compared to 18% for the wild-type RDCs (Fig. 1A, top left). Thus, there must also be a significant change in orientation of the EIN domain relative to the EIC domain in the EI(H189Q) mutant. The overall change in shape, however, as judged by SAXS/WAXS, is quite subtle, since the agreement of the wild-type EI dimer structure9 with the EI(H189Q) data (q ≤ 0. 44 Å−1) is only slightly worse (χ2 ~ 0.9) than with the wild-type data (χ2 ~ 0.5) (see Supplementary Fig. S3). Nevertheless, analysis of the P(r) distribution for the X-ray scattering data suggests that there is a small decrease in Rgyr from 42.1±0.2 Å for wild-type EI9 to 41.3±0.5 Å for the EI(H189Q) mutant.
Figure 1.
(A) Comparison of observed and calculated RDCs for the E. coli EI (wild-type) and EI(H189Q) dimers The experimental RDCs for the EI(H189Q) mutant comprise 20 backbone 1DNH couplings for each subdomain of EIN (i.e. a total of 40×2 RDCs for the dimer) obtained from a 1H-15N TROSY-based ARTSY spectrum15 with EI(H189Q) aligned in a dilute liquid crystalline medium of pf1 phage16 (11 mg/ml). For wild-type EI there are 29 1DNH couplings for each subdomain of EIN (i.e. a total of 58×2 RDCs for the dimer).9 Top right, wild-type RDCs versus wild-type dimer structure;9 top left, EI(H189Q) RDCs versus wild-type EI dimer structure; bottom left, wild-type RDCs versus EI(H189Q) dimer structure (this paper); bottom right, EI(H189Q) RDCs versus EI(H189Q) dimer structure. (B) Comparison of experimental SAXS/WAXS curves (black with gray vertical bars equal to 1 s.d.) recorded for EI(H189Q) with the calculated curves for the final 100 simulated annealing structures in red. The residuals given by are plotted above. The structures were determined by refining against the SAXS/WAXS curve in the range q ≤0.44 Å−1, and the upper end of this range is indicated by the vertical dashed black line. The SAXS/WAXS data were collected at the Advanced Photon Source (Argonne National Laboratory) and processed as described previously.9 The SAXS data extend out to q = 0.22 Å−1 and report on interatomic interactions > 28 Å. The SAXS/WAXS data used in refinement (q ≤ 0.44 Å−1) report on interatomic interactions down to 14 Å, and therefore include additional information related to interdomain interactions.
We therefore set out to solve the solution structure of the EI(H189Q) mutant using a similar conjoined rigid body/torsion angle/Cartesian dynamics simulated annealing strategy driven by RDCs and SAXS/WAXS (up to q = 0.44 Å−1) data that was employed for free EI.9 The calculations were carried out in Xplor-NIH.18,19 Starting with 120 simulated annealing structures previously calculated for wild-type EI,9 120 structures for EI(H189Q) were calculated as follows: the two symmetry-related EIC domains (residues 262–573) were held fixed in space, the EINα (residues 25–142) and EINα/β (residues (1–21 and 147–254) subdomains were treated as separate rigid bodies; the backbone of the linker region connecting the EINα/β domain to the EINα subdomain (residues 22–24 and 143–146) and to the EIC domain (residues 255–261) were given Cartesian degrees of freedom with broad φ/ψ restraints to ensure that these backbone residues lie within the allowed regions of the Ramachandran map, and side chains within the linker region, and side chains at the EINα/EINα/β interface (defined by residues on each subdomain containing an atom within 6 Å of an atom on the other subdomain) and at the EIN/EIC interface (defined by residues on each domain containing an atom within 14.5 Å of an atom on the other domain) were given torsional degrees of freedom. In addition to the experimental RDC and SAXS/WAXS restraints, the target function comprised terms for covalent geometry, a non-bonded repulsion term, C2 symmetry restraints, backbone and side chain torsion angle database potentials of mean force,21 and a gyration volume restraint22 applied only to the EIN domain to ensure reasonable packing at the EINα/EINα/β interface. Calculations were carried out both with the raw experimental RDC restraints, and with 2 Hz random noise added to the experimental RDC restraints (with different random noise added for each individual structure calculation resulting in 120 sets of RDC restraints). The purpose of adding noise was to investigate the effects of errors in the RDCs on the accuracy and precision of the alignment tensor and resulting structures. The results are summarized in Tables 1 and 2, and Figs. 1–3. (The coordinates and experimental data have been deposited in the PDB, accession number 2L5H).
Table 1.
Structural statistics for refinement of the EI(H189Q) mutant structure based on RDCs and SAXS/WAXS
| RDCs with no noise | RDCs with random noisea | |
|---|---|---|
| RDC R-factor (%)b | 19.0±0.4 | 19.6±0.7 |
| RDC DaNH (Hz) | 11.1±0.2 | 11.1±2.4 |
| RDC η | 0.59±0.02 | 0.58±0.06 |
| SAXS/WAXSc | ||
| q = 0.022 → 0.44 Å−1 | 0.58±0.09 | 0.58±0.13 |
| q = 0.022 → 1.0 Å−1 | 0.76±0.16 | 0.78±0.19 |
| Coordinate precision | ||
| EIN domaind | ||
| Cα rms displacement (Å) | 2.2 | 2.7 |
| rotation (deg.) | 5.8 | 7.3 |
| EINα subdomaine | ||
| Cα rms displacement (Å) | 2.1 | 2.5 |
| rotation (deg.) | 3.8 | 5.3 |
2% random noise added to experimental RDCs with a different distribution of random noise for each structure calculated. The values of the RDC R-factor, DaNH and η are reported with respect to the experimental RDCs (with no noise added). The RDC R-factor is calculated as Rinf = [<(Dobs-Dcalc)2>/(2<Dobs2>]1/2 where Dobs and Dcalc are the observed and calculated RDCs, respectively.18
The RDC data comprise 40×2 backbone N-H RDCs for the dimer, 20 for each subdomain of each EIN subunit.
Structures are refined against SAXS/WAXS data for q ≤0.44 Å−1 and the χ2 values reported for all the data (up to q = 1.0 Å−1) are obtained with the parameters from the fits to the data up to q = 0.44 Å−1; these χ2 values therefore report on how well the structures obtained using data up to q = 0.44 Å−1 predict the data from q = 0.44 to 1.0 Å−1.
The EIC domain is superimposed and fixed, and the position of the EIN domain compared.
The EINα/β subdomain is superimposed and fixed, and the position of the EINα subdomain is compared.
Table 2.
Comparison of the positions of the EIN domain and the EINα subdomain
| Cα rms displacement (Å)/rotation (deg) | ||
|---|---|---|
| EI(H189Q) no RDC noise | EI(H189Q) noise added to RDCs | |
| EIN domaina | ||
| EI wild-type | 8.8 Å/26.8° | 8.4 Å/25.6° |
| EI(H189Q) (no RDC noise) | 0.6 Å/1.3° | |
| EINα subdomainb | ||
| EI wild-type | 3.5 Å/9.4° | 3.4 Å/8.9° |
| EI(H189Q) (no RDC noise) | 0.8 Å/1.5° | |
The EIC domain is superimposed and fixed, and the position of the EIN domain compared.
The EINα/β subdomain is superimposed and fixed, and the position of the EINα subdomain is compared.
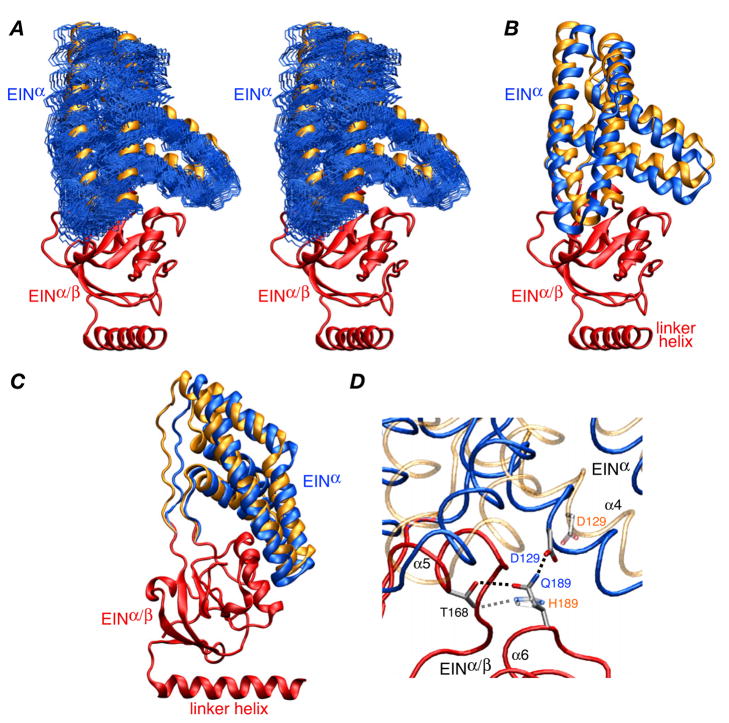
Figure 3.
Structure of the EIN domain in the EI(H189Q) dimer. (A) Stereoview of the final 100 simulated annealing structures (calculated with random noised added to the RDCs) best-fitted to the EINα/β subdomain (red ribbon) showing the precision with which the orientation of the EINα subdomain (backbone atoms in blue) relative to the EINα/β subdomain has been determined. The location of the EINα subdomain in wild-type EI9 is shown as a gold ribbon. (B) and (C) Two views of the EIN domain with the EINα/β subdomain superimposed (red) and the EINα subdomains of the restrained regularized mean structures of the EI(H189Q) mutant and wild-type EI9 in blue and gold, respectively. (D) Detail depicting the EINα/EINα/β interface and the hydrogen bonding interactions involving residues 189 (Gln in the mutant and His in the wild-type). The backbone of the EINα/β subdomain of the mutant and wild-type is superimposed (red tube), and the backbone of the EINα subdomain is shown as blue and transparent gold tubes for the mutant and wild-type, respectively. The side chains of Thr168, His189/Gln189 and Asp129 are displayed, with the mutant opaque and the wild-type transparent; the dashed lines indicate hydrogen bonding interactions.
The calculated EI(H189Q) dimer structures agree well with the experimental RDCs (R-factor of ~19%; Fig. 1A, bottom right) and SAXS/WAXS data (χ2 ~ 0.6 for the q ≤ 0.44 Å−1 data used in refinement; Fig. 1B). Cross-validation is provided by the excellent agreement (χ2 ~ 0.8) with the SAXS/WAXS data up to q = 1 Å−1 using the fitting parameters obtained from refinement against the data up to q = 0.44 Å−1. In addition there is good agreement between the calculated value of 5.83 S for the sedimentation coefficient obtained by hydrodynamic modeling of the EI(H189Q) structure using HYDROPRO23 and the experimentally determined value of 5.73±0.02 S derived from sedimentation velocity experiments at natural isotopic abundance (see Supplementary). As expected, the structure of the EI(H189Q) dimer does not agree with the RDCs for wild-type EI (R-factor = 28%; Fig. 1A, bottom left), and agrees only relatively poorly (χ2 ~ 2.4) with the wild type SAXS/WAXS data (q ≤ 0.44 Å−1). Addition of random noise to the RDC data has no significant effect upon the restrained regularized mean coordinate positions, with Cα rms differences of only 0.6–0.8Å when examining the position of the EINα subdomain relative to the EINα/β subdomain and of the EIN domain relative to EIC domain, but reduces coordinate precision by 20–25%, as expected (Table 2). The uncertainties in the relative domain and subdomain orientations observed in the ensemble of structures calculated with noise added to the RDCs (Table 1) are consistent with the expected uncertainties in the alignment tensor based on the number of measured RDCs.24
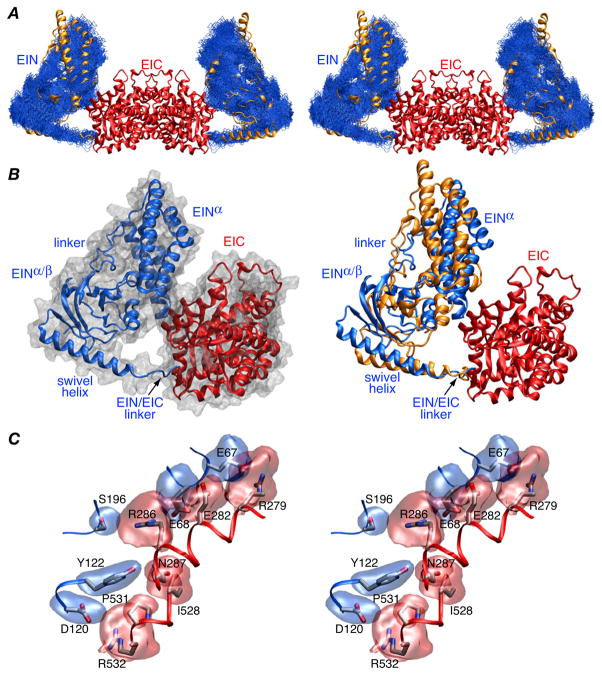
Comparison of the position of the EIN domain relative to the EIC dimerization domain (Fig. 2A, right panel Fig 2B, and Table 2) reveals that the EIN domain is rotated by ~26° and displaced by 8–9 Å relative to its position in wild-type EI. Nevertheless, similar contacts between the EIN and EIC domains involving the same residues are retained (Fig. 2B, left panel and Fig. 2C): there is a salt bridge between Arg279 and Glu67, a hydrogen bond between the carboxylate of Glu282 and the backbone amide of Glu68, and electrostatic interactions between Arg286 and Glu68, Arg286 and Ser196, and Arg532 and Asp120. The accessible surface area buried at the EIN/EIC interface is 435 Å2 which is ~20% larger than that found for wild-type EI.9
Figure 2.
Structure of the EI(H189Q) dimer determined from RDC and SAXS/WAXS data. (A) Stereoview of a best-fit superposition (to the EIC dimerization domain which remains fixed) of the final 100 simulated annealing structures calculated with random noise added to the RDCs with the backbone (N, Cα, C′) atoms of the EIN domain in blue, the EIC domain displayed as a red ribbon, and the position of the EIN domain in wild-type EI9 displayed as a gold ribbon. (B) Ribbon diagram of a single subunit of EI(H189Q) with the EIN and EIC domains in red and blue, respectively, a molecular surface shown in grey in the left panel and the position of the EIN domain in wild-type EI in gold in the right panel. The y axis of the RDC alignment tensor coincides with the C2 symmetry axis of the dimer. (C) Stereoview of the side chain interactions at the EIN/EIC interface in the EI(H189Q) mutant with reweighted atomic probability maps20 for the side chains of EIN and EIC shown in blue and red, respectively (plotted at a threshold of 15% of maximum and calculated from the 100 final simulated annealing structures).
The origin for the change in orientation of the EIN domain relative to the EIC domain can be found at the interface of the EINα and EINα/β subdomains (Fig. 3). There is a rotation of ~9° and a Cα rms displacement of ~3.5 Å of the EINα subdomain with respect to the EINα/β subdomain relative to the wild-type structure (Figs. 3A–C). This can be attributed to the different hydrogen bonding interactions of His189 versus Gln189. Although the side chains of histidine and glutamine are isosteric and both can accept and donate a hydrogen bond, the conformational space available to the linear Gln side chain is not identical to that of the histidine imadozole ring. In wild-type unphosphorylated EI, the Nε atom of His189 accepts a hydrogen bond from the hydroxyl group of Thr168.4 The Nδ atom of His189, which is protonated,4 however, is not appropriately positioned to donate a hydrogen bond to any suitable acceptor group. In the case of Gln189, the carbonyl oxygen of the carboxyamide group is analogous to the Nε2 atom of His and accepts a hydrogen bond from the hydroxyl group of Thr168. However, the side chain amide of Gln189 can now come into close proximity to the carboxylate of Asp129 located on the EINα subdomain with only a small rigid body reorientation of the EINα subdomain. This is readily accomplished by minimal backbone torsion angle changes in the EINα/EINα/β linker regions.
In conclusion we have shown that RDC and SAXS/WAXS data combined with conjoined rigid body/torsion angle/Cartesian simulated annealing refinement techniques can be used to rapidly and accurately probe multiple, relatively small rigid body conformational changes in large multidomain proteins. In the case of EI, the orientations of the EIN subdomains relative to each other and of the EIN domain relative to the EIC dimerization domain vary during the course of the catalytic cycle and these conformational transitions are crucial to function.8,9 The rigid body domain and subdomain conformational transitions observed for the EI(H189Q) mutant demonstrate how easily small changes in side chain interactions can be propagated to much larger domain reorientations and illustrate the positional lability of the EIN domain and its subdomains.
Supplementary Material
Acknowledgments
This work was supported by the intramural program of NIDDK (G.M.C.) and CIT (C.D.S.) and the AIDS Targeted Antiviral Program of the Office of the Director of the NIH (G.M.C.). Use of the shared scattering beamline resource allocated under the PUP-77 agreement between NCI, NIH and the Argonne National Laboratory, is acknowledged.
Footnotes
Supporting Information: Experimental details, TROSY spectrum of EI(H189Q), RDC analysis for an individual EIN domain subunit, and SAXS/WAXS curves for wild type and E(H189Q) EI. This material is available free of charge on the internet at http://pubs.acs.org.
References
- 1.Deutscher J, Francke C, Postma PW. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LiCalsi C, Crocenzi TS, Freire E, Roseman S. J Biol Chem. 1991;266:19519–19527. [PubMed] [Google Scholar]
- 3.Liao DI, Silverton E, Seok YJ, Lee BR, Peterkofsky A, Davies DR. Structure. 1996:861–872. doi: 10.1016/s0969-2126(96)00092-5. [DOI] [PubMed] [Google Scholar]
- 4.Garrett DS, Seok YJ, Liao DI, Peterkofsky A, Gronenborn AM, Clore GM. Biochemistry. 1997;36:2517–2530. doi: 10.1021/bi962924y. [DOI] [PubMed] [Google Scholar]
- 5.Garrett DS, Seok YJ, Peterkofsky A, Clore GM, Gronenborn AM. Biochemistry. 1997;36:4393–4398. doi: 10.1021/bi970221q. [DOI] [PubMed] [Google Scholar]
- 6.Garrett DS, Seok YJ, Peterkofsky A, Gronenborn AM, Clore GM. Nature Struct Biol. 1999;6:166–173. doi: 10.1038/5854. [DOI] [PubMed] [Google Scholar]
- 7.Patel HV, Vyas KA, Savtchenko R, Roseman S. J Biol Chem. 2006;281:17570–17578. doi: 10.1074/jbc.M508965200. [DOI] [PubMed] [Google Scholar]
- 8.Teplyakov A, Lim K, Zhu PP, Kapadia G, Chen CC, Schwartz J, Howard A, Reddy PT, Peterkofsky A, Herzberg O. Proc Natl Acad Sci U S A. 2006;103:16218–16223. doi: 10.1073/pnas.0607587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwieters CD, Suh JY, Grishaev A, Ghirlando R, Takayama Y, Clore GM. J Am Chem Soc. 2010;132:13026–13045. doi: 10.1021/ja105485b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquez J, Reinelt S, Koch B, Engelmann R, Hengstenberg W, Scheffzek K. J Biol Chem. 2006;281:32508–32515. doi: 10.1074/jbc.M513721200. [DOI] [PubMed] [Google Scholar]
- 11.Oberholzer AE, Schneider P, Siebold C, Baumann U, Erni B. J Biol Chem. 2009;284:33169–33176. doi: 10.1074/jbc.M109.057612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberholzer AE, Bumann M, Schneider P, Bachler C, Siebold C, Baumann U, Erni B. J Mol Biol. 2005;346:521–532. doi: 10.1016/j.jmb.2004.11.077. [DOI] [PubMed] [Google Scholar]
- 13.Bax A, Kontaxis G, Tjandra N. Methods Enzymol. 2001;339:127–174. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 14.Svergun DI, Koch MH. Curr Opin Struct Biol. 2002;12:654–660. doi: 10.1016/s0959-440x(02)00363-9. [DOI] [PubMed] [Google Scholar]
- 15.Fitzkee NC, Bax A. J Biomol NMR. 2010;48:65–70. doi: 10.1007/s10858-010-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Clore GM, Starich MA, Gronenborn AM. J Am Chem Soc. 1998;120:10571–10572. [Google Scholar]; (b) Hansen MR, Mueller L, Pardi A. Nature Struct Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 17.Clore GM, Garrett DS. J Am Chem Soc. 1999;121:9008–9012. [Google Scholar]
- 18.Schwieters CD, Clore GM. J Magn Reson. 2001;152:288–302. doi: 10.1006/jmre.2001.2413. [DOI] [PubMed] [Google Scholar]
- 19.Schwieters CD, Kuszewski J, Clore GM. Progr Nucl Magn Reson Spectr. 2006;48:47–62. [Google Scholar]
- 20.(a) Schwieters CD, Clore GM. J Biomol NMR. 2002;23:221–225. doi: 10.1023/a:1019875223132. [DOI] [PubMed] [Google Scholar]; (b) Delano WL, Brünger AT. Proteins. 1994;20:105–123. doi: 10.1002/prot.340200202. [DOI] [PubMed] [Google Scholar]
- 21.Clore GM, Kuszewski J. J Am Chem Soc. 2002;124:2866–2867. doi: 10.1021/ja017712p. [DOI] [PubMed] [Google Scholar]
- 22.Schwieters CD, Clore GM. J Phys Chem B. 2008;112:6070–6073. doi: 10.1021/jp076244o. [DOI] [PubMed] [Google Scholar]
- 23.Garcia De La Torre J, Huertas ML, Carrasco B. Biophys J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zweckstetter M, Bax A. J Biomol NMR. 2002;23:127–137. doi: 10.1023/a:1016316415261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.