Abstract
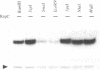
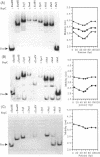
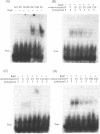
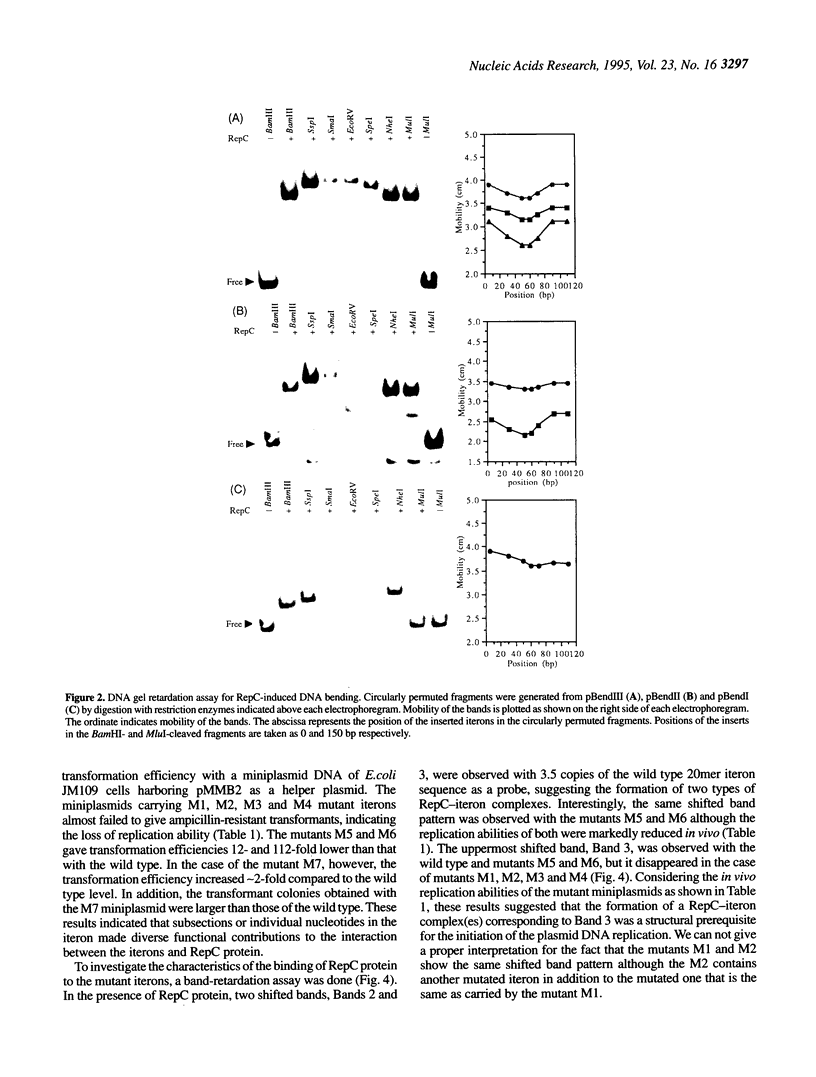
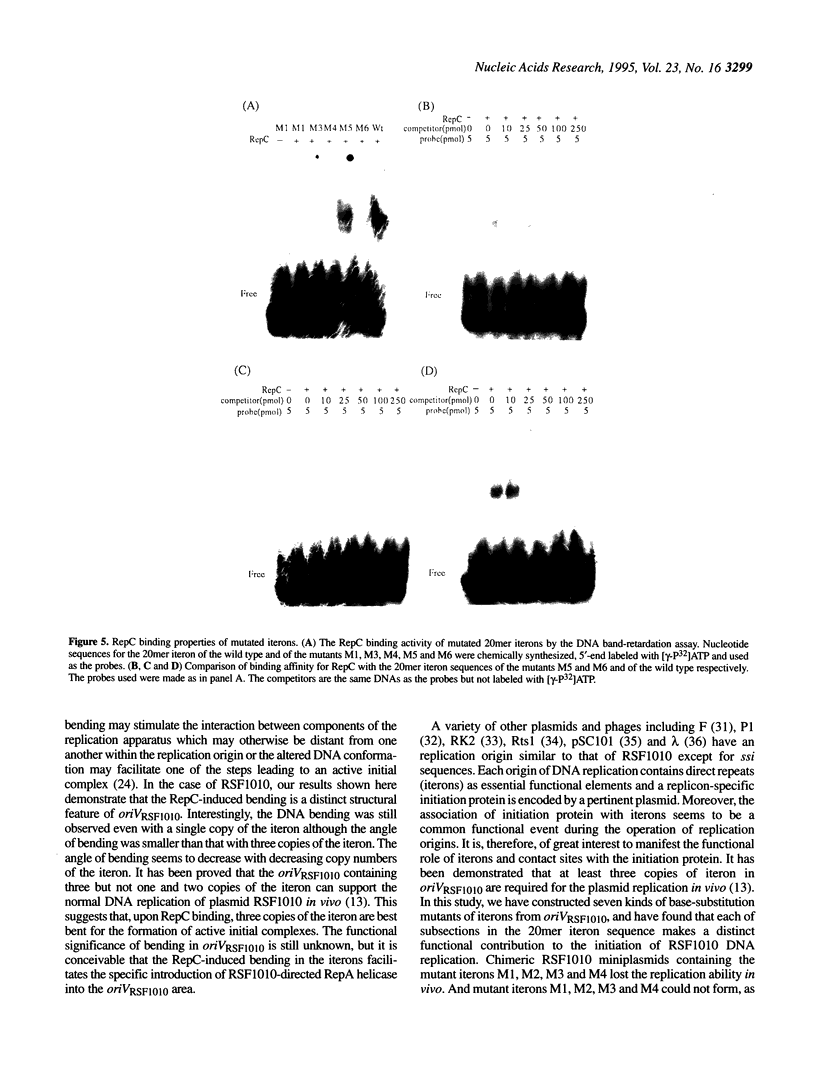
The replication origin of the broad host-range plasmid RSF1010 contains 3.5 copies of a 20mer iteron sequence that bind specifically to the plasmid-encoded initiator, RepC. Here we demonstrated that even a single iteron was bent upon binding of RepC. Moreover, the bending angle seems to become larger along with the increment of the number of iterons. In a mutational analysis of the iteron sequence, we isolated seven kinds of base-substitution mutants of iterons, and estimated the replication activity of these mutants in vivo. We found that each of the subsections in the 20mer iteron sequence made a distinct contribution to the initiation of RSF1010 DNA replication. With the binding assay of RepC and mutated iterons in vitro, we found that the formation of a productive RepC-iteron complex was required for the initiation of plasmid DNA replication.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Snyder K. M., Chattoraj D. K. P1 plasmid replication: replicon structure. J Mol Biol. 1984 Mar 5;173(3):307–324. doi: 10.1016/0022-2836(84)90123-2. [DOI] [PubMed] [Google Scholar]
- Armstrong K. A., Acosta R., Ledner E., Machida Y., Pancotto M., McCormick M., Ohtsubo H., Ohtsubo E. A 37 X 10(3) molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J Mol Biol. 1984 May 25;175(3):331–348. doi: 10.1016/0022-2836(84)90352-8. [DOI] [PubMed] [Google Scholar]
- Chattoraj D. K., Snyder K. M., Abeles A. L. P1 plasmid replication: multiple functions of RepA protein at the origin. Proc Natl Acad Sci U S A. 1985 May;82(9):2588–2592. doi: 10.1073/pnas.82.9.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattoraj D., Cordes K., Abeles A. Plasmid P1 replication: negative control by repeated DNA sequences. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6456–6460. doi: 10.1073/pnas.81.20.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward G., Linder P., Caro L. The nucleotide sequence of replication and maintenance functions encoded by plasmid pSC101. Nucleic Acids Res. 1983 Aug 25;11(16):5645–5659. doi: 10.1093/nar/11.16.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M., Dellis S., Levchenko I., Urh M., Wu F., York D. Regulation of replication of an iteron-containing DNA molecule. Prog Nucleic Acid Res Mol Biol. 1994;48:239–273. doi: 10.1016/s0079-6603(08)60857-0. [DOI] [PubMed] [Google Scholar]
- Gormley E. P., Davies J. Transfer of plasmid RSF1010 by conjugation from Escherichia coli to Streptomyces lividans and Mycobacterium smegmatis. J Bacteriol. 1991 Nov;173(21):6705–6708. doi: 10.1128/jb.173.21.6705-6708.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter N. J., Barth P. T. Characterization of SmSu plasmids by restriction endonuclease cleavage and compatibility testing. J Bacteriol. 1976 Oct;128(1):394–400. doi: 10.1128/jb.128.1.394-400.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashitani A., Greenstein D., Hirokawa H., Asano S., Horiuchi K. Multiple DNA conformational changes induced by an initiator protein precede the nicking reaction in a rolling circle replication origin. J Mol Biol. 1994 Apr 8;237(4):388–400. doi: 10.1006/jmbi.1994.1242. [DOI] [PubMed] [Google Scholar]
- Honda Y., Akioka T., Takebe S., Tanaka K., Miao D., Higashi A., Nakamura T., Taguchi Y., Sakai H., Komano T. Mutational analysis of the specific priming signal essential for DNA replication of the broad host-range plasmid RSF1010. FEBS Lett. 1993 Jun 7;324(1):67–70. doi: 10.1016/0014-5793(93)81534-7. [DOI] [PubMed] [Google Scholar]
- Honda Y., Sakai H., Komano T., Bagdasarian M. RepB' is required in trans for the two single-strand DNA initiation signals in oriV of plasmid RSF1010. Gene. 1989 Aug 1;80(1):155–159. doi: 10.1016/0378-1119(89)90261-8. [DOI] [PubMed] [Google Scholar]
- Honda Y., Sakai H., Komano T. Two single-strand DNA initiation signals located in the oriV region of plasmid RSF1010. Gene. 1988 Sep 7;68(2):221–228. doi: 10.1016/0378-1119(88)90024-8. [DOI] [PubMed] [Google Scholar]
- Kamio Y., Tabuchi A., Itoh Y., Katagiri H., Terawaki Y. Complete nucleotide sequence of mini-Rts1 and its copy mutant. J Bacteriol. 1984 Apr;158(1):307–312. doi: 10.1128/jb.158.1.307-312.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley W., Bastia D. Replication initiator protein of plasmid R6K autoregulates its own synthesis at the transcriptional step. Proc Natl Acad Sci U S A. 1985 May;82(9):2574–2578. doi: 10.1073/pnas.82.9.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Zwieb C., Wu C., Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989 Dec 21;85(1):15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- Koepsel R. R., Khan S. A. Static and initiator protein-enhanced bending of DNA at a replication origin. Science. 1986 Sep 19;233(4770):1316–1318. doi: 10.1126/science.3749879. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- McEachern M. J., Bott M. A., Tooker P. A., Helinski D. R. Negative control of plasmid R6K replication: possible role of intermolecular coupling of replication origins. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7942–7946. doi: 10.1073/pnas.86.20.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M. J., Filutowicz M., Helinski D. R. Mutations in direct repeat sequences and in a conserved sequence adjacent to the repeats result in a defective replication origin in plasmid R6K. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1480–1484. doi: 10.1073/pnas.82.5.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Patel I., Bastia D. Conformational changes in a replication origin induced by an initiator protein. Cell. 1985 Nov;43(1):189–197. doi: 10.1016/0092-8674(85)90023-6. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay G., Chattoraj D. K. Conformation of the origin of P1 plasmid replication. Initiator protein induced wrapping and intrinsic unstacking. J Mol Biol. 1993 May 5;231(1):19–28. doi: 10.1006/jmbi.1993.1253. [DOI] [PubMed] [Google Scholar]
- Murotsu T., Matsubara K., Sugisaki H., Takanami M. Nine unique repeating sequences in a region essential for replication and incompatibility of the mini-F plasmid. Gene. 1981 Nov;15(2-3):257–271. doi: 10.1016/0378-1119(81)90135-9. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Nordström K., Molin S., Aagaard-Hansen H. Partitioning of plasmid R1 in Escherichia coli. II. Incompatibility properties of the partitioning system. Plasmid. 1980 Nov;4(3):332–339. doi: 10.1016/0147-619x(80)90071-2. [DOI] [PubMed] [Google Scholar]
- Papp P. P., Chattoraj D. K., Schneider T. D. Information analysis of sequences that bind the replication initiator RepA. J Mol Biol. 1993 Sep 20;233(2):219–230. doi: 10.1006/jmbi.1993.1501. [DOI] [PubMed] [Google Scholar]
- Ryder K., Silver S., DeLucia A. L., Fanning E., Tegtmeyer P. An altered DNA conformation in origin region I is a determinant for the binding of SV40 large T antigen. Cell. 1986 Mar 14;44(5):719–725. doi: 10.1016/0092-8674(86)90838-x. [DOI] [PubMed] [Google Scholar]
- Scherer G. Nucleotide sequence of the O gene and of the origin of replication in bacteriophage lambda DNA. Nucleic Acids Res. 1978 Sep;5(9):3141–3156. doi: 10.1093/nar/5.9.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E., Bagdasarian M. M., Scholz P., Lurz R., Rückert B., Bagdasarian M. Replication of the broad host range plasmid RSF1010: requirement for three plasmid-encoded proteins. Proc Natl Acad Sci U S A. 1984 Feb;81(3):654–658. doi: 10.1073/pnas.81.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz P., Haring V., Wittmann-Liebold B., Ashman K., Bagdasarian M., Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989 Feb 20;75(2):271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- Smith C. A., Thomas C. M. Nucleotide sequence of the trfA gene of broad host-range plasmid RK2. J Mol Biol. 1984 May 25;175(3):251–262. doi: 10.1016/0022-2836(84)90347-4. [DOI] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada C., Imai M., Yura T. Host control of plasmid replication: requirement for the sigma factor sigma 32 in transcription of mini-F replication initiator gene. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8849–8853. doi: 10.1073/pnas.84.24.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Binding and bending of the lambda replication origin by the phage O protein. EMBO J. 1985 Dec 16;4(13A):3605–3616. doi: 10.1002/j.1460-2075.1985.tb04124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985 Oct 3;317(6036):451–453. doi: 10.1038/317451a0. [DOI] [PubMed] [Google Scholar]