Abstract
Synthetic nanopores have been used to study individual biomolecules in high thoroughput but their performance as sensors does not match biological ion channels. Controlling the translocation times of single-molecule analytes and their non-specific interaction with pore walls remain a challenge. Inspired by the olfactory sensilla of the insect antenna, here we show that coating nanopores with fluid bilayer lipids allows the pore diameters to be fine-tuned in sub-nanometre increments. Incorporation of mobile ligands in the lipid conferred specificity and slowed down the translocation of targeted proteins sufficiently to time-resolve translocation events of individual proteins. The lipid coatings also prevented pores from clogging, eliminated non-specific binding and enabled the translocation of amyloid-beta (Aβ) oligomers and fibrils. Through combined analysis of translocation time, volume, charge, shape and ligand affinity, different proteins were identified.
Nanopores hold tremendous promise for applications such as single-molecule binding assays1–3, portable detection of (bio)warfare agents4–6, and ultra-fast sequencing of DNA or RNA7,8. Nanopore-based experiments provide sub-molecular detail on the composition of individual molecules9 and on the formation of molecular complexes or aggregates1,10. Recording of resistive current pulses during the translocation of single molecules through electrolyte-filled nanopores makes it possible to study their size1,4,6,11–13, conformation14,15, and activity16,17 in situ3,18–23. This technique can characterize hundreds of unlabeled single molecules per second in physiological solutions and yields distributions of measured parameters from these single-molecule investigations3,9. However, several challenges should be addressed. First, there is a need for methods that can reliably fabricate synthetic nanopores on the sub-nanometre scale24 and adjust or actuate pore diameters in situ24,25. Second, better control of translocation times of single-molecule analytes are still needed to achieve complete time resolution of translocation signals and more accurate determination of the amplitude and duration of resistive pulses26–28. Third, methods to control the surface chemistry inside synthetic pores16 may reduce non-specific interactions of analytes with the pore walls1,3,29 and prevent pore clogging3. Finally, low frequency of translocation events at low analyte concentrations30 and the poor specificity of the nanopores for analytes3 need to be improved.
Nature solved most of these challenges in the design of biological nanopores23. Ion channel proteins, for instance, fold into three-dimensional structures with predetermined locations of individual atoms and precisely defined internal diameters that can be actuated by ligand binding or by changes in the environment of the pore31. Many ion channel proteins are specific towards ligands and permeants, have minimal non-specific interactions, and irreversible clogging is rare. However, instability of these proteins limits their sensing applications23.
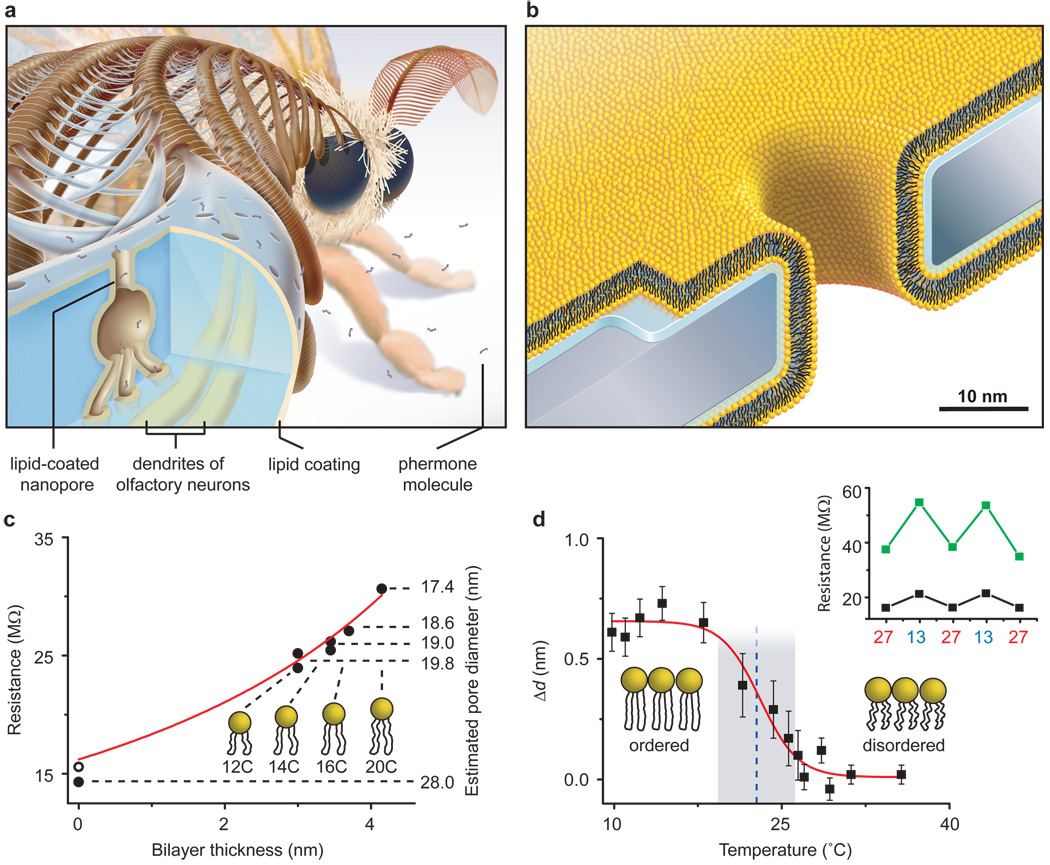
Insects detect pheromones by translocating odorant molecules through lipid-coated nanopores (diameter 6–65 nm) that span their exoskeleton (Fig. 1a)32–34. These lipid coatings are thought to participate in capture, pre-concentration, and subsequent translocation of odorants to specific receptors on dendrites of olfactory neurons in the antennae of insects32,34. Inspired by this design, we explored whether coating synthetic nanopores of comparable diameters with fluid lipid bilayers could provide benefits for nanopore-based, resistive pulse sensing of single proteins while addressing the associated challenges. Coating synthetic nanopores with organic molecules has been shown but these coatings were fixed on the surface of the pore35–37. Here we introduce the concept of fluid coatings
Figure 1. Bioinspired synthetic nanopores with bilayer-coated fluid walls.
a, Cartoon showing a cross-section through one sensillum in the antenna of the silk moth Bombyx mori. Capture, pre-concentration, and translocation of pheromones through the exoskeleton of these sensilla towards dendrites of olfactory neurons is thought to occur via lipid-coated nanopores and pore tubules32–34. b, Cartoon, drawn to scale, showing a synthetic, lipid-coated (yellow) nanopore in a silicon nitride substrate (grey) and the interstitial water layer (blue). c, Nanopore resistance and corresponding open pore diameter as a function of the thickness of the bilayer coating38. Red curve is a best fit of the data to equation (1). Numbers underneath the lipid cartoons refer to the number of carbons in their acyl chains (see Table 1). d, Actuation of nanopore diameters by a change in the thickness of the bilayer coating, Δd, in response to a thermal phase transition of DMPC lipids (see Supplementary Section S1). Blue dotted line and grey shaded region represent the mean value and range of phase transition temperatures reported for DMPC lipids39. Inset: cycling the temperature between 13° and 27° C actuated the pore diameter dynamically as indicated by the larger changes in electrical resistance through a pore with (green squares) than without (back squares) a bilayer.
Advantages of fluid coatings
To create lipid bilayer-coated nanopores (Fig. 1b), we exposed silicon chips that contained a single pore through a silicon nitride window to an aqueous suspension of small unilamellar liposomes40–43. Spreading of these liposomes on the Si3N4 window and on the walls of the nanopore (see Supplementary Sections S1 – S3) created a bilayer coating and reduced the nanopore diameter. The thickness and surface chemistry of this coating can be accurately controlled by the choice of lipids in the liposome preparation. For instance, the bilayer thickness is fine-tuned by the length and the number of double bonds in the hydrocarbon tails of the lipids (Fig. 1c), whereas the surface chemistry is controlled by the nature of their polar head groups (see Supplementary Section S4).
The capability of fine-tuning the diameter of nanopores is illustrated by the red curve in Fig. 1c. This curve resulted from a best fit of the data to a simple physical model that described the electrical resistance through the nanopore, R (Ω), as the sum of four terms: 1) the resistance of the cylindrical nanopore, 2) the access resistance to and from the nanopore31, 3) the resistance of the cylindrical channel through the silicon nitride window that led to the pore (see Supplementary Section S1 for a schematic drawing), and 4) the access resistance to this cylindrical channel. These four resistances in series are represented in sequence by the terms in equation (1) (see Supplementary Section S1 for a derivation):
| (1) |
where ρ(Ω m) represents the resistivity of the electrolyte, lP (m) the length of the cylindrical nanopore, d (m) the thickness of the lipid bilayer (see Table 1), wL (m) the thickness of the interstitial water layer between the bilayer and the silicon nitride wall of the pore44,45, rP (m) the radius of the nanopore, lC (m) the length of the cylindrical channel through the silicon nitride that led to the pore, and rC (m) the radius of this cylindrical channel (see Supplementary Section S1 for values of ρ, lP, rP, lC, and rC).
Table 1.
Lipids used in this work to coat nanopore walls.
| Chemical Name | Abbreviation | Acyl Chainsa | Bilayer Thicknessb (nm) |
|---|---|---|---|
| 1,2-dilauroyl-sn-glycero-3-phosphocholine | DLPC | (12:0) | 3.0 ± 0.1 |
| 1,2-dimyristoyl-sn-glycero-3-phosphocholine | DMPC | (14:0) | 3.4 ± 0.1 |
| 1,2-dipalmitoleoyl-sn-glycero-3-phosphocholine | DΔPPC | (16:1) | 3.6 ± 0.1 |
| 1,2-dieicosenoyl-sn-glycero-3-phosphocholine | DEPC | (20:1) | 4.2 ± 0.1 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine | POPC | (18:1–16:0) | 3.7 ± 0.1 |
| 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) | biotin-PE | (16:0) | - |
For lipids with two identical acyl chains, (c:db) indicates the number of carbons (c) and the number of double bonds (db); for lipids with two different acyl chains, (c1:db1–c2:db2) refer to acyl chains 1 and 2.
Thickness according to Lewis et al38.
Equation (1) shows that this model estimated the effective, open radius of a pore by taking into account the reduction of its radius and increase of its length as a function of the thickness of the bilayer coating and the thickness of the interstitial water layer between the bilayer and the silicon nitride wall of the pore. A fit of the data in Fig. 1c to this model returned a thickness of the water layer of wL = 1.2 ± 0.1 nm (literature values: 0.5 – 1.7 nm)44,45 as the only fitting parameter. The excellent fit of the data to equation (1) (R2 = 0.97, N = 7) and the realistic value for the thickness of the water layer, suggest that self-assembled bilayer coatings make it possible to fine-tune and predict the radius of a cylindrical nanopore in increments of two carbon atoms (albeit in a range limited to lipids that can generate stable supported lipid bilayers).
Since the sensitivity and information content of nanopore-based single-molecule experiments depend strongly on the size of the pore, one particularly desirable feature for nanopore sensing would be the ability to adjust the diameter of a nanopore dynamically to the size of various analytes, in situ. Figure 1d demonstrates that a thermal phase transition of a coating of DMPC lipids (Table 1) from the ordered gel phase (Lβ) to the disordered liquid crystalline phase (Lα) decreased the estimated thickness of the bilayer coating by Δd ≈ 0.7 nm (lit.: 0.9 – 1.1 nm)39,46,47 and made it possible to actuate the diameter of the nanopores dynamically by 1.4 ± 0.1 nm. Figure 1d also shows that the midpoint (dashed blue line) and range (grey area) of the phase transition in the nanopore coating occurred precisely at the reported temperature for DMPC lipids of 23.5 ± 2.3° C 39. Changing the diameter of nanopores by a phase transition of lipids may be a relevant mechanism by which insects regulate their water uptake and evaporative loss through lipid-coated nanopores in their exoskeleton34,48. In the context of synthetic nanopores, this bioinspired capability of changing pore diameters constitutes a novel approach to determine thermal phase transition temperatures of lipid bilayers, in situ.
In addition to fine-tuning and actuating the diameters of nanopores, bilayer coatings provide a straightforward strategy to render nanopore recordings specific for certain analytes by functionalizing the bilayer surface with ligands or receptors. Fig. 2 illustrates that adding defined mole fractions of lipids with desired functional groups (here, biotinylated lipids) during the formulation of liposomes and the subsequent formation of a bilayer coating42 can control the surface density of ligands in and around the pore. These lipid-anchored ligands, which were mobile within the fluid sheet of the lipid bilayer, could concentrate dilute analytes from the bulk solution to specific ligands on the bilayer surface and deliver these analytes to the pore by two-dimensional diffusion (Fig. 2a,b). This same basic principle is thought to occur on the lipid coating of olfactory sensilla in insect antenna, which contributes to the extremely sensitive detection of lipophilic pheromones by insects32,34,49.
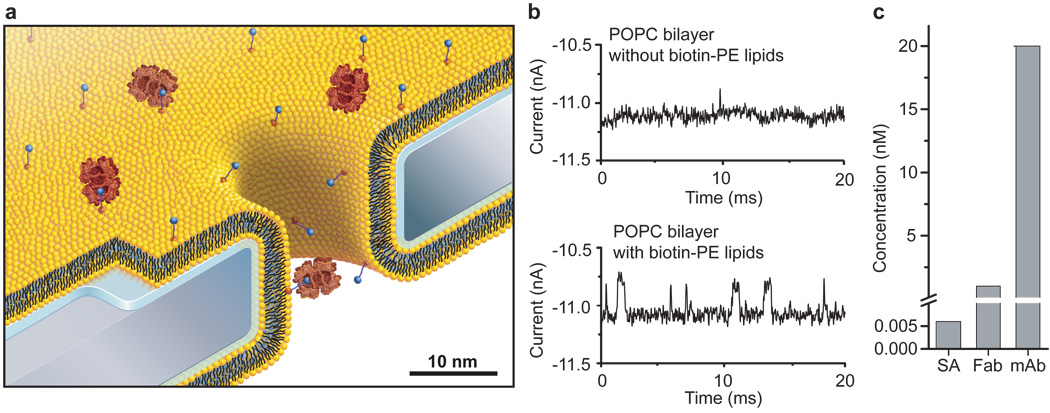
Figure 2. Capture, affinity-dependent pre-concentration, and translocation of specific proteins after binding to ligands on mobile lipid anchors.
a, Cartoon, drawn to scale, illustrating binding of streptavidin (large red) to specific lipid-anchored biotin-PE (blue circles) followed by single molecule translocation of the anchored complex through the nanopore. b, Current versus time traces illustrating capture, pre-concentration, and reduced translocation speed of streptavidin. In the absence of biotin groups, only rare translocation events with short translocation times, td, could be detected in electrolytes containing 6 pM streptavidin (top current trace). In contrast, 0.4 mol% of biotinylated lipids in the lipid coating strongly increased the event frequency and slowed down the translocation speed sufficiently to enable complete time resolution of translocation events (bottom current trace). c, Minimum bulk concentrations of streptavidin, polyclonal anti-biotin Fab fragments, and monoclonal anti-biotin IgG antibodies required to observe at least 30 – 100 translocation events per second.
Pre-concentrating and translocating analytes that are bound to a fluid surface also made it possible to distinguish between different analytes based on their affinity to the displayed ligand (Fig. 2c). For instance, proteins present at picomolar concentrations in the bulk electrolyte solution concentrated at the surface and induced frequent translocation events if they bound with high affinity to lipid-anchored ligands in the bilayer. In contrast, proteins with low affinity to these ligands required more than 300-fold increased bulk concentrations to reach comparable frequencies of time-resolved translocation events (Fig. 2c). In the case of streptavidin, polyclonal anti-biotin Fab fragments and monoclonal anti-biotin IgG antibodies, we found that to reach a frequency of 30 – 100 translocation events per second, a concentration of only 0.006 nM streptavidin was required compared to 1 nM of Fab fragment and 20 nM monoclonal antibody. Control experiments revealed that in the absence of biotinylated lipids in the bilayer coating, or in the presence of excess biotin in solution, the frequency of detectable translocation events for each protein was up to 500-fold lower than in the presence of specific capture sites in the bilayer (Fig. 2b and Supplementary Section S5).
Lipid coating resolves translocation events
The capability of moving captured analytes through pores with fluid walls made it possible to obtain the translocation time, td, through the pore as well as the corresponding amplitude of the resistive pulses, ΔI. This information is unique to the fluid nanopore coatings introduced here; previous reports on nanopore recordings with specific, surface-attached binding groups captured analytes on permanently fixed positions4,5 and did not allow translocation of bound analytes thereby excluding the possibility to determine td or to relate ΔI to the molecular volume of the bound analyte. An additional benefit of translocating analytes that are bound to a lipid anchor emerges if the intrinsic translocation speed of the unbound analyte through a pore is too fast to resolve td and ΔI completely in time – a problem encountered previously by other groups26–28.
Fig 2b and Supplementary Section S5 show that translocation events of individual proteins could not be fully resolved without lipid-anchored capture sites. In contrast, anchoring analytes to lipids during their passage through the pore had the advantage that the translocation speed was dominated by the high viscosity of the bilayer coating rather than the low viscosity of the aqueous electrolyte in the pore50. The resulting, prolonged translocation times enabled time-resolved detection of td (Fig. 3) and ΔI (Fig. 4) combined with accurate, quantitative characterization of individual proteins. Alternative strategies for prolonging the translocation time by increasing the length of the pore or the viscosity of the electrolyte or by reducing the applied voltage have been associated with a reduction of the amplitude of translocation events and reduced the signal to noise ratio28. In contrast, bilayer coatings with fluid capture sites can fine-tune the viscosity of the bilayer and prolong the translocation times of lipid-anchored analytes while the conductivity of the aqueous electrolyte remains unchanged.
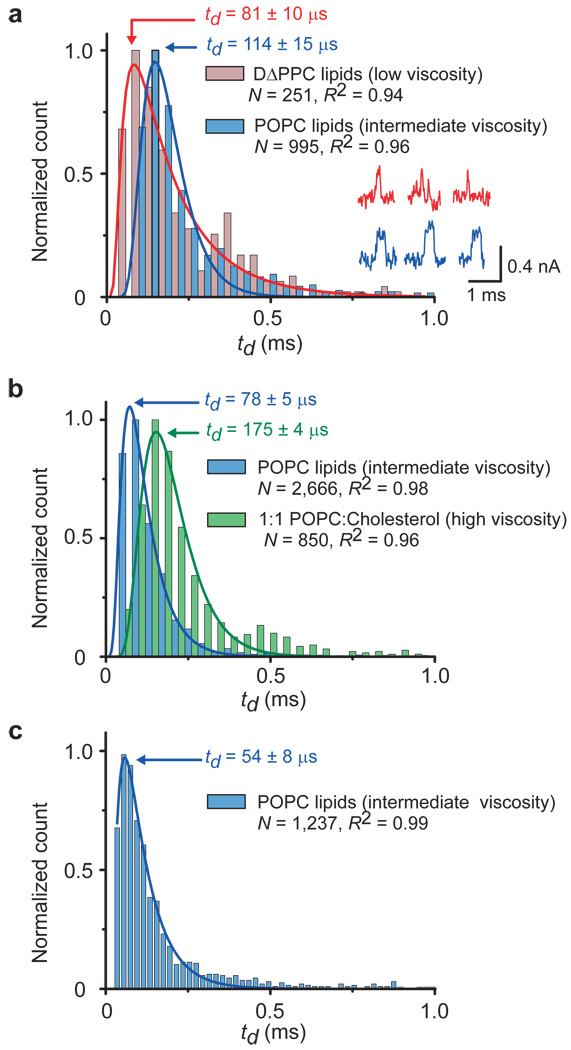
Figure 3. Controlling the translocation times, td, of single lipid-anchored proteins by the viscosity of the bilayer coating and distinguishing proteins by their most probable td values.
a, Distribution of translocation times of streptavidin. Insets: current versus time traces illustrating that td could be prolonged more with intermediate viscosity POPC bilayers (blue current traces) than with low viscosity DΔPPC bilayers (red current traces). b, Translocation of anti-biotin Fab fragments through nanopores with bilayers of intermediate viscosity (POPC) or high viscosity (~49 mol% cholesterol and 50 mol% POPC). c, Translocation of anti-biotin antibodies through a pore with a coating of intermediate viscosity (POPC). Red, blue, and green curves represent a best fit of the corresponding data to a biased diffusion first passage time model14 (equation S10 in Supplementary Section S5). All bilayers contained 0.15 – 0.4 mol% biotin-PE. See Supplementary Sections S7 and S9 for binning methods, errors of td, and measurement errors.
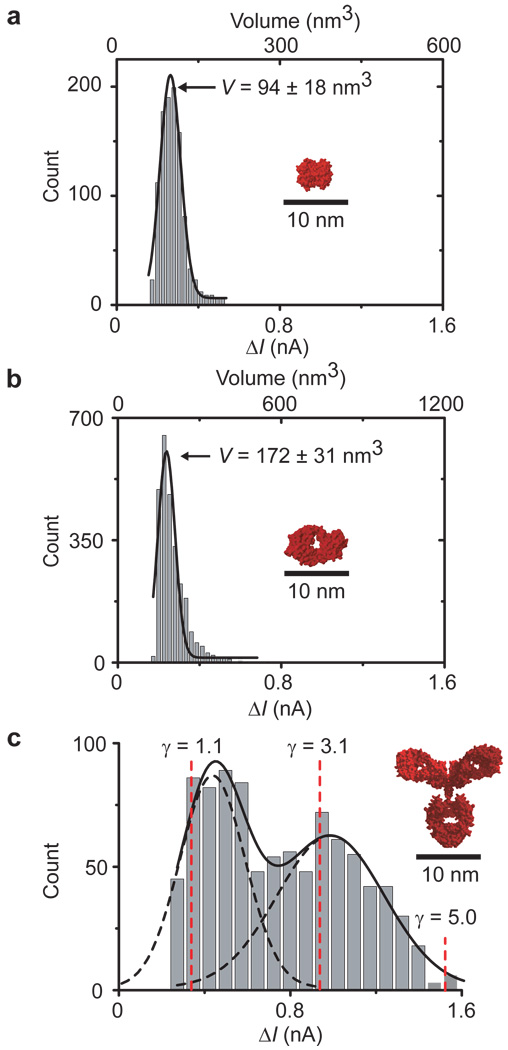
Figure 4. Distribution of ΔI values and corresponding molecular volumes and shape factors of individual proteins translocating through bilayer-coated nanopores with biotinylated lipids.
a–c, Translocation of streptavidin (a), anti-biotin Fab fragments (b) and anti-biotin antibodies (c); the dashed red lines indicate ΔI values that would be expected for IgG antibodies with a volume of 347 nm3 and different shape factors γ; see Supplementary Section S6 for a schematic illustration and discussion of shape factors52,55.
Figure 3a demonstrates that acyl chains with increasing length and saturation could slow down translocation speeds. For instance, POPC lipids with one monounsaturated acyl chain of 18 carbon atoms and a second saturated acyl chain of 16 carbons generated approximately 1.4 times more viscous bilayers than DΔPPC lipids with two monounsaturated acyl chains of 16 carbons. These two bilayer coatings resulted in most frequently observed translocation times for streptavidin of 114 ± 15 µs in the POPC coating compared to 81 ± 10 µs in the DΔPPC coating (Fig. 3a). Translocation speeds could be slowed down even further by adding 50 mol% cholesterol to a POPC bilayer; in this case the most frequently observed translocation time of Fab fragments doubled from 78 ± 5 µs to 175 ± 4 µs (Fig. 3b).
Complete time resolution of translocation events of lipid-anchored proteins allowed us to determine the volume of individual translocating proteins based on accurate acquisition of the amplitude of resistive pulses, ΔI(t). Figure 4 shows amplitude distributions of the resistive pulses for three different biotin-binding proteins. We used equation (2) to estimate the transiently excluded volume of electrolyte, Λ(t) (m3) during the translocation of these three proteins12,13,51.
| (2) |
In this equation, γ(unitless) represents a shape factor52 with a value of 1.5 for spheres, Va (V) is the total applied voltage, and S(rP/dM) is a correction factor that depends on the relative values of rP and the diameter of the molecule, dM. Like most groups, we used a value of 1 for S(rP, dM) for all calculations12,13. Since Λ(t) from the translocation of spheroidal particles is approximately equal to the molecular volume of the particles14,29, we were able to estimate the molecular volumes of streptavidin (94 ± 18 nm3; lit. value: 105 ± 3 nm3)53, Fab fragments (172 ± 31 nm3; lit. value: ~140 nm3)54, and antibodies (308 – 696 nm3; lit. value: 347 ± 15 nm3)55. The distributions of ΔI values for streptavidin (Fig. 4a) and Fab fragments (Fig. 4b) were significantly narrower than the distribution for the antibodies (Fig. 4c). Since control experiments revealed that the broad distribution was not caused by contamination of the antibody sample with other proteins (see Supplementary Section S6), we attribute the broad distribution of ΔI values in Fig. 4c primarily to the complex molecular shape of IgG antibodies (γ ≠ 1.5) compared to the approximately spherical shape (γ ≈ 1.5) of streptavidin and Fab fragments (for a detailed discussion on the proposed effect of molecular shape on ΔI, see Supplementary Section S6).
Determining translocation time and charge of proteins
Figure 3 shows that different proteins moved through the nanopores at different distributed speeds as expected for biased diffusion first passage time processes14. Because we performed the experiments with streptavidin using a different pore (see Supplementary Table S1 for dimensions of pores used for all experiments), a direct comparison of the most frequently observed td values was only possible between Fab fragments (78 ± 5 µs, blue bars in Fig. 3b) and monoclonal antibodies (54 ± 8 µs; Fig. 3c). The observed differences in td values added a third dimension for distinguishing between different proteins in addition to comparing their affinity to specific ligands based on the frequency of translocation events (Fig. 2c) and quantifying their molecular volumes based on ΔI values (Fig. 4a–c).
Since the translocation speed of different lipid-anchored proteins varied, we hypothesized that the fluid nature of the pore walls may minimize non-specific adsorption processes and open the door to determining the net charge of proteins. To test this hypothesis, we developed the simplest possible model that yields a relationship between td of a lipid-anchored protein and the net charge of this protein, |z| × e, based on a model introduced recently by Sexton et al26. Here z (unitless) is the net valency of the overall charge on the protein and e (C) is the elementary charge of an electron. This model assumed that a charged protein experiences an electrophoretic force that is opposed by the viscous drag inside the pore and leads to a constant drift velocity (lP/td) through the pore. It also assumed that the viscous drag of lipid-anchored proteins is determined by the diffusion constant of the lipid anchor, DL (m2 s−1) in the lipid bilayer rather than by the diffusion constant of the protein in the aqueous electrolyte inside the pore lumen50. Based on these assumptions, we derived equation (3) to predict td values theoretically (for a detailed derivation and additional assumptions made, see Supplementary Section S8):
| (3) |
Here kB (J K−1) is the Boltzmann constant, T (K) is temperature and Vp (V) refers to the part of the total applied voltage that drops inside the pore; it does not include the voltage drop due to the access resistance to and from the pore (see Supplementary Section S8).
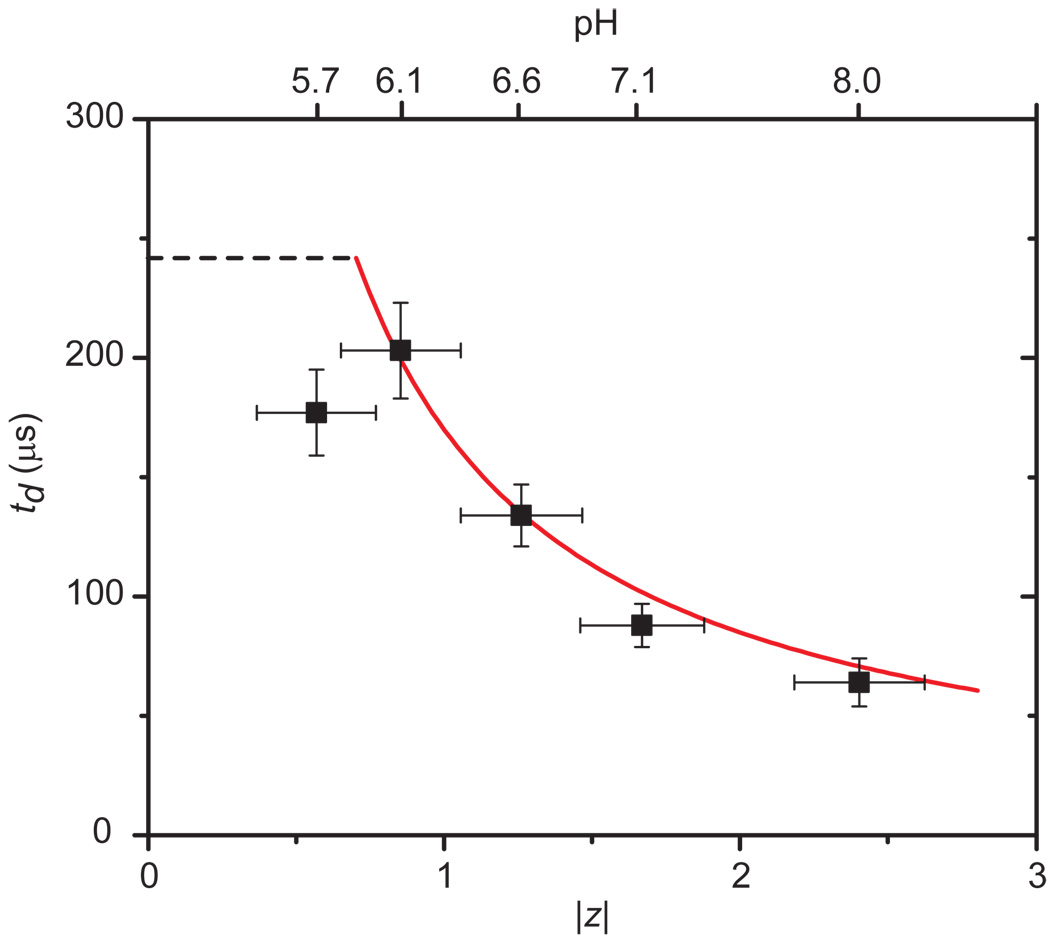
Equation (3) made it possible to compare theoretically predicted td values with experimentally determined values for proteins with known net charge. Figure 5 shows this comparison for translocation events of streptavidin at five different pH values in the recording electrolyte and therefore five different values of |z|. The excellent agreement between the data (black squares) and the predicted td values (red curve) supports the simple model used for the derivation of equation (3).
Figure 5. Comparison of experimental and theoretical values of charge-dependent translocation times of streptavidin.
Experimental values are shown in black squares and the red curve represents the theoretical prediction by equation 3. Dashed black line corresponds to the expected translocation time for streptavidin assuming a translocation event due purely to diffusion in one dimension (td = <lp>2/(2DL), i.e. without an electrophoretic effect. The valance |z| of the net charge of streptavidin was varied by the pH of the electrolyte56. The length of the pore with the bilayer coating was 28 ± 0.2 nm. Note that the red curve is not a best fit to the data; it is the prediction of td as a function of |z| according to equation (3) when all parameters were fixed to their known values.
Additional support for this model stems from a comparison between two bilayer coatings of different viscosity. In one experiment we coated the nanopore with a POPC bilayer and in the other experiment with a DΔPPC bilayer. Before adding streptavidin to the top compartment of the chips, we determined the lateral diffusion coefficient of lipids in the POPC bilayer (DL = 1.13 ± 0.11 nm2 µs−1) and in the DΔPPC bilayer (DL = 1.56 ± 0.16 nm2 µs−1) by fluorescence recovery after photobleaching (FRAP) experiments on the silicon nitride support (see Supplementary Section S2)57. With these DL values and a valence of net charge of |z| = |−1.9 ± 0.4| at pH 7.456, equation (3) predicted a translocation time for streptavidin of 126 ± 29 µs in POPC-coated pores and of 91 ± 21 µs in DΔPPC-coated pores. Experimentally, the most frequently observed translocation time of streptavidin (Fig. 3a) was 114 ± 15 µs through pores with a POPC coating (deviation from the predicted value: −10%) and 81 ± 10 µs through pores with a DΔPPC coating (deviation from the predicted value: −11%). The excellent agreement between the theoretically predicted values of td and the experimentally measured td values as well as the data in Table 2 confirm that translocation times of lipid-anchored analytes were indeed dominated by the viscosity of the bilayer50 and were hence independent of the shape of the proteins (Fig. 3b,c).
Table 2.
Comparison of diffusion coefficients of lipid-anchored proteins within the nanopore, DP, with diffusion coefficients of lipids, DL, in coatings of two different lipid bilayers on three different nanopores.
| Protein | Lipid bilayera |
DLb (nm2 µs−1) |
DPc (nm2 µs−1) |
ΔD % |
|---|---|---|---|---|
| SAd | DΔPPC | 1.56 ± 0.16 | 1.7 ± 0.4 | +9 |
| SAd | POPC | 1.13 ± 0.11 | 1.2 ± 0.3 | +6 |
| SAe | POPC | 1.65 ± 0.17 | 1.9 ± 0.5 | +15 |
| mAbf | POPC | 1.29 ± 0.13 | 2.6 ± 0.7 | +100 |
| Fabf | POPC | 1.27 ± 0.13 | 1.5 ± 0.2 | +18 |
All lipid bilayers also contained 0.15 – 0.4 mol% of biotin-PE.
Values for DL were determined by FRAP as described in Supplementary Section S2.
Values for DP were determined with equation (3) based on the most frequently measured values of td and values of |z| for SA from Sivasankar et al56 and values of |z| for mAb and Fab as determined by capillary electrophoresis (see Supplementary Section S8).
Nanopore dimensions: rP = 10.0 nm, lP = 18 nm
Nanopore dimensions: rP = 10.5 nm, lP = 18 nm
Nanopore dimensions: rP = 16.5 nm, lP = 22 nm
These observations raise the possibility to use td values, in analogy to migration times in electrophoresis, for distinguishing between, and possibly identifying, specific proteins. The agreement between theory and experiment also suggests that determining translocation times of lipid-anchored proteins through a bilayer-coated nanopore makes it possible to determine the net charge of proteins. For instance, at pH 7.4, the measured td values suggest a net charge between −2.9 and −5.3 for the polyclonal anti-biotin Fab fragments and a net charge of −4.2 ± 0.5 for the monoclonal anti-biotin antibodies (see Supplementary Section S8). These values agree well with results from capillary electrophoresis experiments (see Supplementary Section S8). Moreover, for a protein with known charge, translocation experiments combined with equation (3), make it possible to determine – non-optically – the lateral diffusion constants of lipids and therefore the fluidity of bilayers within seconds (Table 2). This attribute might be useful to test therapeutic compounds for their propensity to change membrane fluidity57.
Finally, the agreement between predicted and experimental td values suggests that the measured td values are close to the “true” electrophoretic translocation times. In other words, these measured translocation times represent translocation in the absence of non-specific adsorption of proteins to the bilayer coating or to the silicon nitride substrates. This point is important because all single-molecule translocation experiments with proteins reported so far were hampered by non-specific adsorption of proteins to the nanopore walls with regard to accurate determination of td values1,14,26. In some cases, these interactions increased the translocation times of proteins by several orders of magnitude26.
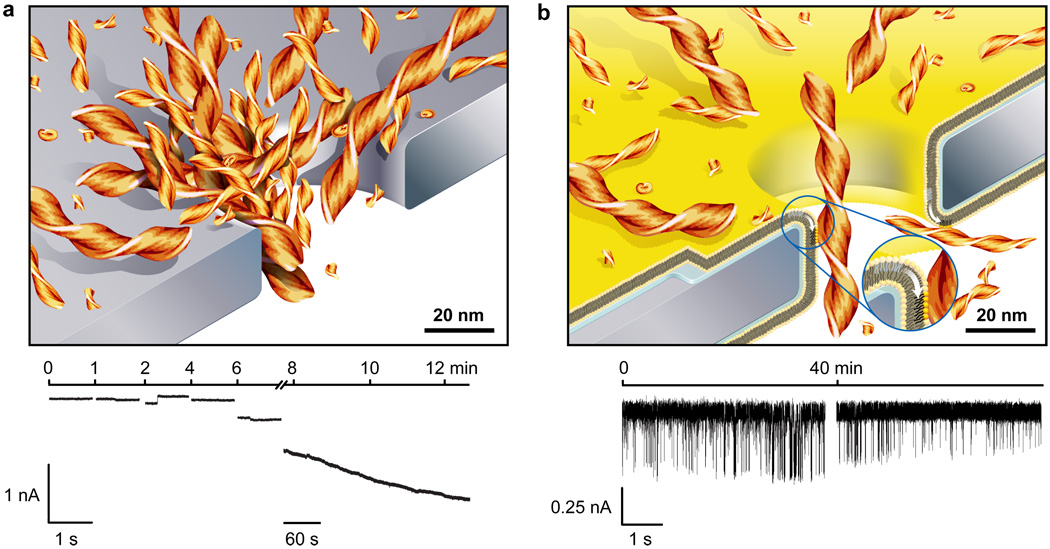
Aggregated Aβ peptides translocate without clogging
Due to the unique capability of fluid bilayer coatings to eliminate non-specific interactions, these pores made it possible to analyze translocation events of molecules that aggregate and have a tendency to clog nanopores. Amyloidogenic peptides, such as Alzheimer’s disease-related amyloid-β (Aβ) peptides58, belong to this category of molecules. The current versus time trace in Fig. 6a shows that a nanopore without a bilayer coating clogged within minutes after addition of Aβ peptides. Despite several attempts, we were never able to detect translocation events from samples of Aβ peptides with uncoated pores. In contrast, Fig. 6b illustrates that coating nanopores with bio-inspired, fluid lipid bilayers incurred non-fouling properties to these pores and made it possible to detect numerous large amplitude translocation events due to the passage of individual Aβ oligomers and fibrils.
Figure 6. Bilayer-coated nanopores resist clogging and enable the monitoring of the aggregation of amyloid-beta (Aβ) peptides.
a, Cartoon illustrating clogging of uncoated nanopores and a typical current versus time trace during clogging of a nanopore by Aβ aggregates. This concatenated current trace shows several 1 s recordings and one 5 min recording. b, Cartoon illustrating translocation of individual Aβ aggregates through a bilayer-coated nanopore with a fluid wall (white arrow in the inset) and a typical current versus time trace of translocation events. The bilayer coating conferred non-fouling properties to these pores and enabled resistive pulse recordings over at least 40 min without clogging. Both recordings are 5 s long, one was taken immediately after addition of the Aβ sample and the other one 40 min later. Aβ (1–40) samples were aggregated for 72 h.
Conclusions
In conclusion, the crucial novelty of lipid-coated, synthetic nanopores is the multifunctional and fluid nature of the self-assembled coating. This singular, bio-inspired strategy addresses many of the unmet challenges in nanopore sensing and is particularly beneficial in the context of single-molecule studies of native proteins. For example, the fluidity of the coating enables capture and concentration of proteins from dilute solutions and permits translocation of lipid-anchored proteins at frequencies that reveal information about their affinity to ligands on these lipid anchors. Fluid coatings also eliminate non-specific adsorption of proteins to the synthetic walls of the pore by translocating captured proteins on top of a fluid, biocompatible lipid bilayer and establish a predictable, quantitative relationship between translocation times and the charge of individual proteins. The viscous character of the fluid coating slows the translocation speed of lipid-anchored proteins and makes it possible to introduce selectivity while resolving translocation events completely in time. These viscous coatings therefore enable accurate quantitative analyses of the molecular volume and qualitative analyses of the shape of individual proteins. The antifouling character of fluid coatings made it possible to translocate aggregated forms of disease-relevant Aβ peptides through the pore without clogging. This capability may open the door for analyses of the diameter, length, and volume from a large number of individual Aβ oligomers and fibrils during their aggregation in situ.
Methods
Lipids and Proteins
We obtained all phospholipids from Avanti Polar Lipids, Inc. We purchased the proteins streptavidin (SA) and monoclonal anti-biotin antibody (mAb, B7653) from Sigma Alrdrich and polyclonal anti-biotin Fab fragments (Fab, 20938) from Rockland Inc.
Nanopores
We used a focused ion beam to fabricate nanopores in a silicon nitride membrane that was supported by a silicon chip (see Supplementary Section S1 for information on the pores)59. Prior to experiments, we cleaned the pore-containing chips for at least 30 min with a fresh mixture of 3:1 (v/v) concentrated sulfuric acid and 30% (v/v) aqueous hydrogen peroxide solution at a temperature of 60 – 70 °C followed by rinsing with deionized water and drying with argon gas. To create separate fluid compartments on either side of the nanopore, we mounted the chip between two pieces of cured polydimethylsiloxane (PDMS) 10. After each experiment, we rinsed the silicon chips for 2 – 3 min successively with the following solvents: water, ethanol, methanol, and chloroform. We stored chips in chloroform between experiments.
Formation of Supported Lipid Bilayers
We formed supported lipid bilayers by fusion of small unilamellar vesicles (SUVs)40–43. We prepared these SUVs as described in Supplementary Section S2. To form the supported lipid bilayer on silicon nitride membranes, we filled the top compartment of the PDMS fluidic setup with 10 – 30 µL of the aqueous solution with the SUVs and the bottom compartment with a 150 mM KCl solution without liposomes. After 5–10 min, we removed excess SUVs by immersing the entire fluidic setup for 5 – 10 min in a large (500 mL) beaker containing deionized water. Before recordings, the fluidic compartments were filled with the desired electrolyte. Each liposome preparation contained 0.8 mol% of the fluorescently-labeled lipid, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rh-PE), for measuring the fluidity of lipid bilayers by fluorescence recovery after photobleaching (FRAP, see Supplementary Section S2).
Electrical Resistance as a Function of Bilayer Thickness
We used Ag/AgCl pellet electrodes (Warner Instruments) to monitor ionic currents through electrolyte-filled nanopores with a patch-clamp amplifier (Axopatch 200B, Molecular Devices Inc.) in voltage clamp mode (i.e., at constant applied voltage). See Supplementary Section S9 for a description of data acquisition methods. We determined the resistance between the electrodes by measuring the current at various applied voltages in the range of ± 0.5 V; the slope of the corresponding current versus voltage plots equaled the inverse of the resistance. To measure the resistance as a function of the bilayer thickness, we formed different lipid bilayers on the same chip by using SUVs composed of DLPC, DMPC, DΔPPC, or DEPC lipids. We cleaned this chip before the formation of each lipid bilayer as described above. The chip used for these experiments contained a nanopore with a diameter of 28 nm and a length of 12 nm (see Supplementary Section S1 for a TEM image) and the recording buffer contained 500 mM KCl and 10 mM HEPES at a pH value of 7.4 ± 0.1. To measure the resistance of nanopores as a function of temperature, we used a feedback-controlled Peltier Cooler from Warner Instruments (see Supplementary Section S1).
Sensing Proteins with biotinylated lipids in the Bilayer
We formed supported lipid bilayers on the silicon chip from SUVs containing 0.15 – 0.4 mol% of biotin-PE, 0.8 mol% Rh-PE, and ~99 mol% POPC. We used an electrolyte containing 2.0 M KCl and 10 mM HEPES with a pH of 7.4 ± 0.1 and performed all current recordings at −0.1 V. To detect SA, we used a nanopore with an area-equivalent diameter of 19.2 nm (see Supplementary Section S1) and a length of 18 nm (before formation of the bilayer), and we added SA to the top compartment at concentrations of 3.2 – 6.2 pM. To detect mAb and Fab, we used a nanopore with an area equivalent diameter of 33.0 nm and a length of 22 nm; we added mAb or Fab to the top compartment at concentrations of mAb or Fab of 0.1 – 50 nM. See Supplementary Section S9 for a description of the resistive-pulse analysis.
Detection of Aggregates of Amyloid-Beta (Aβ) Peptides
See Supplementary Section S10 for a description of Aβ sample preparation. We used a nanopore with a diameter of 96 nm and a length of ~ 275 nm (before bilayer coating), which was either uncoated or coated with a POPC bilayer. We added solutions containing Aβ peptides (residues 1–40) to the top compartment at concentrations of Aβ of 0.1 to 0.2 mg × mL−1. We used an electrolyte containing 70 mM KCl and 10 mM HEPES with a pH of 7.4 ± 0.1 and recorded resistive pulses at +0.2 V.
Supplementary Material
Acknowledgements
The authors thank David Sept and David Talaga for assistance in modeling distributions of translocation times and Yazan N. Billeh for valuable discussions. The authors also thank Daniel J. Estes and Jeffrey D. Uram for their work on the LabView recording software. This work was supported by a National Science Foundation Career Award (M.M., Grant No. 0449088), the National Institute of Health (M.M., Grant No. 1R01GM081705), the Alzheimer’s disease Research Center (JY, 3P50 AG005131), the Alzheimer’s Association (JY, NIRG-08-91651), the National Human Genome Research Institute (JL, Grant No. HG003290 and HG004776), and a Graduate Assistance in Areas of National Need Fellowship (E.C.Y).
Footnotes
Author contributions
E.C.Y, J.Y., and M.M. conceived and designed the experiments, E.C.Y., J.M.J., S.M., P.P. performed the experiments, R.C.R. and J.L. fabricated the nanopores, E.C.Y., J.L., J.Y., and M.M. co-wrote the manuscript and Supplementary Information.
The authors declare no competing financial interests.
Supplementary information accompanies this paper at www.nature.com/naturenanotechnology.
Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Sexton LT, et al. Resistive-pulse studies of proteins and protein/antibody complexes using a conical nanotube sensor. J. Am. Chem. Soc. 2007;129:13144–13152. doi: 10.1021/ja0739943. [DOI] [PubMed] [Google Scholar]
- 2.Movileanu L, Howorka S, Braha O, Bayley H. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nature Biotech. 2000;18:1091–1095. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- 3.Howorka S, Siwy Z. Nanopore analytics: sensing of single molecules. Chem. Soc. Rev. 2009;38:2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 4.Siwy Z, et al. Protein biosensors based on biofunctionalized conical gold nanotubes. J. Am. Chem. Soc. 2005;127:5000–5001. doi: 10.1021/ja043910f. [DOI] [PubMed] [Google Scholar]
- 5.Ding S, Gao CL, Gu LQ. Capturing single molecules of immunoglobulin and ricin with an aptamer-encoded glass nanopore. Anal. Chem. 2009;81:6649–6655. doi: 10.1021/ac9006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uram JD, Ke K, Hunt AJ, Mayer M. Submicrometer pore-based characterization and quantification of antibody-virus interactions. Small. 2006;2:967–972. doi: 10.1002/smll.200600006. [DOI] [PubMed] [Google Scholar]
- 7.Branton D, et al. The potential and challenges of nanopore sequencing. Nature Biotech. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iqbal SM, Akin D, Bashir R. Solid-state nanopore channels with DNA selectivity. Nature Nanotech. 2007;2:243–248. doi: 10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]
- 9.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nature Nanotech. 2010;5:160–165. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uram JD, Ke K, Hunt AJ, Mayer M. Label-free affinity assays by rapid detection of immune complexes in submicrometer pores. Angew. Chem.-Int. Edit. 2006;45:2281–2285. doi: 10.1002/anie.200502862. [DOI] [PubMed] [Google Scholar]
- 11.Robertson JWF, et al. Single-molecule mass spectrometry in solution using a solitary nanopore. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8207–8211. doi: 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han AP, et al. Label-free detection of single protein molecules and protein-protein interactions using synthetic nanopores. Anal. Chem. 2008;80:4651–4658. doi: 10.1021/ac7025207. [DOI] [PubMed] [Google Scholar]
- 13.Ito T, Sun L, Crooks RM. Simultaneous determination of the size and surface charge of individual nanoparticles using a carbon nanotube-based coulter counter. Anal. Chem. 2003;75:2399–2406. doi: 10.1021/ac034072v. [DOI] [PubMed] [Google Scholar]
- 14.Talaga DS, Li JL. Single-molecule protein unfolding in solid state nanopores. J. Am. Chem. Soc. 2009;131:9287–9297. doi: 10.1021/ja901088b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oukhaled G, et al. Unfolding of proteins and long transient conformations detected by single nanopore recording. Phys. Rev. Lett. 2007;98:158101. doi: 10.1103/PhysRevLett.98.158101. [DOI] [PubMed] [Google Scholar]
- 16.Benner S, et al. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nature Nanotech. 2007;2:718–724. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nature Nanotech. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 18.Bayley H, Cremer PS. Stochastic sensors inspired by biology. Nature. 2001;413:226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 19.Nakane JJ, Akeson M, Marziali A. Nanopore sensors for nucleic acid analysis. J. Phys. Condens. Matter. 2003;15:R1365–R1393. [Google Scholar]
- 20.Dekker C. Solid-state nanopores. Nature Nanotech. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 21.Martin CR, Siwy ZS. Learning nature's way: Biosensing with synthetic nanopores. Science. 2007;317:331–332. doi: 10.1126/science.1146126. [DOI] [PubMed] [Google Scholar]
- 22.Movileanu L. Interrogating single proteins through nanopores: challenges and opportunities. Trends Biotechnol. 2009;27:333–341. doi: 10.1016/j.tibtech.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Majd S, et al. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Curr. Opin. Biotechnol. 2010;21:439–476. doi: 10.1016/j.copbio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou X, et al. A biomimetic potassium responsive nanochannel: G-Quadruplex DNA conformational switching in a synthetic nanopore. J. Am. Chem. Soc. 2009;131:7800–7805. doi: 10.1021/ja901574c. [DOI] [PubMed] [Google Scholar]
- 25.Yameen B, et al. Single conical nanopores displaying pH-tunable rectifying characteristics. Manipulating ionic transport with zwitterionic polymer brushes. J. Am. Chem. Soc. 2009;131:2070–2071. doi: 10.1021/ja8086104. [DOI] [PubMed] [Google Scholar]
- 26.Sexton LT, et al. An adsorption-based model for pulse duration in resistive-pulse protein sensing. J. Am. Chem. Soc. 2010;132:6755–6763. doi: 10.1021/ja100693x. [DOI] [PubMed] [Google Scholar]
- 27.Pedone D, Firnkes M, Rant U. Data analysis of translocation events in nanopore experiments. Anal. Chem. 2009;81:9689–9694. doi: 10.1021/ac901877z. [DOI] [PubMed] [Google Scholar]
- 28.Uram JD, Ke K, Mayer M. Noise and bandwidth of current recordings from submicrometer pores and nanopores. ACS Nano. 2008;2:857–872. doi: 10.1021/nn700322m. [DOI] [PubMed] [Google Scholar]
- 29.Fologea D, Ledden B, David SM, Li J. Electrical characterization of protein molecules by a solid-state nanopore. Appl. Phys. Lett. 2007;91 doi: 10.1063/1.2767206. 053901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chun KY, Mafe S, Ramirez P, Stroeve P. Protein transport through gold-coated, charged nanopores: Effects of applied voltage. Chem. Phys. Lett. 2006;418:561–564. [Google Scholar]
- 31.Hille B. Ion Channels of Excitable Membranes. Sunderland: Sinauer Associaties, Inc.; 2001. [Google Scholar]
- 32.Steinbrecht RA. Pore structures in insect olfactory sensilla: A review of data and concepts. Int. J. Insect Morphol. Embryol. 1997;26:229–245. [Google Scholar]
- 33.Zacharuk RY. Antennae and Sensilla. In: Kerkut GA, Gilbert LI, editors. Comparative Insect Physiology Chemistry and Pharmacology. Oxford: Pergamon Press; 1985. [Google Scholar]
- 34.Locke M. Permeability of insect cuticle to water and lipids. Science. 1965;147:295–298. [PubMed] [Google Scholar]
- 35.Nilsson J, Lee JRI, Ratto TV, Letant SE. Localized functionalization of single nanopores. Adv. Mater. 2006;18:427–431. [Google Scholar]
- 36.Wang GL, Zhang B, Wayment JR, Harris JM, White HS. Electrostatic-gated transport in chemically modified glass nanopore electrodes. J. Am. Chem. Soc. 2006;128:7679–7686. doi: 10.1021/ja061357r. [DOI] [PubMed] [Google Scholar]
- 37.Wanunu M, Meller A. Chemically modified solid-state nanopores. Nano Lett. 2007;7:1580–1585. doi: 10.1021/nl070462b. [DOI] [PubMed] [Google Scholar]
- 38.Lewis BA, Engelman DM. Lipid bilayer thickness varies linearly with acyl chain-length in fluid phosphatidylcholine vesicles. J. Mol. Biol. 1983;166:211–217. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- 39.Caffrey M, Hogan J. LIPIDAT: A database of lipid phase transition temperatures and enthalpy changes. DMPC data subset analysis. Chem. Phys. Lipids. 1992;61:1–109. doi: 10.1016/0009-3084(92)90002-7. [DOI] [PubMed] [Google Scholar]
- 40.Watts TH, Brian AA, Kappler JW, Marrack P, McConnell HM. Antigen presentation by supported planar membranes containing affinity-purified I-Ad. Proc. Natl. Acad. Sci. U. S. A. 1984;81:7564–7568. doi: 10.1073/pnas.81.23.7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cremer PS, Boxer SG. Formation and spreading of lipid bilayers on planar glass supports. J. Phys. Chem. B. 1999;103:2554–2559. [Google Scholar]
- 42.Reimhult E, Hook F, Kasemo B. Intact vesicle adsorption and supported biomembrane formation from vesicles in solution: Influence of surface chemistry, vesicle size, temperature, and osmotic pressure. Langmuir. 2003;19:1681–1691. [Google Scholar]
- 43.Sackmann E. Supported membranes: Scientific and practical applications. Science. 1996;271:43–48. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- 44.Miller CE, Majewski J, Gog T, Kuhl TL. Characterization of biological thin films at the solid-liquid interface by X-ray reflectivity. Phys. Rev. Lett. 2005;94 doi: 10.1103/PhysRevLett.94.238104. [DOI] [PubMed] [Google Scholar]
- 45.Bayerl TM, Bloom M. Physical-properties of single phospholipid-bilayers adsorbed to micro glass-beads - a new vesicular model system studied by H-2-nuclear magnetic-resonance. Biophys. J. 1990;58:357–362. doi: 10.1016/S0006-3495(90)82382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tokumasu F, Jin AJ, Dvorak JA. Lipid membrane phase behaviour elucidated in real time by controlled environment atomic force microscopy. J. Electron Microsc. 2002;51:1–9. doi: 10.1093/jmicro/51.1.1. [DOI] [PubMed] [Google Scholar]
- 47.Schuy S, Janshoff A. Thermal expansion of microstructured DMPC bilayers quantified by temperature-controlled atomic force microscopy. ChemPhysChem. 2006;7:1207–1210. doi: 10.1002/cphc.200600013. [DOI] [PubMed] [Google Scholar]
- 48.Gibbs AG. Lipid melting and cuticular permeability: new insights into an old problem. J. Insect Physiol. 2002;48:391–400. doi: 10.1016/s0022-1910(02)00059-8. [DOI] [PubMed] [Google Scholar]
- 49.Adam G, Delbrueck M. Reduction of Dimensionality in Biological Diffusion Processes. In: Rich A, Davidson N, editors. Structural Chemistry and Molecular Biology. San Francisco: W. H. Freeman and Company; 1968. pp. 198–215. [Google Scholar]
- 50.Gambin Y, et al. Lateral mobility of proteins in liquid membranes revisited. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2098–2102. doi: 10.1073/pnas.0511026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grover NB, Naaman J, Ben-sasson S, Doljansk F, Nadav E. Electrical sizing of particles in suspensions. 2. Experiments with rigid spheres. Biophys. J. 1969;9:1415–1425. doi: 10.1016/S0006-3495(69)86462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grover NB, Naaman J, Ben-sasson S, Doljansk F. Electrical sizing of particles in suspensions. I.Theory. Biophys. J. 1969;9:1398–1414. doi: 10.1016/S0006-3495(69)86461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neish CS, Martin IL, Henderson RM, Edwardson JM. Direct visualization of ligandprotein interactions using atomic force microscopy. Br. J. Pharmacol. 2002;135:1943–1950. doi: 10.1038/sj.bjp.0704660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janeway CA. Immunobiology: the immune system in health and disease. 5th edn. New York: Garland Publishing; 2001. [Google Scholar]
- 55.Schneider SW, Larmer J, Henderson RM, Oberleithner H. Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflugers Arch. 1998;435:362–367. doi: 10.1007/s004240050524. [DOI] [PubMed] [Google Scholar]
- 56.Sivasankar S, Subramaniam S, Leckband D. Direct molecular level measurements of the electrostatic properties of a protein surface. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12961–12966. doi: 10.1073/pnas.95.22.12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Majd S, Mayer M. Hydrogel stamping of arrays of supported lipid bilayers with various lipid compositions for the screening of drug-membrane and protein-membrane interactions. Angew. Chem.-Int. Edit. 2005;44:6697–6700. doi: 10.1002/anie.200502189. [DOI] [PubMed] [Google Scholar]
- 58.Capone R, et al. Amyloid-beta-induced ion flux in artificial lipid bilayers and neuronal cells: Resolving a controversy. Neurotox. Res. 2009;16:1–13. doi: 10.1007/s12640-009-9033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, et al. Ion-beam sculpting at nanometre length scales. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.