Abstract
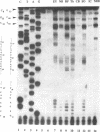
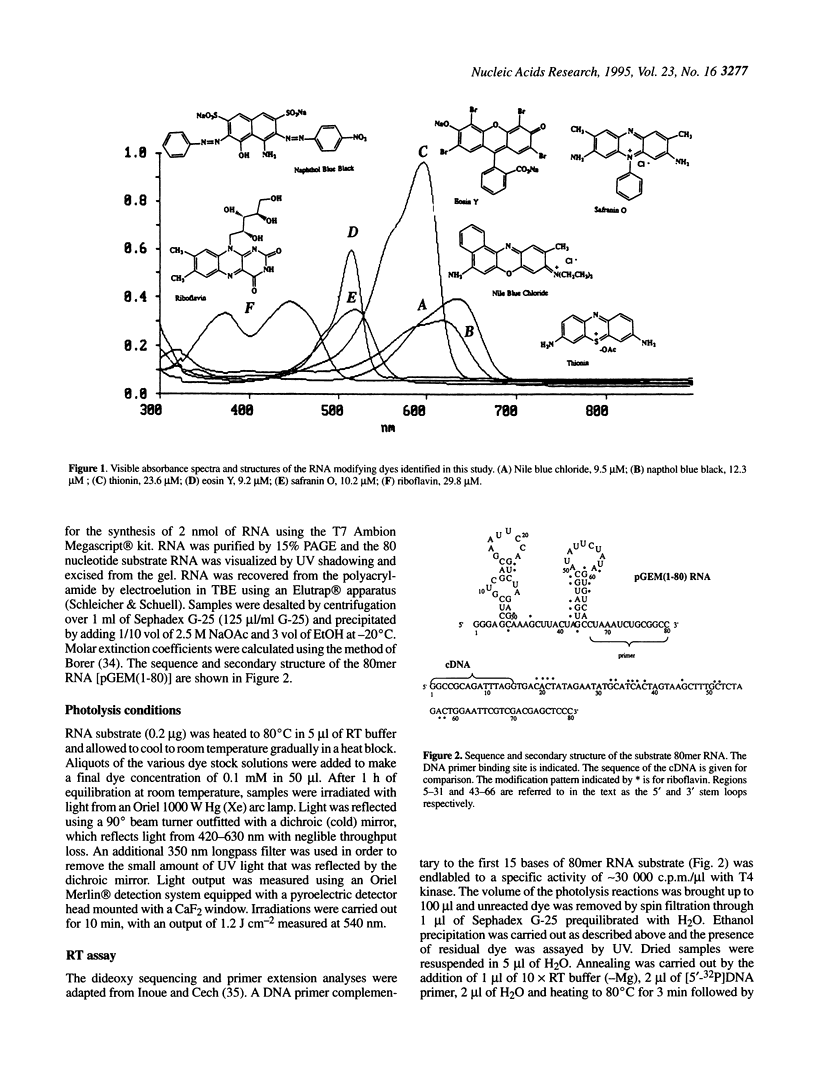
Oxidative base damage in DNA and metabolic defects in the recognition and removal of such damage play important roles in mutagenesis and human disease. The extent to which cellular RNA is a substrate for oxidative damage and the possible biological consequences of RNA base oxidation, however, remain largely unexplored. Since oxidatively modified RNA may contribute to the high mutability of retroviral genomic DNA, we have been interested in developing methods for the sequence specific detection of such damage. We show here that a primer extension assay using AMV reverse transcriptase (RT) can be used to reveal oxidatively damaged sites in RNA. This finding extends the currently known range of RNA modifications detectable with AMV reverse transcriptase. Analogous assays using DNA polymerases to detect base damage in DNA substrates appear to be restricted to lesions at thymine. Oxidative base damage in the absence of any detectable chain breaks was produced by dye photosensitization of RNA. Six out of 20 dyes examined were capable of producing RT detectable lesions. RT stops were seen predominantly at purines, although many pyrimidine sites were also detected. Dye specific photofootprints revealed by RT analysis suggests differential dye binding to the RNA substrate. Some of the photoreactive dyes described here may have potential utility in RNA structural analysis, particularly in the identification of stem-loop regions in complex RNAs.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barta A., Steiner G., Brosius J., Noller H. F., Kuechler E. Identification of a site on 23S ribosomal RNA located at the peptidyl transferase center. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3607–3611. doi: 10.1073/pnas.81.12.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M. M., Sternberg D. W., Parada L. F., Chao M. V. K-252a inhibits nerve growth factor-induced trk proto-oncogene tyrosine phosphorylation and kinase activity. J Biol Chem. 1992 Jan 5;267(1):13–16. [PubMed] [Google Scholar]
- Breimer L. H., Lindahl T. DNA glycosylase activities for thymine residues damaged by ring saturation, fragmentation, or ring contraction are functions of endonuclease III in Escherichia coli. J Biol Chem. 1984 May 10;259(9):5543–5548. [PubMed] [Google Scholar]
- Buchko G. W., Cadet J., Berger M., Ravanat J. L. Photooxidation of d(TpG) by phthalocyanines and riboflavin. Isolation and characterization of dinucleoside monophosphates containing the 4R* and 4S* diastereoisomers of 4,8-dihydro-4-hydroxy-8-oxo-2'-deoxy-guanosine. Nucleic Acids Res. 1992 Sep 25;20(18):4847–4851. doi: 10.1093/nar/20.18.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J., Berger M., Decarroz C., Wagner J. R., van Lier J. E., Ginot Y. M., Vigny P. Photosensitized reactions of nucleic acids. Biochimie. 1986 Jun;68(6):813–834. doi: 10.1016/s0300-9084(86)80097-9. [DOI] [PubMed] [Google Scholar]
- Cheng K. C., Cahill D. S., Kasai H., Nishimura S., Loeb L. A. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G----T and A----C substitutions. J Biol Chem. 1992 Jan 5;267(1):166–172. [PubMed] [Google Scholar]
- Clark J. M., Beardsley G. P. Template length, sequence context, and 3'-5' exonuclease activity modulate replicative bypass of thymine glycol lesions in vitro. Biochemistry. 1989 Jan 24;28(2):775–779. doi: 10.1021/bi00428a054. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epe B., Hegler J. Oxidative DNA damage: endonuclease fingerprinting. Methods Enzymol. 1994;234:122–131. doi: 10.1016/0076-6879(94)34083-8. [DOI] [PubMed] [Google Scholar]
- Floyd R. A., West M. S., Eneff K. L., Schneider J. E. Methylene blue plus light mediates 8-hydroxyguanine formation in DNA. Arch Biochem Biophys. 1989 Aug 15;273(1):106–111. doi: 10.1016/0003-9861(89)90167-7. [DOI] [PubMed] [Google Scholar]
- Fraga C. G., Shigenaga M. K., Park J. W., Degan P., Ames B. N. Oxidative damage to DNA during aging: 8-hydroxy-2'-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes R. C., LeClerc J. E. Sequence dependence for bypass of thymine glycols in DNA by DNA polymerase I. Nucleic Acids Res. 1986 Jan 24;14(2):1045–1061. doi: 10.1093/nar/14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchakdjian M., Bodepudi V., Shibutani S., Eisenberg M., Johnson F., Grollman A. P., Patel D. J. NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn).dA(anti) alignment at lesion site. Biochemistry. 1991 Feb 5;30(5):1403–1412. doi: 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy G., Polte T., Rooney T., Hogan M. E. A photochemical method to map ethidium bromide binding sites on DNA: application to a bent DNA fragment. Biochemistry. 1990 Jan 30;29(4):981–988. doi: 10.1021/bi00456a021. [DOI] [PubMed] [Google Scholar]
- LEPECQ J. B., YOT P., PAOLETTI C. INTERACTION DU BROMHYDRATE D''ETHIDIUM (BET) AVEC LES ACIDES NUCL'EIQUES (A. N.). ETUDE SPECTROFLUORIM'ETRIQUE. C R Hebd Seances Acad Sci. 1964 Sep 7;259:1786–1789. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Moore P., Strauss B. S. Sites of inhibition of in vitro DNA synthesis in carcinogen- and UV-treated phi X174 DNA. Nature. 1979 Apr 12;278(5705):664–666. doi: 10.1038/278664a0. [DOI] [PubMed] [Google Scholar]
- Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C-->T.A transversions in simian kidney cells. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller E., Boiteux S., Cunningham R. P., Epe B. Enzymatic recognition of DNA modifications induced by singlet oxygen and photosensitizers. Nucleic Acids Res. 1990 Oct 25;18(20):5969–5973. doi: 10.1093/nar/18.20.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W., Crothers D. M. Interactions of heteroaromatic compounds with nucleic acids. 1. The influence of heteroatoms and polarizability on the base specificity of intercalating ligands. Eur J Biochem. 1975 May;54(1):267–277. doi: 10.1111/j.1432-1033.1975.tb04137.x. [DOI] [PubMed] [Google Scholar]
- Müller W., Crothers D. M. Interactions of heteroaromatic compounds with nucleic acids. 1. The influence of heteroatoms and polarizability on the base specificity of intercalating ligands. Eur J Biochem. 1975 May;54(1):267–277. doi: 10.1111/j.1432-1033.1975.tb04137.x. [DOI] [PubMed] [Google Scholar]
- Müller W., Crothers D. M., Waring M. J. A non-intercalating proflavine derivative. Eur J Biochem. 1973 Nov 1;39(1):223–234. doi: 10.1111/j.1432-1033.1973.tb03120.x. [DOI] [PubMed] [Google Scholar]
- Pavlov Y. I., Minnick D. T., Izuta S., Kunkel T. A. DNA replication fidelity with 8-oxodeoxyguanosine triphosphate. Biochemistry. 1994 Apr 19;33(15):4695–4701. doi: 10.1021/bi00181a029. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette J. G., Hearst J. E. Termination sites of the in vitro nick-translation reaction on DNA that had photoreacted with psoralen. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5540–5544. doi: 10.1073/pnas.80.18.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A. L., Lippard S. J. Sequence-dependent termination of in vitro DNA synthesis by cis- and trans-diamminedichloroplatinum (II). Proc Natl Acad Sci U S A. 1985 Jul;82(14):4616–4619. doi: 10.1073/pnas.82.14.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P., Essigmann J. M. Possible role for thymine glycol in the selective inhibition of DNA synthesis on oxidized DNA templates. Cancer Res. 1985 Dec;45(12 Pt 1):6113–6118. [PubMed] [Google Scholar]
- Rubin C. M., Schmid C. W. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980 Oct 24;8(20):4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. E., Jr, Phillips J. R., Pye Q., Maidt M. L., Price S., Floyd R. A. Methylene blue and rose bengal photoinactivation of RNA bacteriophages: comparative studies of 8-oxoguanine formation in isolated RNA. Arch Biochem Biophys. 1993 Feb 15;301(1):91–97. doi: 10.1006/abbi.1993.1119. [DOI] [PubMed] [Google Scholar]
- Schneider J. E., Price S., Maidt L., Gutteridge J. M., Floyd R. A. Methylene blue plus light mediates 8-hydroxy 2'-deoxyguanosine formation in DNA preferentially over strand breakage. Nucleic Acids Res. 1990 Feb 11;18(3):631–635. doi: 10.1093/nar/18.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman L. H. Structure and function of Escherichia coli formylmethionine transfer RNA: loss of methionine acceptor activity by modification of a specific guanosine residue in the acceptor stem of formylmethionine transfer RNA from Escherichia coli. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3594–3597. doi: 10.1073/pnas.69.12.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani S., Takeshita M., Grollman A. P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991 Jan 31;349(6308):431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- Singer B., Fraenkel-Conrat H. Dye-catalyzed photoinactivation of tobacco mosaic virus ribonucleic acid. Biochemistry. 1966 Jul;5(7):2446–2450. doi: 10.1021/bi00871a039. [DOI] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Swanstrom R., Hallick L. M., Jackson J., Hearst J. E., Bishop J. M. Interaction of psoralen derivatives with the RNA genome of Rous sarcoma virus. Virology. 1981 Sep;113(2):613–622. doi: 10.1016/0042-6822(81)90189-6. [DOI] [PubMed] [Google Scholar]
- Swanstrom R., Hallick L. M., Jackson J., Hearst J. E., Bishop J. M. Interaction of psoralen derivatives with the RNA genome of Rous sarcoma virus. Virology. 1981 Sep;113(2):613–622. doi: 10.1016/0042-6822(81)90189-6. [DOI] [PubMed] [Google Scholar]
- Tanious F. A., Veal J. M., Buczak H., Ratmeyer L. S., Wilson W. D. DAPI (4',6-diamidino-2-phenylindole) binds differently to DNA and RNA: minor-groove binding at AT sites and intercalation at AU sites. Biochemistry. 1992 Mar 31;31(12):3103–3112. doi: 10.1021/bi00127a010. [DOI] [PubMed] [Google Scholar]
- Vuister G. W., Kim S. J., Wu C., Bax A. NMR evidence for similarities between the DNA-binding regions of Drosophila melanogaster heat shock factor and the helix-turn-helix and HNF-3/forkhead families of transcription factors. Biochemistry. 1994 Jan 11;33(1):10–16. doi: 10.1021/bi00167a002. [DOI] [PubMed] [Google Scholar]
- White S. A., Draper D. E. Single base bulges in small RNA hairpins enhance ethidium binding and promote an allosteric transition. Nucleic Acids Res. 1987 May 26;15(10):4049–4064. doi: 10.1093/nar/15.10.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W. D., Ratmeyer L., Zhao M., Strekowski L., Boykin D. The search for structure-specific nucleic acid-interactive drugs: effects of compound structure on RNA versus DNA interaction strength. Biochemistry. 1993 Apr 20;32(15):4098–4104. doi: 10.1021/bi00066a035. [DOI] [PubMed] [Google Scholar]
- Wood M. L., Dizdaroglu M., Gajewski E., Essigmann J. M. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990 Jul 31;29(30):7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- Youvan D. C., Hearst J. E. A sequence from Drosophila melanogaster 18S rRNA bearing the conserved hypermodified nucleoside am psi: analysis by reverse transcription and high-performance liquid chromatography. Nucleic Acids Res. 1981 Apr 10;9(7):1723–1741. doi: 10.1093/nar/9.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youvan D. C., Hearst J. E. Reverse transcriptase pauses at N2-methylguanine during in vitro transcription of Escherichia coli 16S ribosomal RNA. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3751–3754. doi: 10.1073/pnas.76.8.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]