Abstract
Influenza virus is reliant on numerous host cell functions during its replication cycle. RNA interference technology, applied on a genome-wide level, has identified human host factors that are necessary for efficient virus replication and provides new insight into how influenza virus interacts with its host at the molecular level.
Keywords: Influenza virus, RNA interference, virus-host interaction
1. Introduction
All viruses are dependent on the host cell machinery for their replication. This is particularly true of RNA viruses, such as influenza virus, due to their small genomes and therefore limited coding capacity. Over seventy years since the first isolation of a human influenza virus, and tremendous advances in molecular biology, we now have a fairly good understanding of how each influenza virus-encoded protein functions during the replication cycle [1]. In comparison, our knowledge of how the host cell facilitates virus replication is, at best, limited. If one considers that the 10 or 11 viral proteins must associate, either directly or indirectly, with hundreds of cellular factors during the course of the virus life cycle, it becomes clear that we still have much to learn. The molecular details of how influenza virus interacts with its host cell would provide a more complete picture of the influenza virus replication cycle and perhaps allow us to address questions concerning species-specificity and pathogenicity. Moreover, knowledge of host factors that are required for influenza virus replication may be applied to drug discovery. There are currently two classes of antiviral drugs approved for influenza; the adamantanes, which target the M2 ion channel and the neuraminidase inhibitors [2]. However there are increasing levels of resistance to both of these drug classes and new anti-influenza drugs are being actively sought. An alternative strategy to the traditional virus-directed drug is to instead target a cellular protein that is required by the virus. The advantage of these drugs would be reduced chances of drug-resistance arising, and since viruses may share dependence on certain cellular pathways, there is the possibility of achieving broad-spectrum antiviral activity. The caveat is obviously the need to ensure that inhibition of the cellular protein is not detrimental to the host. With an acute disease such as influenza, where drug administration is likely to be short-term, there is perhaps more flexibility with this approach than there is with chronic viral infections.
The lack of data on how influenza virus interacts with its host at the molecular level is partly due to us only having recent access to the full human genome sequence. With this information in hand, more comprehensive analyses can now be envisioned, and improved genome annotation will further advance these efforts. Another major development in recent years is that of RNA interference (RNAi) technology. This allows one to downregulate the mRNAs of specific genes and to therefore query the participation of the encoded protein in a functional assay. By using algorithms to design small interfering RNAs (siRNAs) that target each gene in the human genome, it is possible to interrogate the entire genome in so-called genome-wide RNAi screens. This approach has recently been applied to influenza virus, with the goal of identifying cellular proteins that are required for influenza virus replication. In this review we will describe and discuss the outcomes of the five studies that performed RNAi screens for influenza virus [3-7].
2. Approaches to identifying virus-host interactions in the pre-RNAi era
2.1. Influenza virus-host protein interactions
Before RNAi, a number of other approaches were used to identify host proteins and functional pathways that are involved during the influenza virus replication cycle. The most widely-used techniques are those that uncover novel protein-protein interactions between a viral protein and its cellular binding partners. The yeast-two-hybrid (Y2H) assay has been utilized extensively in this regard and some of the best-characterized influenza-host interactions, such as that between NP and karyopherin-alpha proteins [8] and NS1 and CPSF30 [9], were first revealed via this approach. The discovery aspect of the Y2H assay was sometimes hindered by the lack of an annotated human genome and advances were hampered if the function or even the identity of an interacting protein was unknown. Nowadays this is becoming less of a concern and Y2H was recently employed by Shapira et al. [6] to identify interacting partners of the 10 major proteins of influenza A/PR/8/34 (H1N1) virus. They identified 135 interactions between the 10 viral proteins and 87 human proteins and the same procedure for influenza A/Udorn/(H3N2) virus revealed 81 interactions with 66 human proteins, with 56 of these partners shared by the two virus strains. There appears to be a high degree of connectivity in this protein interaction network, with 24 of the human proteins interacting with at least two of the PR8 virus proteins and 51 interactions occurring between the 87 human proteins. Thus, this systematic approach reveals more complexity than would otherwise be appreciated if one were studying only one viral protein. However, the Y2H system only identifies binary interactions and does not allow one to detect interactions in the context of virus infected cells or specific viral protein complexes.
Another common approach is to isolate protein complexes from cells via various affinity purification techniques using tagged viral protein constructs. The identity of the isolated proteins is determined by mass spectrometry and database searching which matches the sequence of the peptides to the protein sequence in the database. This proteomics approach was used to identify 41 human proteins that interact with the influenza viral ribonucleoprotein (vRNP) complex which is composed of NP, PB1, PB2, and PA proteins [10]. In this case the NP was fused to a STREP tag and the proteins were associated with a virus-like RNA to recapitulate the true vRNP. Co-transfection of these components into HEK 293T cells reconstitutes a functional vRNP complex and therefore associated human proteins could be involved with viral RNA synthesis in either a proviral or antiviral manner. A similar strategy was used for the polymerase complex alone, where either the PA [10] or PB1 [11] proteins were fused to the TAP tag and co-expressed with wild-type versions of the other two viral polymerase proteins. These studies identified 3 and 10 interacting human proteins, respectively. While none of these were common to both studies, a number were confirmed by western blot with specific antibodies or were shown to co-localize with the viral replication machinery in influenza virus infected cells [11]. Two of these factors, HSP90 and RanBP5/importin beta 3, have been shown to play a functional role in polymerase assembly and nuclear transport. RanBP5 interacts with PB1 alone or in complex with PA and is required for importing the PB1-PA dimer into the nucleus [12]. HSP90 is a known PB2 and PB1-interacting host protein and is thought to act as a chaperone for assembly of the trimeric viral polymerase complex [13, 14]. It has been shown that RNAi-mediated knockdown of RanBP5 results in decreased viral replication and also that HSP90 inhibitors can inhibit influenza virus growth [12, 15]. This supports the concept that identification of interacting host proteins which function to facilitate virus replication, can lead to novel antiviral strategies.
While successful at identifying novel interactions, the described proteomics approaches are still somewhat artificial and ideally one would want to incorporate the affinity tag into the viral genome such that the tagged protein is expressed in the natural context of the virus life-cycle. This would also allow one to isolate interacting partners at different stages of infection which would assist in identifying transient protein interactions and help to determine their functional role. The segmented genome of influenza virus has made this approach challenging, as fusion of any ORF with an N- or C-terminal tag will disrupt the packaging signals that are required for genome incorporation into the budding virion [16]. Work to define these signals has paved the way to overcoming this problem by duplicating the necessary sequence on the termini of the vRNA segments. Using the plasmid-based reverse genetics system [17, 18], replication competent influenza viruses that express STREP-tagged PB2 proteins have been generated [19], and provide a useful tool for analyzing the nature of interactions with the viral polymerase at different stages of the influenza virus life-cycle. Another recombinant influenza virus that could be used for proteomic purposes is one which expresses an NS1-GFP fusion [20], where the NS1 and NEP ORFs are separated by a protease cleavage site. Similar strategies applied to all influenza viral proteins, and use of sophisticated proteomics techniques would greatly enhance our ability to detect host interacting proteins and to map their functional role in virus replication. One additional proteomics approach that has been used is that of identifying the host proteins that are incorporated into influenza virions [21]. In this case, highly purified virus preparations were subjected to analysis by mass spectrometry and 36 host proteins were identified along with the known viral components of the virion. The presence of these cellular components, either in the core of the virus particle or in the lipid envelope, was confirmed with specific antibodies. While it is unclear if these are all specifically packaged, one assumes that any interactions between the host and viral components will control the incorporation of the host protein into the virion and may imply a role for that factor in the virus life-cycle.
2.2. Host transcriptional responses to influenza virus infection
Differences in host transcription in response to influenza virus can also be used to identify cellular signaling pathways that are activated or inhibited by the virus. DNA microarrays that cover the entire cellular transcriptome can provide a global picture of this response in an unbiased manner. Geiss et al. [22] performed a comparison of live influenza virus infection versus inactivated virus and were thereby able to identify the cellular responses that are replication-dependent and those that are replication-independent. The former includes genes involved in protein synthesis, transcriptional regulation, mRNA processing and transport, cytokine and growth factor signaling, and the ubiquitin pathway. Microarray analyses have also been used to examine host responses to influenza virus infection in animal models, which can identify cellular pathways that may be associated with pathogenicity and can provide important information on the kinetics of the host response to infection and how this corresponds with the disease. Such studies have been performed in mice [23-25], ferrets [26, 27], chickens [28], and also non-human primates [29-31]. A particular strength of microarrays is that pairwise comparisons of different viruses can reveal functional characteristics of these viruses that can be related to differences in the viral genomes. This has been used to determine gene signatures that distinguish pathogenic from non-pathogenic influenza viruses and also to reveal functional roles for viral proteins or consequences of specific mutations [25, 32, 33].
2.3. Alternative approaches with a direct functional readout
While protein interaction and microarray data reveal important information regarding how the host cellular network interacts with the virus, neither assay is able to directly identify host functions that are required by the virus. Some necessary functions have been uncovered through the use of specific inhibitors (e.g. PI3 kinase [34], MAP kinases [35], protein kinase C [36]) but a wider screen with a functional readout of virus replication is required to properly address this question. In addition to RNAi (which will be covered in the next section), other approaches have been used to identify host factors that facilitate influenza virus replication. The first study showed that transfected vRNP complexes could replicate in yeast and therefore single-gene deletion libraries were screened for those in which decreased virus replication was observed [37]. The authors characterized the mammalian homologue of one hit, Tat-SF1, which they showed is required for vRNA-NP complex formation. The second study was performed in mammalian MDCK cells and used a technique known as Random Homozygous Gene Pertubation (RHGP) [38] in which integration of a vector sequence into the genome disrupts gene expression. Cells that survived challenge with influenza virus were isolated and the disrupted gene sequenced. 110 genes were identified and were enriched for those associated with nucleic acid and protein metabolism, intracellular protein trafficking and signal transduction. The requirement of four of these host factors (MRPL42, COX5A, TAPT1, SLC25A25) for optimal influenza virus replication in human cells was confirmed via siRNA knockdown.
3. Genome-wide RNAi screens applied to influenza virus for the identification of required host factors
3.1. Description of the screening approaches and results
Five RNAi screens for influenza virus have been reported to date and they are summarized in Table 1. The first genome-wide RNAi screen for host factors required for influenza virus replication was performed in Drosophila cells with a genome-wide library of dsRNAs by Hao et al. in 2008 [7]. The authors generated a modified influenza virus, which was able to infect Drosophila DL1 cells, by replacing the hemagglutinin (HA) gene with the glycoprotein of vesicular stomatitis virus (VSV-G) and the neuraminidase (NA) gene with Renilla luciferase. Using this model system they were able to capture the stages from uncoating to viral protein expression and identify cellular factors involved in these steps of the viral replication cycle. Out of 176 primary hits the authors could confirm 110 Drosophila genes with alternative dsRNAs. After cell viability assays to exclude toxic effects 104 genes remained as hits. Furthermore, for three of these genes (ATP6V0D1, COX6A1, NXF1) Hao et al. could demonstrate that the human homologues are required for efficient influenza virus replication in human cells, validating the Drosophila screen as a model system.
TABLE 1. Description of the published RNAi screens performed with influenza virus.
| RNAi screens with influenza virus | Brass et al. (2009) | Hao et al. (2008) | Karlas et al. (2010) | König et al. (2010) | Shapira et al. (2009) |
|---|---|---|---|---|---|
| Host cell | U2OS (Human) | DL1 (Drosophila) | A549 (Human) | A549 (Human) | HBEC (Human) |
| Virus | PR8 | rWSNa | WSN | rWSNb | PR8 |
| Readout | Surface HA expression | Luciferase activity | 1st cycle: NP expression 2nd cycle: luciferase activity | Luciferase activity | Luciferase activity |
| si/dsRNA source | Dharmacon | Ambion | Qiagen | Qiagen | Dharmacon |
| Length of RNAi treatment | 72hr | 48hr | 48hr | 48hr | 72hr |
| Time of assay readout | 12hr PIc | 24hr PI | 24hr PI | 12, 24, 36hr PI | 48hr PI |
| Virus stages captured | Attachment until HA surface trafficking | Uncoating until protein expression | Attachment until budding/release | Attachment until protein expression | Attachment until budding/release |
| Genes targeted | 17,877 | 13,071 | 22,843 | 19,628 | 1,745d |
| Primary gene hits | 312 | 176 | 287 | 295 | 220 |
| Validated hitse | 129 | ND | 168 | 219 | ND |
Recombinant WSN virus in which the HA gene was replaced by VSV G and the NA gene by Renilla luciferase.
Recombinant WSN virus in which the HA gene was replaced by Renilla luciferase.
Post infection.
Select genes were screened based on microarray and protein interaction data.
Confirmed with at least 2 individual siRNA per gene and with wild-type influenza virus. ND, Not determined based on criteria for validation
Since then three different groups have performed genome-wide RNAi screens for influenza virus host factors in human cells. Brass and colleagues [5] performed their genome-wide siRNA screen in human U2OS cells and infected them with influenza A/PR/8/34 virus. At 12h post infection (p.i.) they stained for viral HA on the surface and analyzed replication efficiency by microscopy-based quantification of the HA signal. In their primary screen they identified 312 pools of siRNAs that led to a decrease in influenza virus replication. By testing the different siRNAs from these pools separately, and requiring that two or more siRNAs per gene were effective, the authors confirmed 129 required host factors and four restriction factors. They analyzed one of the restriction factors, IFITM3, and its paralogs IFITM1 and IFITM2 in more detail and could show that the IFITM proteins represent a previously unknown group of influenza virus restriction factors. Karlas et al. [4] chose a different approach. They used human A549 cells, transfected them with the genome-wide library of siRNAs and infected them with influenza virus A/WSN/33. Their read-out was divided into two parts: Firstly, they stained for viral nucleoprotein (NP) at 24h p.i. and quantified NP levels by a microscopy-based approach. Secondly, they transferred the supernatants onto reporter cells bearing an influenza virus-specific luciferase reporter for quantification of released virus. They were therefore able to cover all stages of the viral replication cycle. Their primary screen, using pools of siRNAs, resulted in 287 primary hits. 168 hits were confirmed in subsequent analysis by two or more individual siRNAs per gene, using the two different influenza virus strains A/WSN/33 and a pandemic H1N1 virus A/Hamburg/04/2009. A subset of siRNAs were also tested for their impact on the replication of the highly pathogenic H5N1 virus A/Vietnam/1203/2004 and they were found to reduce H5N1 replication by two orders of magnitude. These data suggest that influenza A viruses of different subtypes share a dependency on at least certain host factors. Furthermore, the authors provided additional functional evidence for the requirement of two of their hits; the cell cycle regulator p27, and CLK1, a kinase that regulates splicing by phosphorylating the splicing factor SF2/ASF [39, 40]. They show that influenza virus replication is reduced significantly in p27-/- mice and that the CLK1 inhibitor TG003 could prevent virus replication. The third genome-wide RNAi screen in human cells by König et al. [3] employed yet another approach. This group used human A549 cells like Karlas et al. [4] but the virus used in the screen and the read-out of the assay were different. They generated a reporter virus encoding Renilla luciferase in place of the HA ORF. This allowed for direct quantification of replication efficiency in the infected cells by measuring luciferase activity at different time points post infection. Since the recombinant virus does not encode HA, late stages of the replication cycle, like budding, were not covered by this assay. König et al. identified 295 factors that were required for efficient replication of the reporter virus. Of these 295 factors, 219 were confirmed in an assay using the wildtype influenza virus A/WSN/33. A subset of these hits were further studied in different functional assays and classified as entry or post entry factors. For a number of genes König and colleagues provide data showing that compound inhibitors of the identified cellular factors can result in inhibition of influenza virus replication. As an example KN-93, an inhibitor of the calcium/calmodulin-dependent kinase CAMK2B, inhibited virus growth by more than two orders of magnitude. Together with the results on the CLK1 inhibitor from Karlas et al. [4], these findings prove that it is possible to find anti-influenza virus compounds that target host cell proteins.
Shapira and colleagues [6] chose an integrative genomics approach to shed light on the interactions of influenza virus with its host cell. First, they performed a systematic yeast two-hybrid screen to identify interactors of each of the ten major viral proteins. Second, they analyzed the transcriptional responses of human bronchial epithelial cells (HBECs) to influenza virus infection, transfection of viral RNA, infection with an influenza virus lacking the NS1 gene (ΔNS1) or treatment with interferon beta (IFNβ). Integration of these data sets resulted in a list of 1745 candidate genes comprised of 1056 genes that are transcriptionally regulated by influenza virus infection, 259 direct interactors of influenza virus proteins and their first neighbors, as well as 504 predicted candidates from pathways that were overrepresented in the interaction and transcription data sets. Using siRNA pools to mediate knockdown, the 1745 candidate genes were then tested for their impact on influenza virus replication and IFN production upon ΔNS1 infection or viral RNA transfection. Among the 616 genes that affected virus replication and/or IFN production, 220 were required for efficient virus replication. It should be noted that these 220 factors have not been confirmed with individual siRNAs.
3.2. Comparison of data from all influenza virus RNAi screens
At first glance there is surprisingly little overlap of identified factors between the different screens. In fact, only one gene (COPA) is present in the hitlist of all four screens performed in human cells. When looking at the four genome-wide screens, which comprise three screens in human cells and one in Drosophila cells, three genes are shared: ARCN1, ATP6AP1 and COPG. This low number of overlapping genes is similar to what has been observed for the different RNAi screens for HIV [41-44]. Most likely, differences in the assays such as different cell lines, viruses, siRNA libraries, and readout techniques account for this. However, these screens can provide us with important information on which pathways and cellular functions are crucial for the virus. Furthermore, we now have a list of genes that should be studied in more detail to better understand the virus-host interplay. It is important to note that each group applied different criteria for their hit selection and therefore comparisons of hitlists are generally biased. In order to identify the most promising candidate genes we have listed the primary hits separately from the validated hits for each screen (Supp. Table 1, tabs 1, 2). We considered a gene validated if at least two individual siRNAs resulted in a significant reduction of wild-type influenza virus replication according to the criteria of each screen. Since Shapira et al. [6] did not further validate their pools of siRNAs and Hao et al. [7] only validated three hits with a wild-type influenza virus, we listed their hits under primary hits but did not include them in our analysis of validated hits. The criteria for validated hits were fulfilled by 129 factors from the study by Brass and colleagues [5], 168 from Karlas et al. [4] and 219 from König and colleagues [3]. In total, 478 genes have been validated and 34 of them were confirmed by two or all three studies (Supp. Table 1, tab 4; Fig. 1). When the primary hits of all five screens are compared 86 out of 1182 host cell factors are shared between at least two screens (Supp. Table 1, tab 3). When looking at the 34 genes, two groups of host factors clearly stand out: the genes encoding the vacuolar ATPase subunits (ATP6V1A, ATP6V1B2, ATP6V0C, ATP6V0B, ATP6AP1, ATP6V0D1, ATP6AP2) and the COPI proteins (ARCN1, COPA, COPB1, COPB2, COPG). The strong requirement of the virus for the endosomal vATPase did not come as a surprise and had been described previously [45, 46]. The vATPase is required for the acidification of the endosome and influenza virus relies on a low endosomal pH for successful entry into the host cell. In contrast, it was an unexpected finding that the virus relies heavily on the COPI proteins. The described function of COPI proteins is to form a coat on vesicles that mediate retrograde transport from Golgi cisternae to the endoplasmic reticulum (ER) [47]. It has also been described that COPI proteins localize to endosomal membranes and might be involved in endosomal trafficking [48, 49]. However, this function is not understood in detail. Initial studies on the role of COPI in influenza virus infection by Brass et al. [5] and König et al. [3] suggest an involvement of COPI at two different stages of the replication cycle: firstly entry, and secondly trafficking of HA at a late stage of the virus [50] life cycle. However, much more work is required to unravel the role of COPI for influenza virus.
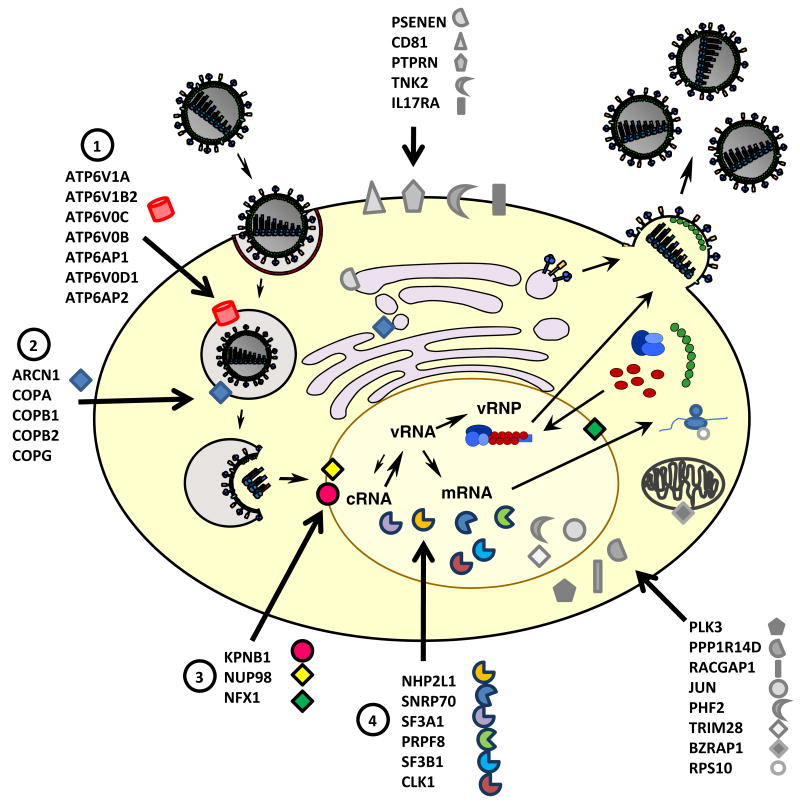
Figure 1. Graphical representation of the 34 best validated host factors identified in the different RNAi screens.
Analysis of the validated hits from the RNAi screens by Brass et al. [5], Karlas et al. [4] and König et al. [3] resulted in a list of 34 genes that were identified in at least two screens. These 34 host cell factors are shown with regards to their localization within the cell and in context of the viral replication cycle. The four different groups of host proteins with known function in the viral life-cycle are depicted in colored symbols ((1) components of the vATPase complex, (2) COPI proteins, (3) nucleo-cytoplasmic transport factors, (4) members of the splicing machinery); the host factors with as yet unknown function are illustrated with grey symbols.
A third group of host factors among the 34 genes comprises CLK1, SNRP70, SF3A1, SF3B1, PRPF8 and NHP2L1. They have been described to function in the splicing of cellular pre-mRNAs and are probably required for splicing of the viral mRNAs. Although it was known before that the virus uses the cellular splicing machinery, the specific involvement of the identified host genes was not known. Follow-up studies on these factors will further improve our understanding of the virus-host interplay at the stage of viral mRNA processing. The members of a fourth group, NUP98, KPNB1 and NXF1, function in the nuclear transport of mRNAs and proteins. The cellular function of NXF1 is to export spliced mRNAs [50] and influenza virus is known to use this transport route for the export of the viral mRNAs [51]. In addition, the viral NS1 protein has been described to interact with NXF1, which might to contribute to viral interference with the transport of cellular mRNAs [52]. Since KPNB1 and NUP98 are known to be required for the nuclear import of cellular proteins, they are probably used by the virus for the import of viral proteins or vRNPs. Previous studies have demonstrated the involvement of several karyopherin family members such as KPNA1, KPNA2, KPNA5 and KPNB3 in the nuclear import of influenza virus proteins [10, 12, 53-55]. However, KPNB1 and NUP98 have not been described in influenza virus nuclear import, so their exact role remains to be determined. It will be interesting to see if the different host factors are required for transport of different viral proteins and if NUP98 and KPNB1 interact directly with viral components. For the remaining genes on the list of validated 34 host factors, their function in the viral replication cycle is less obvious. For CD81, a member of the tetraspanin protein family, König et al. [3] mapped its requirement to the entry step and this protein is also known to be present in influenza virions [21]. König et al. [3] also tested PPP1R14D, a subunit of protein phosphatase 1, in several functional assays and found that it is a post-entry factor. At this point, we can only speculate on the functions of the remaining host factors. It could be envisioned that some of the factors like the tyrosine kinase TNK2, the cytokine receptor IL17RA or the protein tyrosine phosphatase PTPRN are engaged in signaling cascades that are important for the virus but more work is required to understand the interaction of the virus with these host factors. It should also be noted that for some of these genes only very limited information on their cellular function is available. As an example we only know that PHF2 contains a zinc finger-like PHD (plant homeodomain) finger and a hydrophobic and highly conserved domain. It has been suggested that this protein could be involved in regulation of transcription but much work needs to be done to unravel its function for influenza virus.
In summary, these 34 host factors that were validated by at least two groups, have the strongest evidence from the different screening approaches, and therefore represent promising candidates for future studies.
4. The contribution of RNAi screens to our knowledge of influenza virus dependency on the host cell
4.1. Identification of cellular pathways and complexes to explore in the context of influenza virus infection
Besides looking at the individual genes that were found to be critical for influenza virus replication it is also important to highlight the cellular functions, pathways and complexes that have been identified in the different RNAi screens. At this level the different screens show a much higher degree of overlap than at the level of individual genes. A gene ontology (GO) analysis using the DAVID database [56] on all 478 validated hits (Supp. Table 1 tab 2) showed that kinase-mediated signaling was the most overrepresented GO category, followed by splicing/processing of pre-mRNAs and intracellular transport, in particular nucleocytoplasmic transport. Furthermore, the COPI complex, as well as the vacuolar ATPases, were highly enriched amongst all validated genes. Additional cellular functions and pathways that were overrepresented comprise protein kinase C (PKC) signaling, apoptosis induction, the ubiquitin machinery as well as protein translation. It has been described before that activation of PKC signaling is crucial for influenza virus replication, specifically for virus entry [36, 57]. Using PKC inhibitors as well as a dominant-negative mutant of PKC-β, Sieczkarski et al. [36] demonstrated that influenza virus was trapped in late endosomes if PKC signaling was blocked. Furthermore, it has been shown that binding of HA to the cell can trigger PKC activation [58]. However, we do not have a detailed understanding of this signaling event and it is unknown how activation of PKC facilitates completion of the entry process. With this additional line of evidence for the importance of PKC signaling for influenza virus we should aim for a better comprehension of the interaction of the virus with the PKC pathway. Moreover, as a kinase, PKC might represent an interesting drug target candidate for novel anti-influenza compounds.
Since the GO analysis on the validated hits did not include genes that were identified in the Drosophila screen [7] or the integrative genomics study by Shapira and colleagues [6] we also performed a GO analysis on the 86 overlapping genes of the primary hits from all five RNAi screens. Whereas most of the GO categories highlighted above were also found for the 86 overlapping primary hits there were some interesting differences. Protein translation was found to be the most overrepresented GO category and translation initiation was listed fourth, emphasizing the role of translation in this analysis more than in that with the validated hits. In contrast, kinase-mediated signaling was less prominent compared to the analysis for the validated hits. Interestingly, positive regulation of metabolic processes and cell proliferation also scored much higher for the overlapping primary hits compared to the validated hits.
4.2. Limitations of the RNAi approach
One must recognize that, as with all techniques, there are limitations to the RNAi approach and that it cannot provide an all-encompassing list of required cellular factors. Additional limitations will no doubt be realized as genome-wide RNAi screens are used for more and more applications but some that we are already aware of are the following: 1) The knockdown efficiency for the targeted genes is not equivalent due to siRNAs with varying efficacies, and targets with differences in mRNA and protein stabilities. Moreover, changes in protein expression levels between cell types can account for differences observed between assays performed in different cells. 2) The screens are not able to identify host factors that are also required for cell growth and survival. 3) Some host factors have redundant functions and therefore single gene knockdown experiments are unlikely to capture these host factors. 4) Assay design will affect the composition of host factors that are identified. 5) Bioinformatic data analysis is limited by the quantity and quality of the available data i.e. protein interactions, accurate gene annotation etc. In summary, while we strive to eliminate false-positives as much as possible, there will always be a number of false-negatives associated with RNAi screens.
5. Perspectives and future directions
In the immediate future it will be important to perform a more comprehensive cross-comparison of all influenza virus RNAi screens. As noted above and by similar reviews [59-61], comparison of the published hits from each screen shows limited overlap when analyzed at the gene level. Differences in the assay conditions, and in particular the nature of the siRNA libraries, is the likely cause for the lack of agreement. However, if one analyzes the data at the level of cellular function rather than at the level of the gene name, more concordance is observed as discussed in section 4.1. A simple explanation is that the different siRNA libraries probably have variable knockdown efficiencies for targets in a particular pathway. While one screen may identify protein X as being important, another may identify protein Y. At the gene level, these screens do not agree, but if proteins X and Y are members of the same biological pathway, the screens do agree that this functional pathway is required for influenza virus replication. Another factor to consider is that while all screens start off with an unbiased approach, each introduces different layers of bias with the necessity to establish criteria by which to select hits. We have attempted to overcome this in part by noting the overlap in gene hits from primary screen data as well as validated data, but to truly address the bias the raw data from all screens should be analyzed using the same criteria. Such analysis is critical to provide the influenza field with information that they can pursue with confidence.
Further supply of data on host protein interactions and gene expression profiles will also help to filter the current list of required factors such that more weight will be given to those with more lines of evidence. Some of this evidence will come from further functional analyses of the factors identified in these RNAi screens. This includes defining the stage of the virus life-cycle that the host factor is involved in, determining if the requirement extends to other influenza virus strains (including those that are not associated with human infections), and whether the lack of this host factor in an in vivo setting (e.g. knock-out mouse) results in protection from influenza disease. More focused RNAi screens can also be envisioned, in which specific events in the viral replication cycle are assessed (e.g. entry, vRNP nuclear import). This will aid in identifying roles for the cellular factors during the virus life-cycle and allow us to define networks of virus-host interactions that have distinct functions. RNAi knockdown of combinations of host factors, particularly those known to partake in the same signaling pathway, may also be used to establish a clear requirement for these biological functions. This is useful if the single gene knockdown does not give a strong phenotype, or if redundant cellular functions exist.
While we have focused this review on those factors that facilitate influenza virus replication, RNAi screens also have the potential to identify host factors that restrict virus replication. In the assays this is observed as enhanced replication in the absence of the antiviral host factor. While most of the influenza RNAi studies made mention of such factors, only Brass et al. [5] confirmed and characterized the antiviral activity of their hits. They report that a family of interferon-inducible transmembrane proteins (IFITM-1, -2, -3) inhibits an early stage of influenza virus replication and that IFITMs can also restrict replication of Dengue virus and West Nile virus. Thus RNAi screens can reveal important information regarding the innate immune defense pathways of the cell, and in particular the functions of interferon-stimulated genes which remain largely uncharacterized. As the expression of many antiviral proteins is induced in response to virus infection (e.g. via interferon signaling), optimal detection of these factors in RNAi screens requires an assay design that is not necessarily suitable for the simultaneous detection of proviral factors. Therefore, for further expanding our knowledge of influenza virus restriction factors, RNAi screens that are specifically designed for this purpose must be employed.
All of these analyses will help to define host factors that are essential for influenza virus replication and identify those that could serve as targets for antiviral drug development. For those with enzymatic functions, it will be important to establish that the enzymatic activity is required for their proviral role. Available small molecule inhibitors can be used to test this, or mutant proteins that lack enzymatic activity and which act in a dominant negative manner could be examined for their ability to block influenza virus replication. This should expand our current repertoire of proteins that are considered as targets for influenza antiviral drugs, and hopefully re-invigorate the drug pipeline. An additional advantage of pursuing host factors as drug targets is that many viruses may share a dependence on the same host factor. For instance RNAi screens have been performed for several other RNA viruses (HIV-1, West Nile virus, Dengue virus, hepatitis C virus) and a similar comparison between all these screens can uncover common pathways and cellular factors. For example, the vATPases are also required by Dengue virus and West Nile virus [62, 63], and hepatitis C virus shares a requirement for COPI proteins [64, 65] and of course CD81 with influenza virus. This presents us with the opportunity to develop host-directed drugs with broad-spectrum antiviral activity. Such drugs would be beneficial for treating infections caused by viruses for which no drugs exist as well as those that have developed resistance against current antiviral therapies.
Supplementary Material
Acknowledgments
We would like to thank Dr. Ben Hale (Mount Sinai School of Medicine, NY) for helpful discussions on the manuscript. S.S. was supported by a fellowship from the German Research Foundation DFG. The Shaw laboratory is supported by NIH grants R21AI083673 and HHSN272200900032C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Palese P, Shaw ML. Orthomyxoviridae:The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1647–1689. [Google Scholar]
- 2.De Clercq E. Antiviral agents active against influenza A viruses. Nat Rev Drug Discov. 2006;5:1015–1025. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.König R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 5.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Neill RE, Palese P. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology. 1995;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 9.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 10.Mayer D, Molawi K, Martinez-Sobrido L, Ghanem A, Thomas S, Baginsky S, Grossmann J, Garcia-Sastre A, Schwemmle M. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J Proteome Res. 2007;6:672–682. doi: 10.1021/pr060432u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorba N, Juarez S, Torreira E, Gastaminza P, Zamarreno N, Albar JP, Ortin J. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics. 2008;8:2077–2088. doi: 10.1002/pmic.200700508. [DOI] [PubMed] [Google Scholar]
- 12.Deng T, Engelhardt OG, Thomas B, Akoulitchev AV, Brownlee GG, Fodor E. Role of ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. J Virol. 2006;80:11911–11919. doi: 10.1128/JVI.01565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Momose F, Naito T, Yano K, Sugimoto S, Morikawa Y, Nagata K. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J Biol Chem. 2002;277:45306–45314. doi: 10.1074/jbc.M206822200. [DOI] [PubMed] [Google Scholar]
- 14.Naito T, Momose F, Kawaguchi A, Nagata K. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J Virol. 2007;81:1339–1349. doi: 10.1128/JVI.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chase G, Deng T, Fodor E, Leung BW, Mayer D, Schwemmle M, Brownlee G. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology. 2008;377:431–439. doi: 10.1016/j.virol.2008.04.040. [DOI] [PubMed] [Google Scholar]
- 16.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. Genome packaging in influenza A virus. J Gen Virol. 2010;91:313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 17.Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rameix-Welti MA, Tomoiu A, Dos Santos Afonso E, van der Werf S, Naffakh N. Avian Influenza A virus polymerase association with nucleoprotein, but not polymerase assembly, is impaired in human cells during the course of infection. J Virol. 2009;83:1320–1331. doi: 10.1128/JVI.00977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci U S A. 2010;107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. Cellular proteins in influenza virus particles. PLoS Pathog. 2008;4:e1000085. doi: 10.1371/journal.ppat.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiss GK, An MC, Bumgarner RE, Hammersmark E, Cunningham D, Katze MG. Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events. J Virol. 2001;75:4321–4331. doi: 10.1128/JVI.75.9.4321-4331.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kash JC, Tumpey TM, Proll SC, Carter V, Perwitasari O, Thomas MJ, Basler CF, Palese P, Taubenberger JK, Garcia-Sastre A, Swayne DE, Katze MG. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Su YA, Hu P, Yang J, Zheng B, Wu P, Peng J, Tang Y, Zhang L. Signature patterns revealed by microarray analyses of mice infected with influenza virus A and Streptococcus pneumoniae. Microbes Infect. 2006;8:2172–2185. doi: 10.1016/j.micinf.2006.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fornek JL, Gillim-Ross L, Santos C, Carter V, Ward JM, Cheng LI, Proll S, Katze MG, Subbarao K. A single-amino-acid substitution in a polymerase protein of an H5N1 influenza virus is associated with systemic infection and impaired T-cell activation in mice. J Virol. 2009;83:11102–11115. doi: 10.1128/JVI.00994-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowe T, Leon AJ, Crevar CJ, Carter DM, Xu L, Ran L, Fang Y, Cameron CM, Cameron MJ, Banner D, Ng DC, Ran R, Weirback HK, Wiley CA, Kelvin DJ, Ross TM. Modeling host responses in ferrets during A/California/07/2009 influenza infection. Virology. 2010;401:257–265. doi: 10.1016/j.virol.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cameron CM, Cameron MJ, Bermejo-Martin JF, Ran L, Xu L, Turner PV, Ran R, Danesh A, Fang Y, Chan PK, Mytle N, Sullivan TJ, Collins TL, Johnson MG, Medina JC, Rowe T, Kelvin DJ. Gene expression analysis of host innate immune responses during Lethal H5N1 infection in ferrets. J Virol. 2008;82:11308–11317. doi: 10.1128/JVI.00691-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reemers SS, van Leenen D, Koerkamp MJ, van Haarlem D, van de Haar P, van Eden W, Vervelde L. Early host responses to avian influenza A virus are prolonged and enhanced at transcriptional level depending on maturation of the immune system. Mol Immunol. 2010;47:1675–1685. doi: 10.1016/j.molimm.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Baskin CR, Garcia-Sastre A, Tumpey TM, Bielefeldt-Ohmann H, Carter VS, Nistal-Villan E, Katze MG. Integration of clinical data, pathology, and cDNA microarrays in influenza virus-infected pigtailed macaques (Macaca nemestrina) J Virol. 2004;78:10420–10432. doi: 10.1128/JVI.78.19.10420-10432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cilloniz C, Shinya K, Peng X, Korth MJ, Proll SC, Aicher LD, Carter VS, Chang JH, Kobasa D, Feldmann F, Strong JE, Feldmann H, Kawaoka Y, Katze MG. Lethal influenza virus infection in macaques is associated with early dysregulation of inflammatory related genes. PLoS Pathog. 2009;5:e1000604. doi: 10.1371/journal.ppat.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baskin CR, Bielefeldt-Ohmann H, Tumpey TM, Sabourin PJ, Long JP, Garcia-Sastre A, Tolnay AE, Albrecht R, Pyles JA, Olson PH, Aicher LD, Rosenzweig ER, Murali-Krishna K, Clark EA, Kotur MS, Fornek JL, Proll S, Palermo RE, Sabourin CL, Katze MG. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc Natl Acad Sci U S A. 2009;106:3455–3460. doi: 10.1073/pnas.0813234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kash JC, Basler CF, Garcia-Sastre A, Carter V, Billharz R, Swayne DE, Przygodzki RM, Taubenberger JK, Katze MG, Tumpey TM. Global host immune response: pathogenesis and transcriptional profiling of type A influenza viruses expressing the hemagglutinin and neuraminidase genes from the 1918 pandemic virus. J Virol. 2004;78:9499–9511. doi: 10.1128/JVI.78.17.9499-9511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geiss GK, Salvatore M, Tumpey TM, Carter VS, Wang X, Basler CF, Taubenberger JK, Bumgarner RE, Palese P, Katze MG, Garcia-Sastre A. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc Natl Acad Sci U S A. 2002;99:10736–10741. doi: 10.1073/pnas.112338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrhardt C, Marjuki H, Wolff T, Nurnberg B, Planz O, Pleschka S, Ludwig S. Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell Microbiol. 2006;8:1336–1348. doi: 10.1111/j.1462-5822.2006.00713.x. [DOI] [PubMed] [Google Scholar]
- 35.Pleschka S, Wolff T, Ehrhardt C, Hobom G, Planz O, Rapp UR, Ludwig S. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat Cell Biol. 2001;3:301–305. doi: 10.1038/35060098. [DOI] [PubMed] [Google Scholar]
- 36.Sieczkarski SB, Brown HA, Whittaker GR. Role of protein kinase C betall in influenza virus entry via late endosomes. J Virol. 2003;77:460–469. doi: 10.1128/JVI.77.1.460-469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naito T, Kiyasu Y, Sugiyama K, Kimura A, Nakano R, Matsukage A, Nagata K. An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis. Proc Natl Acad Sci U S A. 2007;104:18235–18240. doi: 10.1073/pnas.0705856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sui B, Bamba D, Weng K, Ung H, Chang S, Van Dyke J, Goldblatt M, Duan R, Kinch MS, Li WB. The use of Random Homozygous Gene Perturbation to identify novel host-oriented targets for influenza. Virology. 2009;387:473–481. doi: 10.1016/j.virol.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bullock AN, Das S, Debreczeni JE, Rellos P, Fedorov O, Niesen FH, Guo K, Papagrigoriou E, Amos AL, Cho S, Turk BE, Ghosh G, Knapp S. Kinase domain insertions define distinct roles of CLK kinases in SR protein phosphorylation. Structure. 2009;17:352–362. doi: 10.1016/j.str.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagiwara M. Alternative splicing: a new drug target of the post-genome era. Biochim Biophys Acta. 2005;1754:324–331. doi: 10.1016/j.bbapap.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 41.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 42.König R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 44.Bushman FD, Malani N, Fernandes J, D'Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, König R, Bandyopadhyay S, Ideker T, Goff SP, Krogan NJ, Frankel AD, Young JA, Chanda SK. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guinea R, Carrasco L. Concanamycin A blocks influenza virus entry into cells under acidic conditions. FEBS Lett. 1994;349:327–330. doi: 10.1016/0014-5793(94)00695-4. [DOI] [PubMed] [Google Scholar]
- 46.Guinea R, Carrasco L. Requirement for vacuolar proton-ATPase activity during entry of influenza virus into cells. J Virol. 1995;69:2306–2312. doi: 10.1128/jvi.69.4.2306-2312.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beck R, Rawet M, Wieland FT, Cassel D. The COPI system: molecular mechanisms and function. FEBS Lett. 2009;583:2701–2709. doi: 10.1016/j.febslet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 48.Aniento F, Gu F, Parton RG, Gruenberg J. An endosomal beta COP is involved in the pH-dependent formation of transport vesicles destined for late endosomes. J Cell Biol. 1996;133:29–41. doi: 10.1083/jcb.133.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitney JA, Gomez M, Sheff D, Kreis TE, Mellman I. Cytoplasmic coat proteins involved in endosome function. Cell. 1995;83:703–713. doi: 10.1016/0092-8674(95)90183-3. [DOI] [PubMed] [Google Scholar]
- 50.Reed R, Cheng H. TREX, SR proteins and export of mRNA. Curr Opin Cell Biol. 2005;17:269–273. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 51.Read EK, Digard P. Individual influenza A virus mRNAs show differential dependence on cellular NXF1/TAP for their nuclear export. J Gen Virol. 2010;91:1290–1301. doi: 10.1099/vir.0.018564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci U S A. 2007;104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang P, Palese P, O'Neill RE. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol. 1997;71:1850–1856. doi: 10.1128/jvi.71.3.1850-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gabriel G, Herwig A, Klenk HD. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 2008;4:e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, Cusack S, Simorre JP, Hart DJ. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol. 2007;14:229–233. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- 56.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 57.Hoffmann HH, Palese P, Shaw ML. Modulation of influenza virus replication by alteration of sodium ion transport and protein kinase C activity. Antiviral Res. 2008;80:124–134. doi: 10.1016/j.antiviral.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arora DJ, Gasse N. Influenza virus hemagglutinin stimulates the protein kinase C activity of human polymorphonuclear leucocytes. Arch Virol. 1998;143:2029–2037. doi: 10.1007/s007050050439. [DOI] [PubMed] [Google Scholar]
- 59.Mehle A, Doudna JA. A host of factors regulating influenza virus replication. Viruses. 2010:566–573. doi: 10.3390/v2020566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Min JY, Subbarao K. Cellular targets for influenza drugs. Nat Biotechnol. 2010;28:239–240. doi: 10.1038/nbt0310-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe T, Watanabe S, Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.