Abstract
Aims/hypothesis
Homozygous staggerer (sg/sg) mice, which have decreased and dysfunctional Rorα (also known as Rora) expression in all tissues, display a lean and dyslipidaemic phenotype. They are also resistant to (high fat) diet-induced obesity. We explored whether retinoic acid receptor-related orphan receptor (ROR) α action in skeletal muscle was involved in the regulation of glucose metabolism.
Methods
We used a three-armed genomic approach, including expression profiling, ingenuity analysis and quantitative PCR validation to identify the signalling pathway(s) in skeletal muscle that are perturbed in sg/sg mice. Moreover, western analysis, functional insulin and glucose tolerance tests, and ex vivo glucose uptake assays were used to phenotypically characterise the impact of aberrant v-AKT murine thymoma viral oncogene homologue (AKT) signalling.
Results
Homozygous and heterozygous (sg/sg and sg/+) animals exhibited decreased fasting blood glucose levels, mildly improved glucose tolerance and increased insulin sensitivity. Illumina expression profiling and bioinformatic analysis indicated the involvement of RORα in metabolic disease and phosphatidylinositol 3-kinase–AKT signalling. Quantitative PCR and western analysis validated increased AKT2 (mRNA and protein) and phosphorylation in sg/sg mice in the basal state. This was associated with increased expression of Tbc1d1 and Glut4 (also known as Slc2a4) mRNA and protein. Finally, in agreement with the phenotype, we observed increased (absolute) levels of AKT and phosphorylated AKT (in the basal and insulin stimulated states), and of (ex vivo) glucose uptake in skeletal muscle from sg/sg mice relative to wild-type littermates.
Conclusions/interpretation
We propose that Rorα plays an important role in regulation of the AKT2 signalling cascade, which controls glucose uptake in skeletal muscle.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-011-2046-3) contains supplementary material, which is available to authorised users.
Keywords: AKT, Glucose tolerance, Metabolism, Nuclear hormone receptor, Retinoic acid receptor-related orphan receptor alpha
Introduction
Retinoic acid receptor-related orphan receptor (ROR) alpha is a member of the nuclear hormone receptor (NR) superfamily. NRs function as ligand-dependent DNA binding factors that translate hormonal, nutritional and pathophysiological signals into gene regulation. In the context of metabolism, NRs control lipid and glucose homeostasis in an organ- or tissue-specific manner [1–4]. It has been demonstrated that liver x receptor agonists and oxygenated sterols function as RORα/γ ligands that can suppress receptor activity [5].
Several studies on RORα [2–4, 6, 7] have revealed a role for this NR in fatty acid homeostasis [8]. Homozygous staggerer (sg/sg) mice, which have a global Rorα (also known as Rora) defect, display decreased and dysfunctional RORα (1 and 4) levels in all tissues [7, 9–11]. Moreover, sg/sg mice have a lean and dyslipidaemic phenotype, and are similar to Rorα-deficient mice (sg operates as a non-functional null allele) [12]. These mice display hypoalphalipoproteinaemia, decreased serum triacylglycerol and HDL-cholesterol [3], vulnerability to atherosclerosis [3], decreased fat depots and resistance to diet-induced obesity [9].
Dysfunctional NR signalling has been associated with dyslipidaemia, insulin resistance, diabetes and obesity [6, 7]. For example, the dietary lipid-dependent NRs (peroxisome proliferator-activated receptor-gamma, liver x receptor alpha etc.) in skeletal muscle regulate insulin-stimulated glucose disposal, and lipid and energy homeostasis [13–15]. Muscle is a major mass tissue that accounts for around 40% of the total body mass and energy expenditure, and is a major site of glucose disposal. Hence muscle has an important role in regulating insulin sensitivity and is increasingly recognised as an important target for combating type 2 diabetes.
Type 2 diabetes is a complex metabolic disease characterised by glucose intolerance and insulin resistance, which result in decreased insulin-mediated glucose utilisation in peripheral tissues [16]. In this context, investigation of the role of the orphan NR, RORα, in the regulation of glucose metabolism has not been completely analysed in the literature. Accordingly, we surveyed the functional role of RORα in glucose tolerance and insulin sensitivity, with particular attention to skeletal muscle.
We observed that homozygous (sg/sg) and heterozygous (sg/+) mice exhibited mildly improved glucose tolerance and increased insulin sensitivity. Expression profiling identified perturbations in the v-AKT murine thymoma viral oncogene homologue (AKT) signalling cascade and in genes involved in the control of glucose homeostasis. For example, we observed enhanced (absolute) levels of: (1) Akt2 mRNA in the basal state; and (2) phosphorylated AKT (pAKT) (in the basal and insulin stimulated states); and (3) (ex vivo) glucose uptake in skeletal muscle from sg/sg mice relative to wild-type littermates. In summary, RORα plays an important role in regulation of the AKT signalling cascade.
Methods
Animals
Heterozygous sg/+ mice, B6.C3(Cg)-Rora sg/J, were obtained from Jackson laboratory (Bar Harbor, ME, USA) and maintained by backcrossing to C57BL/6 J. Feeding has been described previously [9]. Male wild-type and homozygous (sg/sg) mice were fed with a normal diet until 14 weeks and fasted for 6 h by transferring to a food-free holding cage with access to water. Animals were killed and tissues collected and immediately frozen in liquid nitrogen. All animal procedures were approved by The University of Queensland Animal Ethics Committee.
Glucose tolerance test
Blood glucose measurements were obtained from the tail vein of 12 h fasted animals (14–16 weeks old) following glucose challenge, using a blood glucose testing system (Accu-chek Performa; Roche Diagnostics, Castle Hill, NSW, Australia) [17].
Insulin tolerance test and insulin stimulation of AKT phosphorylation
Blood glucose measurements were obtained from 6 h fasted animals (14–16 weeks old) following insulin challenge, as previously described [17]. For measurement of skeletal muscle pAKT, 6 h fasted mice were administered insulin or saline as described for insulin tolerance test, killed by CO2 asphyxiation after 10 min and quadriceps muscle excised and snap-frozen.
Microarray analysis
Microarray and statistical analysis were as previously described [17] except that GeneSpring GX v7.3.1 (Agilent, Santa Clara, CA, USA) was used for analysis.
RNA extraction, cDNA synthesis and quantitative PCR
Total RNA extraction, cDNA synthesis and quantitative PCR were performed as previously described [2, 9]. Primers are listed in the Electronic supplementary material (ESM) Table 1.
Protein extraction and western blot analysis
Protein extraction and western analysis was performed as previously described [17] with the following modifications. The lysis buffer contained 50 mmol/l Tris (pH 8.0), 150 mmol/l NaCl, 1% (vol./vol.) Triton X-100, 1% (wt/vol.) NP40 and 0.1% (wt/vol.) SDS, and tissue was centrifuged at 2,000×g. Membranes were blocked with either 5% (wt/vol.) dried skimmed milk or 5% (wt/vol.) BSA (for phospho-specific antibodies). The anti-GLUT4 (number 62375; Abcam, Cambridge, MA, USA) antibody was used at 1:5,000.
2-Deoxy-d-glucose uptake assay
Extensor digitorum longus (EDL) muscles were dissected from mice (3–7 months old) following cervical dislocation and 2-deoxy-d-glucose uptake assay was performed with or without 10 nmol/l insulin stimulation (Actrapid; Nova Nordisk, Baulkham Hills, NSW, Australia) as previously described [18, 19].
Indirect calorimetry
Respiratory exchange ratio (RER) was measured using a monitoring system (Oxymax Comprehensive Lab Animal System; Columbus Instruments, Columbus, OH, USA). Mice were housed individually in metabolic cages on a 12 h day–night cycle. Following an initial 14 h acclimatisation period, measurements were taken every 10 min for 24 h on normal diet (D12450B; Research Diets, New Brunswick, NJ, USA). After 24 h measurement, food was then withdrawn and measurements were taken for a further 6 h.
Statistical analysis
Results were analysed by unpaired t test or one-way ANOVA with Bonferroni’s post-hoc test, or by two-way ANOVA with Bonferroni’s post hoc test; Prism 5 software (GraphPad, San Diego, CA, USA) was used.
Results
Staggerer mice present decreased fasting glucose levels, improved glucose tolerance and increased insulin sensitivity
We have previously demonstrated that homozygous staggerer (sg/sg) mice (with decreased and dysfunctional RORα levels) display a lean phenotype. Here, we used sg/sg mice to elucidate the role of RORα in glucose tolerance and insulin sensitivity. We examined male wild-type and homozygous sg/sg mice fed a normal diet until 14 weeks and observed significantly lower blood glucose levels in homozygous sg/sg than in wild-type mice (n = 7–11) after 6 and 12 h of fasting (Fig. 1a, b). Plasma insulin levels in the sg/sg, relative to wild-type mice were also decreased (but did not attain significance, p = 0.08) (Fig. 1c). We further examined systemic glucose metabolism and performed intraperitoneal glucose tolerance tests. In the sg/sg mice, glucose clearance was mildly improved following a glucose challenge (Fig. 1d, e), with no significant differences in insulin secretion after 10 min (data not shown). The sg/sg mice also displayed significantly enhanced responsiveness to administration of intraperitoneal insulin, indicating enhanced whole-body insulin action compared with wild-type mice (Fig. 1f, g).
Fig. 1.
a Starting blood glucose levels of 6 h fasted male homozygous sg/sg (black column), heterozygous sg/+ (het) (striped column) and wild-type (white column) mice. These mice were used for insulin tolerance test (f, g); n = 5–9 per group, mean ± SEM; **p < 0.01 by one-way ANOVA with Bonferroni’s post hoc test. b Starting blood glucose levels of 12 h fasted mouse groups as above (a). These mice were used for GTT (d, e); n = 6–11 per group, mean ± SEM; **p < 0.01 by one-way ANOVA with Bonferroni’s post hoc test. c Plasma insulin concentrations of 12 h fasted male sg/sg wild-type littermate control mice on a regular chow diet; n = 4 per group, mean ± SEM. d, e Glucose tolerance tests. Blood glucose concentrations were measured at various times (d) after i.p. administration of glucose (t = 0) to 12 h fasted male sg/sg (black squares), sg/+ (het) (circles) and wild-type (Wt) (white squares) littermate control mice; n = 6–11 per group, mean ± SEM; *p < 0.05 by two-way ANOVA with Bonferroni’s post hoc test. e Mean AUC from glucose tolerance test; mean ± SEM; *p < 0.05 by one-way ANOVA with Bonferroni’s post hoc test. f, g Insulin tolerance tests. Blood glucose concentrations were measured and expressed as percentage of starting glucose (t = 0) at various times (f) after i.p. administration of insulin (t = 0) to 6 h fasted male sg/sg (black squares), sg/+ (het) (circles) and wild-type littermate control (white squares) mice; n = 5–9 per group, mean ± SEM; *p < 0.05, **p < 0.01 and ***p < 0.001 by two-way ANOVA with Bonferroni’s post hoc test. g Mean AUC from insulin tolerance test; mean ± SEM; *p < 0.05, **p < 0.01 by one-way ANOVA with Bonferroni’s post-hoc test
Homozygous sg/sg mice are characteristically lean. They display decreased adipose mass and body weight [9]. In contrast, the size:weight ratio of heterozygote (sg/+, haploinsufficient) mice is comparable to that of their wild-type counterparts (ESM Fig. 1a, b). The heterozygous sg/+ mice provided a tool to investigate whether the improvement in insulin sensitivity observed in sg/sg mice involved differential adiposity. Therefore we investigated whether glucose clearance and/or insulin sensitivity were altered in sg/+ mice. We observed significant improvement in glucose tolerance and insulin sensitivity compared with wild-type mice (Fig. 1d–g), indicating that the increased insulin sensitivity was independent of the animal’s degree of adiposity (and weight).
We also explored whether the improvement in insulin sensitivity involved a shift to (more insulin-sensitive) type I/slow fibres and/or oxidative type IIa/x fibres. Accordingly, we examined the four myosin heavy chain genes, Myh1, -2, -4 and -7 (in wild-type and sg/sg mice), which identify and classify types I, IIa, IIb and IIx fibres, respectively (ESM Fig. 2a–d). In addition, we also examined mRNA expression of the genes encoding type I and II troponin I's (Tnni1, Tnni2) (ESM Fig. 2e, f). There were no significant shifts in the mRNA expression of genes encoding any of these fibre type markers. This suggests that improved insulin sensitivity was independent of transitions in muscle fibre types.
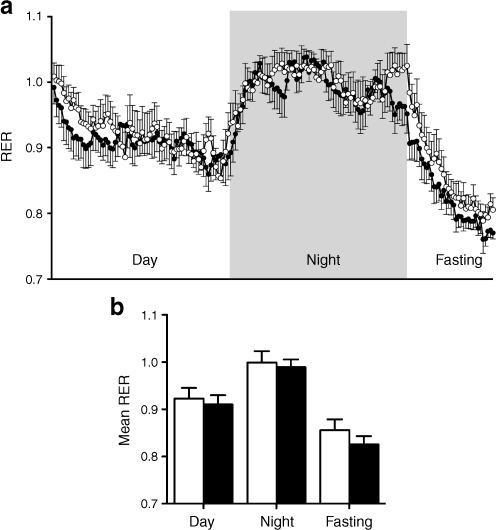
Whole-body metabolic analysis indicated that RER in wild-type and sg/sg mice was similar (in the fed or dark, and fasted phases), suggesting that these lines use fats and carbohydrates for energy to a similar degree in the dark and light phases (Fig. 2a, b). The sg/sg mice are characteristically lean [9] and resistant to diet-induced obesity [9]. We also showed (ESM Fig. 1c, d) that on the regular chow diet the wild-type and sg/sg littermates ate similar quantities of food. O2 consumption in the sg/sg relative to wild-type mice was significantly increased. These observations are consistent with previous reports that demonstrated a 30 to 70% higher metabolic rate (depending on temperature) and higher endogenous levels of norepinephrine in (cold-reared) sg/sg mice relative to control mice [20], and increased β 2- AR (also known as Adrb2) mRNA expression in brown adipose tissue from sg/sg mice [9]. In summary, the sg/sg mice displayed mildly improved glucose tolerance and increased insulin sensitivity.
Fig. 2.
CLAMS Oxymax metabolic monitoring of sg/sg (black circles and columns) and wild-type littermate control (white circles and columns) mice. a RER measurement over 12 h (day) and 12 h (night) with food and 6 h fasting. b Mean ± SEM for RER measurement in specified phases (a). n = 8–11 per group
Expression profiling of skeletal muscle from sg/sg mice identified differential expression of genes involved in AKT signalling, and in cardiovascular and metabolic disease
To probe the mechanisms underlying improved insulin sensitivity, we performed expression profiling analysis using BeadArray chips (46 K; Illumina Sentrix; Illumina, San Diego, CA, USA). Muscle (quadriceps) RNA from three male sg/sg mice and their wild-type littermates was utilised for analysis. Statistically significant differential up- and downregulation of annotated genes was determined on the basis of a p value cut-off of p < 0.05 and a fold change cut-off of 1.2 as shown in Tables 1 and 2, respectively (for the complete list, see ESM Fig. 3a, b, c). All differentially expressed genes were analysed using Ingenuity Pathway Analysis (www.ingenuity.com) to examine their putative function and associated signalling pathways.
Table 1.
Upregulated annotated genes in sg/sg mice
| Gene symbol | Gene name | Genbank ID | Fold change | p value |
|---|---|---|---|---|
| Plekhh1 | Pleckstrin homology domain containing, family H (with Myth4 domain) member 1 | XM_126961.4 | 1.97 | 0.002 |
| Cidea | Cell death-inducing DNA fragmentation factor, alpha subunit-like effector a | NM_007702.1 | 1.69 | 0.039 |
| Dnase1 | Deoxyribonuclease 1 | NM_010061.2 | 1.64 | 0.005 |
| Ddit4 | DNA-damage-inducible transcript 4 | NM_029083.1 | 1.57 | 0.018 |
| Synj2 | Synaptojanin 2 | AK038038 | 1.56 | 0.008 |
| Tph1 | Tryptophan hydroxylase 1 | NM_009414.1 | 1.50 | 0.003 |
| Cdkn1a | Cyclin-dependent kinase inhibitor 1a (p21) | NM_007669.2 | 1.49 | 0.026 |
| Egln3 | EGL nine homologue 3 | AK033509 | 1.49 | 0.044 |
| Gng2 | Guanine nucleotide binding protein (G protein), gamma 2 subunit | NM_010315.2 | 1.48 | 0.000 |
| Klk1b26 | Kallikrein 1-related petidase b26 | NM_010644.2 | 1.44 | 0.012 |
| Akt2 | Thymoma viral proto-oncogene 2 | NM_007434.2 | 1.44 | 0.036 |
| Tns3 | Tensin-3 | NM_001083587 | 1.44 | 0.027 |
| Mt2 | Metallothionein 2 | NM_008630.1 | 1.43 | 0.024 |
| Igh-1a | Immunoglobulin heavy chain 1a | XM_354704.1 | 1.43 | 0.044 |
| Bcl2 | B cell leukaemia/lymphoma 2 | NM_009741.2 | 1.43 | 0.016 |
| Chrna9 | Cholinergic receptor, nicotinic, alpha polypeptide 9 | XM_132045.2 | 1.40 | 0.030 |
| Snrk | SNF related kinase | NM_133741.1 | 1.40 | 0.050 |
| Acot11 | Acyl-CoA thioesterase 11 | NM_025590.4 | 1.39 | 0.033 |
| Slc25a25 | Solute carrier family 25, member 25 | NM_146118.2 | 1.39 | 0.040 |
| Syt12 | Synaptotagmin 12 | NM_134164.2 | 1.38 | 0.009 |
| Foxo1 | Forkhead box o1 | NM_019739.2 | 1.37 | 0.040 |
| Cdkn1a | Cyclin-dependent kinase inhibitor 1a (p21) | NM_007669.2 | 1.36 | 0.035 |
| Tsc22d1 | TSC22 domain family, member 1 | NM_009366.3 | 1.36 | 0.015 |
| Tsc22d3 | TSC22 domain family, member 3 | NM_010286.2 | 1.36 | 0.013 |
| Igfbp3 | Insulin-like growth factor binding protein 3 | NM_008343.1 | 1.35 | 0.015 |
| Notch4 | Notch gene homologue 4 (Drosophila) | NM_010929.1 | 1.34 | 0.010 |
| Car2 | Carbonic anhydrase 2 | NM_009801.3 | 1.34 | 0.005 |
| Mt3 | Metallothionein 3 | NM_013603.1 | 1.32 | 0.002 |
| Tbrg4 | Transforming growth factor beta regulated gene 4 | NM_134011.1 | 1.32 | 0.043 |
| Tbc1d1 | TBC1 domain family, member 1 | NM_019636.1 | 1.31 | 0.004 |
| Map3k6 | Mitogen-activated protein kinase 6 | NM_016693 | 1.31 | 0.046 |
| Sox13 | SRY-box containing gene 13 | NM_011439.1 | 1.31 | 0.016 |
| Shcbp1 | Shc SH2-domain binding protein 1 | NM_011369.1 | 1.30 | 0.033 |
| Lmcd1 | LIMand cysteine-rich domains 1 | NM_144799.1 | 1.29 | 0.009 |
| Syn2 | Synapsin II | NM_013681.1 | 1.29 | 0.017 |
| Ercc5 | Excision repair cross-complementing rodent repair deficiency, complementation group 5 | NM_011729.1 | 1.29 | 0.040 |
| Syt3 | Synaptotagmin 3 | NM_016663.2 | 1.29 | 0.035 |
| Car4 | Carbonic anhydrase 4 | NM_007607.1 | 1.28 | 0.003 |
| Fgfr2 | Fibroblast growth factor receptor 2, transcript variant 1 | NM_010207.1 | 1.28 | 0.013 |
| Dlk1 | Delta-like 1 homologue (Drosophila) | NM_010052.1 | 1.27 | 0.005 |
| Itga10 | Integrin, alpha 10 | XM_112192 | 1.27 | 0.026 |
| Klk1b26 | Kallikrein 1-related petidase b26 | NM_010644.2 | 1.26 | 0.013 |
| Plin3 | Perilipin 3 | NM_025836.3 | 1.26 | 0.004 |
| Prom1 | Prominin 1 | NM_008935.1 | 1.26 | 0.002 |
| Zbtb32 | Zinc finger and BTB domain containing 32 | NM_021397.2 | 1.26 | 0.041 |
| Sh3bp2 | SH3-domain binding protein 2 | NM_011893 | 1.26 | 0.012 |
| Car14 | Carbonic anhydrase 14 | NM_011797.1 | 1.25 | 0.001 |
| Olfr420 | Olfactory receptor 420 | NM_146305.1 | 1.25 | 0.033 |
| Fkbp5 | FK506 binding protein 5 | NM_010220.2 | 1.25 | 0.029 |
| Lao1 | l-amino acid oxidase 1 | NM_133892.3 | 1.25 | 0.017 |
Table 2.
Downregulated annotated genes in sg/sg mice
| Gene symbol | Gene name | Genbank ID | Fold change | p value |
|---|---|---|---|---|
| Cpeb2 | Cytoplasmic polyadenylation element binding protein 2 | NM_175937 | −1.79 | 0.007 |
| Opn1mw | Opsin 1 (cone pigments), medium-wave-sensitive (colour blindness, deutan) | NM_008106.1 | −1.70 | 0.026 |
| Stab2 | Stabilin 2 | NM_138673.1 | −1.69 | 0.000 |
| Sfrp2 | Secreted frizzled-related sequence protein 2 | NM_009144.1 | −1.65 | 0.025 |
| Col7a1 | Procollagen, type VII, alpha 1 | NM_007738.1 | −1.64 | 0.001 |
| Pla2g7 | Phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) | NM_013737.2 | −1.52 | 0.006 |
| Hspa1a | Heat shock protein 1a | NM_010479 | −1.50 | 0.034 |
| Cadm1 | Cell adhesion molecule 1, transcript variant 3 | NM_018770.3 | −1.50 | 0.011 |
| Cadm1 | Cell adhesion molecule 1, transcript variant 3 | NM_018770.3 | −1.50 | 0.026 |
| Cldn10 | Claudin 10, transcript variant 2 | NM_021386 | −1.47 | 0.011 |
| Cited4 | Cbp/P300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 | NM_019563 | −1.45 | 0.046 |
| Pogk | Pogo transposable element with KRAB domain | NM_175170 | −1.45 | 0.007 |
| Tiam1 | T cell lymphoma invasion and metastasis 1 | NM_009384.1 | −1.44 | 0.002 |
| Ptx3 | Pentaxin related gene | NM_008987.2 | −1.38 | 0.042 |
| Phex | Phosphate regulating gene with homologies to endopeptidases on the X chromosome | NM_011077.1 | −1.37 | 0.046 |
| Ifitm6 | Interferon induced transmembrane protein 6 | XM_133956.3 | −1.36 | 0.036 |
| Gne | Glucosamine | NM_015828 | −1.36 | 0.015 |
| Fbn1 | Fibrillin 1 | AK080935 | −1.34 | 0.020 |
| Pon1 | Paraoxonase 1 | NM_011134.1 | −1.34 | 0.020 |
| Exph5 | Exophilin 5 | NM_176846.3 | −1.34 | 0.021 |
| Rasd2 | RASD family, member 2 | XM_204287.3 | −1.33 | 0.005 |
| Adamts1 | A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 1 | NM_009621 | −1.33 | 0.026 |
| Timd4 | T cell immunoglobulin and mucin domain containing 4 | NM_178759.3 | −1.33 | 0.032 |
| Cd209d | CD209 antigen-like protein d | NM_130904.2 | −1.33 | 0.027 |
| Neto2 | Neuropilin (NRP) and tolloid (TLL)-like 2 | XM_134498 | −1.33 | 0.050 |
| Tekt1 | Tektin 1 | NM_011569.1 | −1.33 | 0.002 |
| Retnlg | Resistin like gamma | NM_181596.2 | −1.32 | 0.047 |
| Avil | Advillin | NM_009635.2 | −1.32 | 0.003 |
| Fgf10 | Fibroblast growth factor 10 | NM_008002.3 | −1.31 | 0.036 |
| Hist1h4b | Histone 1, H4b | NM_178193 | −1.31 | 0.016 |
| Jph2 | Junctophilin 2 | NM_021566.1 | −1.30 | 0.042 |
| Arhgap20 | Rho Gtpase activating protein 20 | NM_175535 | −1.30 | 0.017 |
| Fancl | Fanconi anaemia, complementation group l | NM_025923.2 | −1.30 | 0.027 |
| Cyhr1 | Cysteine and histidine rich 1 | NM_019396 | −1.30 | 0.037 |
| Tinag | Tubulointerstitial nephritis antigen | NM_012033.2 | −1.30 | 0.017 |
| Dguok | Deoxyguanosine kinase | AK003639 | −1.29 | 0.005 |
| Rora | Rar-related orphan receptor alpha | NM_013646.1 | −1.29 | 0.018 |
| Stat1 | Signal transducer and activator of transcription 1 | BC057690.1 | −1.29 | 0.026 |
| Csf-1 | Colony stimulating factor 1 | X05010 | −1.28 | 0.047 |
| Ubtf | Upstream binding transcription factor, RNA polymerase i | NM_011551 | −1.28 | 0.020 |
| Atxn7l2 | Ataxin 7 like protein 2 | NM_175183.2 | −1.27 | 0.023 |
| Arhgap20 | Rho Gtpase activating protein 20 | NM_175535 | −1.27 | 0.038 |
| Cd209a | CD209a antigen | NM_133238.2 | −1.27 | 0.023 |
| Aqp9 | Aquaporin 9 | NM_022026.2 | −1.26 | 0.009 |
| Dpysl3 | Dihydropyrimidinase-like 3 | NM_009468.1 | −1.26 | 0.021 |
| Enpp2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 | NM_015744 | −1.26 | 0.000 |
| Ache | Acetylcholinesterase | NM_009599.2 | −1.26 | 0.016 |
| Cul9 | Cullin 9 | NM_001081335 | −1.26 | 0.039 |
| Ube2d1 | Ubiquitin-conjugating enzyme E2D 1, UBC4/5 homologue (yeast) | NM_145420.1 | −1.26 | 0.027 |
| Astn2 | Astrotactin 2, transcript variant 1 | NM_207109.1 | −1.26 | 0.049 |
The phosphatidylinositol 3-kinase (PI3K)–AKT signalling pathway was the primary pathway to be upregulated (ESM Fig. 4c) in sg/sg mice. We focused on the PI3K–AKT pathway to better understand the role of RORα (and the effect of RORα deficiency) on skeletal muscle carbohydrate metabolism. Importantly, using Ingenuity Pathway Analysis, this pathway also had the highest ranking association with genes that are significantly upregulated in bead-array (Illumina) analysis. For example, the PI3K–AKT pathway had the highest ratio (4.4 × 10−2), i.e. calculation of the number of genes in a pathway that are differentially expressed (in a significant manner) divided by the total number of known genes in that pathway. Second, it displayed a very significant p value (1.1 × 10−4), i.e. confidence of the association between the pathway and the differentially expressed genes.
This is consistent with a phenotype displaying improved insulin sensitivity. The majority of differentially expressed genes were associated with metabolic disease, diabetes, dyslipidaemia and obesity (ESM Fig. 5).
Akt2 expression is increased in staggerer (sg/sg) mice
The Ingenuity analysis identified PI3K–AKT signalling as the most significantly increased pathway. In the context of further elucidating the mechanisms involved in the control of glucose tolerance in the skeletal muscle of sg/sg mice, we used quantitative PCR to validate the differential expression of mRNAs encoding Akt in skeletal muscle, liver and adipose tissue of male sg/sg mice relative to wild-type littermates (see Fig. 3a–l).
Fig. 3.
Quantitative PCR of Akt1, Akt2 (variant [var] 1 and var 2) and Akt3 mRNA in (a–d) quadriceps muscle, (e–h) liver and (i–l) epididymal adipose tissue of male sg/sg mice (black columns) relative to wild-type littermates (white columns). n = 7–8 per group, ***p < 0.001 by t test. m Schematic representation of genomic structure of Akt2 var 1 and Akt2 var 2, and use of alternative promoter. Excerpt from MGI mouse genome browser at website as cited [45]. n–p Quantitative PCR of Akt1, Akt2 var1 and Ak2 var 2 mRNA in quadriceps muscle of male wild-type (white columns), heterozygous sg/+ (striped columns) and homozygous sg/sg mice (black columns) (3–7 months old). n = 10–12 per group, *p < 0.05, **p < 0.01 and ***p < 0.001 by one-way ANOVA with Bonferroni’s post hoc test. All bar graph values mean ± SEM, with relative mRNA expression normalised against 18S rRNA
We observed significantly increased levels of Akt1 and Akt2 (variant 2) mRNA, but not of Akt3 mRNA in muscle from (fasted) sg/sg mice (Fig. 3a–d). Interestingly, two variants of Akt2 have been described in the NCBI database (ncbi.nlm.nih.gov/nucleotide, accessed 17 December 2010). Variants 1 and 2 are produced by alternate promoter usage and result in exon 1 encoding different 5′ untranslated region sequences (Fig. 3m). Variant 2, but not variant 1, mRNA was increased in staggerer mice (Fig. 3b, c). Notably, no significant changes in Akt1, Akt2 (variants 1 and 2) or Akt3 mRNA expression were observed in liver or adipose tissue (Fig. 3e–i).
Levels of Rorα mRNA in the muscle of heterozygous staggerer mice are approximately half of those in wild-type mice (ESM Fig. 1b). To understand further the effect of Rorα expression on transcription of Akt isoforms, we analysed the expression of Akt1, Akt2 variant 1 and Akt2 variant 2 mRNAs in the heterozygote (RORα haplo-insufficient mice). The expression of Akt2 variant 2 (but not variant 1) mRNA in quadriceps muscle in heterozygous mice was intermediate between wild-type and homozygous staggerer (sg/sg) mice (Fig. 3o, p). In contrast Akt1 mRNA expression was similar to that of wild-type mice (Fig. 3n).
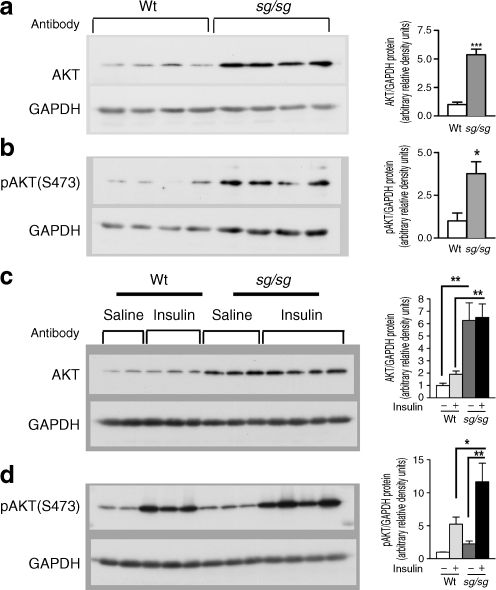
We also determined by western blot analysis whether changes in mRNA expression correlated with changes in the protein levels of total AKT (Fig. 4a) and phosphorylated (serine amino acid residue 473 [ser473]) AKT (Fig. 4b) in quadriceps muscle of wild-type and sg/sg mice in the basal (unstimulated) state. We observed a significant increase in total AKT protein (normalised against glyceraldehyde-3-phosphate dehydrogenase [GAPDH] protein) in the skeletal muscle from sg/sg mice relative to wild-type littermate pairs (Fig. 4a), corresponding with the mRNA expression data. In addition, levels of the active phosphorylated (ser473) AKT species was also significantly increased (normalised against GAPDH) (Fig. 4b) in the basal condition.
Fig. 4.
Western blot analyses of (a) AKT and (b) pAKT relative to GAPDH in skeletal muscle (quadriceps) of male sg/sg and wild-type (WT) littermate control mice, with densitometry analysis of western blots; n = 4 per group, mean ± SEM; *p < 0.05 and ***p < 0.001 by t test. c Western blot analyses (representative blot from two experiments) of AKT and (d) pAKT relative to GAPDH in skeletal muscle (quadriceps) of male wild-type (Wt) and sg/sg littermate mice treated with saline and insulin (0.5 U/kg i.p. for 10 min). Densitometry was analysed using both experiments; n = 4–6 per group, mean ± SEM; *p < 0.05 and **p < 0.01 two-way ANOVA with Bonferroni’s post hoc test
Aberrant expression in sg/sg mice of genes involved in regulation of glucose tolerance and metabolic disease
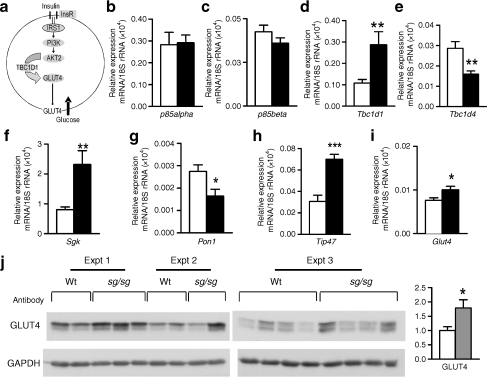
The nodes of the insulin signalling pathway (underscoring the central role of AKT2) are summarised in Fig. 5a. We used quantitative PCR to analyse mRNA expression of the p85α (also known as Pik3r1) and p85β (also known as Pik3r2) regulatory subunits of PI3K. However, no significant differences in expression were observed in the muscle of male sg/sg mice (Fig. 5b, c).
Fig. 5.
a Schematic representation of the insulin-mediated glucose disposal pathway in skeletal muscle. InsR, insulin receptor. b–i Quantitative PCR of mRNA from skeletal muscle (quadriceps) of male sg/sg (black columns) mice relative to wild-type (Wt) littermates (white columns) for genes as labelled. Relative mRNA expression was normalised against 18S rRNA; n = 7–8 per group, mean ± SEM; *p < 0.05, **p < 0.01 and ***p < 0.001, by t test. j Western blot analyses of GLUT4 in skeletal muscle (quadriceps) from male sg/sg and wild-type (WT) littermate control mice, with densitometry analysis (grey, sg/sg; white, wild-type littermate control) using both western blots; n = 8–9 per group, mean ± SEM; *p < 0.05 by t test
We also validated the differential expression of Tbc1d1 (which encodes a Rab GTPase-activating protein, and the paralogue Tbc1d4 [also known as As160]) and several other genes in the sg/sg mice (relative to wild-type littermates). These genes have been implicated in AKT signalling and the regulation of glucose tolerance and/or insulin sensitivity. For example, TBC1D1 has recently been identified (as an AKT substrate) in the insulin-stimulated AKT cascade that facilitates GLUT4 translocation [21] and regulates glucose uptake [22]. The Tbc1d1 and Tbc1d4 mRNA were significantly induced (about threefold) and decreased, respectively (Fig. 5d, e) in sg/sg mice. Serum- and glucocorticoid-inducible kinase 1 (Sgk1) was similarly induced by about threefold (Fig. 5f). Serum- and glucocorticoid-inducible kinase 1 modulates salt-dependent glucose uptake and mediates several PI3K responses, with emerging evidence suggesting it interacts with AKT kinases and mammalian transducer of regulated cAMP response element-binding protein 2 [23]. Polymorphisms (and increases) of paraoxonase 1 are associated with glucose intolerance and type 2 diabetes, and, consistent with the phenotype, was decreased by about twofold (Fig. 5g). Finally, Tip47 (also known as Plin3) in skeletal muscle has been shown to maintain insulin sensitivity [24–26]; its mRNA was significantly induced by about 2.5-fold (Fig. 5h).
With regard to glucose transporters, we observed no changes in expression of Glut1 (also known as Slc2a1) mRNA (data not shown); however, we did observe a small but significant increase in Glut4 (also known as Slc2a4) mRNA expression (Fig. 5i). The increase in Glut4 mRNA was reflected in a significant, approximately twofold increase in the level of GLUT4 protein after western blot analysis of sg/sg mice, relative to wild-type littermates (n = 9; Fig. 5j).
The differential expression of these genes in muscle of sg/sg mice was consistent with improved insulin sensitivity, and with increased AKT levels and AKT phosphorylation. These analyses thus revealed that several components of the insulin signalling pathway are modulated in the sg/sg phenotype, these modulations being associated with decreased blood glucose and improved insulin signalling.
Staggerer mice display significantly elevated 'absolute levels' of pAKT and total AKT protein in the basal and insulin-stimulated states, and have enhanced ex vivo glucose uptake
We subsequently examined whether sg/sg mice display increased insulin-mediated AKT phosphorylation. Whole-cell extracts from quadriceps muscles were isolated from saline- and insulin-injected mice were analysed by western blot analysis using antibodies specific to AKT and pAKT. We observed significantly elevated 'absolute levels' of total AKT protein (in the basal and insulin-stimulated states) in quadriceps muscle from sg/sg mice (normalised against GAPDH) (Fig. 4c). We also observed significantly elevated 'absolute levels' of insulin-stimulated phosphorylation of AKT (ser473) in sg/sg mice (normalised against GAPDH) (Fig. 4d).
We next investigated whether the improved insulin sensitivity and elevated 'absolute' levels of AKT and pAKT in sg/sg mice resulted in increased ex-vivo glucose uptake in EDL muscle (type II fast twitch glycolytic muscle) (Fig. 6). In keeping with the increase in AKT level and phosphorylation, EDL muscle of sg/sg mice showed significantly higher rates of 2-deoxy-d-glucose uptake than wild-type mice in the basal and insulin-stimulated state.
Fig. 6.
2-Deoxy-d-[ 3H]glucose uptake assays of control and insulin-treated EDL muscle explants from male wild-type (Wt) and sg/sg mice (3–7 months old); n = 7–10 per group, mean ± SEM; *p < 0.05 and **p < 0.01 By two-way ANOVA with Bonferroni’s post hoc test
Discussion
A recent review [8] underscored the diverse roles of RORs in development, immunity, circadian rhythm, lipid homeostasis and metabolism. Moreover, (dysfunctional) levels of this NR are associated with ataxia, autoimmune disease, dyslipidaemia, atherosclerosis and resistance to diet-induced obesity. The sg/sg mice are Rorα-deficient, sg/sg is a non-functional null allele and sg/sg mice do not express functional RORα [12]. Moreover, sg/sg mice display identical phenotypes to the Rorα-knockout mice, which do not produce RORα protein [12, 27].
The pathophysiological phenotypes in sg/sg mice are consistent with changes in the expression of genes such as Srebp-1c (also known as Srebf1), Apoa1, Apoc3 and IκBα (also known as Nfkbia) [4, 7, 9, 28]. We have previously shown that sg/sg mice have significantly reduced Rorα1 and Rorα4 expression [9], and display increased expression of Lipin 1 and Pgc-1α (also known as Ppargc1a), and decreased expression of Srebp-1c. The differential expression of these genes is consistent with the ability of sg/sg mice to resist diet-induced obesity and hepatic triacylglycerol accumulation [9].
To date, the role of RORα in the control of insulin sensitivity and glucose uptake remains obscure. The links and interactions between fatty acid and carbohydrate homeostasis in whole-body physiology, and in the pathophysiological development of glucose intolerance and insulin resistance underscore the need to examine the role of this NR in carbohydrate metabolism. Consequently, we examined the outcomes of aberrant RORα function in muscle, a tissue that accounts for the bulk of insulin-stimulated glucose disposal [29]. The (male) sg/sg mice present with significantly reduced fasting serum glucose and improved insulin sensitivity relative to wild-type littermates. Our results differ from those of Kang et al. who showed a lack of serum glucose phenotype in sg/sg compared with their wild-type mice [30]. The nature of this difference is still not known. Recently it has been suggested that, in mice, a 6 h fast provides a more physiologically relevant baseline, increased sensitivity and fewer confounding variables for the evaluation of serum glucose and glucose tolerance [31, 32].
Expression profiling identified differential regulation of genes in the skeletal muscle of sg/sg mice. Similarly, bi-directional changes in gene expression have been observed in liver of sg/sg mice [30]. In the analysis reported here, the upregulated subset of genes was associated with cardiovascular and metabolic disease, cell death, infection, immunity and inflammation. This is consistent with the defined role of RORα in atherosclerosis, lipid homeostasis, fat deposition, obesity and inflammation [8, 33]. Interestingly, the primary signalling pathway upregulated in muscle was the PI3K–AKT pathway, a central control point for insulin-stimulated glucose uptake. Two RNA variants of Akt2 have been identified. We showed that Akt2 variant 2 was highly expressed in epididymal fat and (quadricep) skeletal muscle, but not in liver of wild-type mice. We observed significant differential expression of Akt1 and Akt2 variant 2 in quadriceps of sg/sg mice, indicating a specific role for these signalling molecules in skeletal muscle.
Interestingly, we had previously demonstrated that transgenic muscle-specific overexpression of dominant-negative RORα1 leads to decreased Akt2 mRNA expression, glucose intolerance, decreased insulin-stimulated phosphorylation of AKT and glucose uptake [17]. In this study, we observed that decreased (and dysfunctional) expression of Rorα (1 and 4) led to increased Akt2 mRNA expression and improved insulin sensitivity, and increased levels of basal and insulin-stimulated phosphorylation of AKT and of glucose uptake.
Notably in this context, the staggerer strain has a deletion of the fifth exon encoding the ligand binding domain. The resulting frame shift encodes 'non-sense RNA' and introduces a premature stop codon. However, further analysis by northern blot [34] and RT-PCR [9] revealed greater than tenfold decreases in Rorα1 and Rorα4 mRNA expression. Moreover, northern blot analysis [12] was not able to detect the transcript in homozygous sg/sg mice. Gold et al. [34] state that this is consistent with ‘degradation of the mutant mRNA through the non-sense mediated decay pathway’. In agreement with this, sg/sg mice do not produce the RORα protein. Although initially thought to have a dominant-negative form of RORα, sg/sg mice are in fact Rorα-deficient. In contrast to this, our previous study [17] used a construct that abundantly/ectopically expresses a stable truncated Rorα mRNA transcript with an in-frame stop codon. The truncated Rorα ∆DE includes the encoded amino acids 1–235 [17], but lacks the ligand binding domain and part of the hinge region. McBroom et al. [35] and Lau et al. [2] reported that deletion of this segment preserved DNA recognition and binding, suppressed transactivation and operated in a dominant-negative manner. In addition, Hamilton et al. [11] reported that the staggerer mutation is located in a similar position and produces a non-functional ligand binding domain.
This apparent paradox in ROR function has a precedence in the NR field and there are a number of factors to consider. In the first instance, our previous transgenic mouse model overexpressed a dominant-negative Rorα1 transcript, specifically in skeletal muscle [17]. In comparison, the present study investigated sg/sg mice, which lack Rorα1 and Rorα4 expression in all tissues. Second, in mice lacking NR, ‘bona fide’ target genes are often derepressed, resulting in paradoxically increased expression [36]. We suggest the data are consistent with decreased (and dysfunctional) RORα expression leading to derepression and increase of Akt2 mRNA expression. This is consistent with our observation that RORα is selectively recruited to the Akt2 promoter and that dominant-negative overexpression represses Akt2 mRNA expression [17]. More importantly, these studies clearly identify a link between Rorα expression, Akt2 and glucose tolerance.
Previous studies [37, 38] have demonstrated transcriptional regulation of Rev-erbα (also known as Nr1d1) by RORα and decreased Rev-erbα mRNA expression in sg/sg mice [37]. However, this did not explain the increased expression of Akt2 mRNA in the sg/sg mice. For example, we also observe decreased expression of Rev-erbα in sg/sg mice; however, there was no decrease in REV-ERBα protein (ESM Fig. 6). This is consistent with another report [39], which showed ‘dissociation between mRNA and protein levels’.
In the context of improved insulin sensitivity, glucose uptake and increased AKT levels, we also observed increased Tbc1d1 and Glut4 expression. This is entirely consistent with Peck and colleagues [21], who very recently showed that activated AKT phosphorylates TBC1 domain family, member 1 (TBC1D1) and enhances glucose uptake. We also used quantitative PCR to validate the differential expression of several other mRNAs, including Pon1 and Sgk1, that are associated with type 2 diabetes, insulin secretion and metabolic syndrome [40, 41] and also appeared in several categories, similarly to Akt2.
We were particularly interested in AKT2, as this protein is an integral component of the signalling pathway controlling insulin signalling. For example, several investigations have reported that AKT2 is a major regulator of insulin-mediated glucose disposal in skeletal muscle. Aberrant Akt2 gene expression in rodents is associated with insulin resistance, type 2 diabetes-like syndrome, age-dependent lipodystrophy [42, 43] and phosphorylation of TBC1D1.
The threefold increase in expression of Tbc1d1 mRNA, initially appears paradoxical. Mutations in Tbc1d1 are associated with morbid obesity, for example, the R125W mutation in humans. However, the functional outcomes of these mutations have not yet been characterised. In the context of improved insulin sensitivity, glucose uptake and increased AKT level (and phosphorylation), we also observed increased Tbc1d1 and Glut4 expression. These observations are consistent with the latest studies on Tbc1d1 function. Peck and colleagues recently demonstrated that AKT phosphorylation of TBC1D1 (similar to AKT substrate, 160 kDa/TBC1 domain family, member 4) enables GLUT4 translocation [21]. Second, the obesity-related mutant R125W Tbc1d1 decreases insulin-stimulated glucose transport [22]. Both groups suggest that impaired glucose uptake provides a potential mechanism for the associated obesity. The differential (and increased) production of this Rab GTPase-activating protein, coupled to increased pAKT level in sg/sg mice, is interesting from the perspective of a potential contribution to resistance to diet-induced obesity in sg/sg mice via increased glucose uptake.
Interestingly, and consistent with increased insulin sensitivity, sg/sg mice showed enhanced insulin-mediated AKT phosphorylation and glucose uptake (and increased GLUT4 protein levels) in skeletal muscle. Recently, Duez et al. [44] showed that overexpression of Rorα4 in 3T3-L1 cells inhibits pre-adipocyte differentiation, Glut4 transcription and insulin-induced glucose uptake. In agreement with this, we have shown here that the GLUT4 protein level was increased in muscle from sg/sg mice.
In conclusion, the NR, RORα, is involved in regulation of AKT production and signalling, and glucose uptake. This study extends previous evidence that RORα performs a critical role in the regulation of adiposity. Further studies are required to examine whether this NR can be therapeutically exploited.
Electronic supplementary material
Below is the link to the electronic supplementary materials.
List of quantitative PCR primers (PDF 107 kb)
a Body weight (g) of 15-week-old fed male wild-type (wt; white column), heterozygous sg/+ (striped column) and homozygous sg/sg (black column) mice; n = 5–6 per group, mean ± SEM; *p < 0.05, **p < 0.01 and ***p < 0.001 by one-way ANOVA with Bonferroni’s post hoc test. b Quantitative PCR of wild-type Rorα (primers specific for wild-type Rorα mRNA only), Rorα1 (primers detect wild-type and sg Rorα1 mRNA isoforms) and Rorα4 (primers detect wild-type and sg Rorα4 mRNA isoforms) mRNA in quadriceps muscle of male sg/sg (black columns), heterozygous sg/+ (striped columns) and wild-type (white columns) mice; n = 10–12 per group, 3 to 7 months old, mean ± SEM; ***p < 0.001 by one-way ANOVA with Bonferroni’s post-hoc test. c CLAMS measurements of mean food intake during day (12 h) and night (12 h) by wild-type (white columns) and sg/sg (black columns) mice; n = 8–11 per group, mean ± SEM. d Oxygen consumption (VO2) during day (12 h), night (12 h) and fasting (6 h) by wild-type (white circles) and sg/sg (black circles) mice, with bar chart showing mean VO2 during day, night and fasting of wild-type (white column) and sg/sg (black column) mice; n = 8–11 per group, mean ± SEM; **p < 0.01 by two-way ANOVA with Bonferroni’s post hoc test (PDF 43 kb)
Quantitative PCR of (a–d) myosin heavy chain mRNA as labelled from quadriceps muscle of male wild-type (white columns) and homozygous sg/sg (black columns) mice, n = 10–12 per group, 3–7 months old, mean ± SEM. e Tnni1 and Tnni2 (f) in quadriceps muscle of wild-type (white columns) and sg/sg (black columns) mice; n = 7–8 per group, mean ± SEM. Relative mRNA expression was normalised against 18S rRNA (PDF 644 kb)
ESM Fig. 3. All up- and downregulated genes in sg/sg. Differentially expressed genes from the Illumina BeadArray analysis comparing sg/sg with wild-type mice. All genes had significant difference in expression above 1.24-fold. The fold change value presented is relative to wild-type mice
PDF 2,338 kb
PDF 2,414 kb
PDF 1,890 kb
Ingenuity pathway analysis of differentially expressed genes. Differentially expressed genes from the Illumina BeadArray analysis comparing sg/sg with wild-type mice were analysed via Ingenuity pathway analysis to reveal the top ten: (a) functions associated with upregulated genes; (b) functions associated with downregulated genes; (c) pathways associated with upregulated genes; and (d) pathways associated with upregulated genes. p values represent Fisher's exact test with the threshold at p < 0.05 (PDF 1.01 mb)
Functional annotation of genes that were up- or downregulated in sg/sg mice and associated with metabolic disorders. Differentially expressed genes from the Illumina BeadArray analysis comparing sg/sg to wild-type mice were analysed via Ingenuity pathway analysis. Differentially expressed genes with known associations to metabolic disorders are shown. Up arrows, upregulation; down arrows, downregulation (PDF 69 kb)
a Quantitative PCR of Rev-erbα mRNA in quadriceps muscle of wild-type (white column) and sg/sg (black column) mice; n = 7–8 per group, mean ± SEM; **p < 0.01 by t test. b Western blot of REV-ERBα (Affinity BioReagents, Golden, CO, USA, diluted 1:1000; antibody not commercially available) in quadriceps muscle of wild-type (WT) and sg/sg mice (PDF 331 kb)
Acknowledgements
G.E.O Muscat is a Principal Research Fellow and M.J. Watt a Senior Research Fellow of the National Health and Medical Research Council of Australia (NHMRC). This study was supported by project grants to G.E.O Muscat from the NHMRC and by Diabetes Australia Research Trust.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- AKT
v-AKT murine thymoma viral oncogene homologue
- EDL
Extensor digitorum longus
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- NR
Nuclear hormone receptor
- pAKT
Phosphorylated AKT
- PI3K
Phosphatidylinositol 3-kinase
- RER
Respiratory exchange ratio
- ROR
Retinoic acid receptor-related orphan receptor
- ser473
Serine amino acid residue 473
- TBC1D1
TBC1 domain family, member 1
References
- 1.Schmitz-Peiffer C. Signalling aspects of insulin resistance in skeletal muscle: mechanisms induced by lipid oversupply. Cell Signal. 2000;12:583–594. doi: 10.1016/S0898-6568(00)00110-8. [DOI] [PubMed] [Google Scholar]
- 2.Lau P, Nixon SJ, Parton RG, Muscat GE. RORalpha regulates the expression of genes involved in lipid homeostasis in skeletal muscle cells: caveolin-3 and CPT-1 are direct targets of ROR. J Biol Chem. 2004;279:36828–36840. doi: 10.1074/jbc.M404927200. [DOI] [PubMed] [Google Scholar]
- 3.Mamontova A, Seguret-Mace S, Esposito B, et al. Severe atherosclerosis and hypoalphalipoproteinemia in the staggerer mouse, a mutant of the nuclear receptor RORalpha. Circulation. 1998;98:2738–2743. doi: 10.1161/01.cir.98.24.2738. [DOI] [PubMed] [Google Scholar]
- 4.Raspe E, Duez H, Gervois P, et al. Transcriptional regulation of apolipoprotein C-III gene expression by the orphan nuclear receptor RORalpha. J Biol Chem. 2001;276:2865–2871. doi: 10.1074/jbc.M004982200. [DOI] [PubMed] [Google Scholar]
- 5.Kumar N, Solt LA, Conkright JJ, et al. The benzenesulfoamide T0901317 [N-(2, 2, 2-trifluoroethyl)-N-[4-[2, 2, 2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-alpha/gamma inverse agonist. Mol Pharmacol. 2010;77:228–236. doi: 10.1124/mol.109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raichur S, Lau P, Staels B, Muscat GE. Retinoid-related orphan receptor gamma regulates several genes that control metabolism in skeletal muscle cells: links to modulation of reactive oxygen species production. J Mol Endocrinol. 2007;39:29–44. doi: 10.1677/jme.1.00010. [DOI] [PubMed] [Google Scholar]
- 7.Vu-Dac N, Gervois P, Grotzinger T, et al. Transcriptional regulation of apolipoprotein A-I gene expression by the nuclear receptor RORalpha. J Biol Chem. 1997;272:22401–22404. doi: 10.1074/jbc.272.36.22401. [DOI] [PubMed] [Google Scholar]
- 8.Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORα, regulates gene expression that controls lipid metabolism: Staggerer (sg/sg) mice are resistant to diet-induced obesity. J Biol Chem. 2008;283:18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 10.Matysiak-Scholze U, Nehls M. The structural integrity of ROR alpha isoforms is mutated in staggerer mice: cerebellar coexpression of ROR alpha1 and ROR alpha4. Genomics. 1997;43:78–84. doi: 10.1006/geno.1997.4757. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton BA, Frankel WN, Kerrebrock AW, et al. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- 12.Dussault I, Fawcett D, Matthyssen A, Bader JA, Giguere V. Orphan nuclear receptor ROR alpha-deficient mice display the cerebellar defects of staggerer. Mech Dev. 1998;70:147–153. doi: 10.1016/S0925-4773(97)00187-1. [DOI] [PubMed] [Google Scholar]
- 13.Smith AG, Muscat GE. Skeletal muscle and nuclear hormone receptors: implications for cardiovascular and metabolic disease. Int J Biochem Cell Biol. 2005;37:2047–2063. doi: 10.1016/j.biocel.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Smith AG, Muscat GE. Orphan nuclear receptors: therapeutic opportunities in skeletal muscle. Am J Physiol Cell Physiol. 2006;291:C203–C217. doi: 10.1152/ajpcell.00476.2005. [DOI] [PubMed] [Google Scholar]
- 15.Muscat GE, Wagner BL, Hou J, et al. Regulation of cholesterol homeostasis and lipid metabolism in skeletal muscle by liver X receptors. J Biol Chem. 2002;277:40722–40728. doi: 10.1074/jbc.M206681200. [DOI] [PubMed] [Google Scholar]
- 16.Matthaei S, Stumvoll M, Kellerer M, Haring HU. Pathophysiology and pharmacological treatment of insulin resistance. Endocr Rev. 2000;21:585–618. doi: 10.1210/er.21.6.585. [DOI] [PubMed] [Google Scholar]
- 17.Raichur S, Fitzsimmons RL, Myers SA, et al. Identification and validation of the pathways and functions regulated by the orphan nuclear receptor, ROR alpha1, in skeletal muscle. Nucleic Acids Res. 2010;38:4296–4312. doi: 10.1093/nar/gkq180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brozinick JT, Jr, Birnbaum MJ. Insulin, but not contraction, activates Akt/PKB in isolated rat skeletal muscle. J Biol Chem. 1998;273:14679–14682. doi: 10.1074/jbc.273.24.14679. [DOI] [PubMed] [Google Scholar]
- 19.Watt MJ, Dzamko N, THomas WG, et al. CNTF reverses obesity-induced insulin resistance by activating skeletal muscle AMPK. Nat Med. 2006;12:541–548. doi: 10.1038/nm1383. [DOI] [PubMed] [Google Scholar]
- 20.Bertin R, Guastavino JM, Portet R. Effects of cold acclimation on the energetic metabolism of the staggerer mutant mouse. Physiol Behav. 1990;47:377–380. doi: 10.1016/0031-9384(90)90157-Y. [DOI] [PubMed] [Google Scholar]
- 21.Peck GR, Chavez JA, Roach WG, et al. Insulin-stimulated phosphorylation of the Rab GTPase-activating protein TBC1D1 regulates GLUT4 translocation. J Biol Chem. 2009;284:30016–30023. doi: 10.1074/jbc.M109.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An D, Toyoda T, Taylor EB, et al. TBC1D1 regulates insulin- and contraction-induced glucose transport in mouse skeletal muscle. Diabetes. 2010;59:1358–1365. doi: 10.2337/db09-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang F, Gorlach A, Vallon V. Targeting SGK1 in diabetes. Expert Opin Ther Targets. 2009;13:1303–1311. doi: 10.1517/14728220903260807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sztalryd C, Bell M, Lu X, et al. Functional compensation for adipose differentiation-related protein (ADFP) by Tip47 in an ADFP null embryonic cell line. J Biol Chem. 2006;281:34341–34348. doi: 10.1074/jbc.M602497200. [DOI] [PubMed] [Google Scholar]
- 25.Wolins NE, Quaynor BK, Skinner JR, et al. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes. 2006;55:3418–3428. doi: 10.2337/db06-0399. [DOI] [PubMed] [Google Scholar]
- 26.Bell M, Wang H, Chen H, et al. Consequences of lipid droplet coat protein downregulation in liver cells: abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes. 2008;57:2037–2045. doi: 10.2337/db07-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinmayr M, Andre E, Conquet F, et al. staggerer phenotype in retinoid-related orphan receptor alpha-deficient mice. Proc Natl Acad Sci USA. 1998;95:3960–3965. doi: 10.1073/pnas.95.7.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delerive P, Monte D, Dubois G, et al. The orphan nuclear receptor ROR alpha is a negative regulator of the inflammatory response. EMBO Rep. 2001;2:42–48. doi: 10.1093/embo-reports/kve007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baron AD, Brechtel G, Wallace P, Edelman SV. Rates and tissue sites of non-insulin- and insulin-mediated glucose uptake in humans. Am J Physiol. 1988;255:E769–E774. doi: 10.1152/ajpendo.1988.255.6.E769. [DOI] [PubMed] [Google Scholar]
- 30.Kang HS, Angers M, Beak JY, et al. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genomics. 2007;31:281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- 31.Andrikopoulos S, Blair AR, Deluca N, Fam BC, Proietto J. Evaluating the glucose tolerance test in mice. Am J Physiol Endocrinol Metab. 2008;295:E1323–E1332. doi: 10.1152/ajpendo.90617.2008. [DOI] [PubMed] [Google Scholar]
- 32.McGuinness OP, Ayala JE, Laughlin MR, Wasserman DH. NIH experiment in centralized mouse phenotyping: the Vanderbilt experience and recommendations for evaluating glucose homeostasis in the mouse. Am J Physiol Endocrinol Metab. 2009;297:E849–E855. doi: 10.1152/ajpendo.90996.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boukhtouche F, Mariani J, Tedgui A. The "CholesteROR" protective pathway in the vascular system. Arterioscler Thromb Vasc Biol. 2004;24:637–643. doi: 10.1161/01.ATV.0000119355.56036.de. [DOI] [PubMed] [Google Scholar]
- 34.Gold DA, Gent PM, Hamilton BA. ROR alpha in genetic control of cerebellum development: 50 staggering years. Brain Res. 2007;1140:19–25. doi: 10.1016/j.brainres.2005.11.080. [DOI] [PubMed] [Google Scholar]
- 35.McBroom LD, Flock G, Giguere V. The nonconserved hinge region and distinct amino-terminal domains of the ROR alpha orphan nuclear receptor isoforms are required for proper DNA bending and ROR alpha-DNA interactions. Mol Cell Biol. 1995;15:796–808. doi: 10.1128/mcb.15.2.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner BL, Valledor AF, Shao G, et al. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol Cell Biol. 2003;23:5780–5789. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delerive P, Chin WW, Suen CS. Identification of Reverb(alpha) as a novel ROR(alpha) target gene. J Biol Chem. 2002;277:35013–35018. doi: 10.1074/jbc.M202979200. [DOI] [PubMed] [Google Scholar]
- 38.Raspe E, Mautino G, Duval C, et al. Transcriptional regulation of human Rev-erbalpha gene expression by the orphan nuclear receptor retinoic acid-related orphan receptor alpha. J Biol Chem. 2002;277:49275–49281. doi: 10.1074/jbc.M206215200. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Lazar MA. Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol Cell Biol. 2008;28:2213–2220. doi: 10.1128/MCB.01608-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedrich B, Weyrich P, Stancakova A, et al. Variance of the SGK1 gene is associated with insulin secretion in different European populations: results from the TUEF, EUGENE2, and METSIM studies. PLoS ONE. 2008;3:e3506. doi: 10.1371/journal.pone.0003506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwab M, Lupescu A, Mota M, et al. Association of SGK1 gene polymorphisms with type 2 diabetes. Cell Physiol Biochem. 2008;21:151–160. doi: 10.1159/000113757. [DOI] [PubMed] [Google Scholar]
- 42.Cho H, Mu J, Kim JK, et al. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 43.Garofalo RS, Orena SJ, Rafidi K, et al. Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKB beta. J Clin Invest. 2003;112:197–208. doi: 10.1172/JCI16885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duez H, Duhem C, Laitinen S, et al. Inhibition of adipocyte differentiation by RORalpha. FEBS Lett. 2009;583:2031–2036. doi: 10.1016/j.febslet.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 45.Mouse Genome Informatics. MGI Mouse Genome Browser on build 37. Available from http://gbrowse.informatics.jax.org/cgi-bin/gbrowse/mouse_current/?start=28376571;stop=28425845;ref=7, accessed 20 January 2011
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of quantitative PCR primers (PDF 107 kb)
a Body weight (g) of 15-week-old fed male wild-type (wt; white column), heterozygous sg/+ (striped column) and homozygous sg/sg (black column) mice; n = 5–6 per group, mean ± SEM; *p < 0.05, **p < 0.01 and ***p < 0.001 by one-way ANOVA with Bonferroni’s post hoc test. b Quantitative PCR of wild-type Rorα (primers specific for wild-type Rorα mRNA only), Rorα1 (primers detect wild-type and sg Rorα1 mRNA isoforms) and Rorα4 (primers detect wild-type and sg Rorα4 mRNA isoforms) mRNA in quadriceps muscle of male sg/sg (black columns), heterozygous sg/+ (striped columns) and wild-type (white columns) mice; n = 10–12 per group, 3 to 7 months old, mean ± SEM; ***p < 0.001 by one-way ANOVA with Bonferroni’s post-hoc test. c CLAMS measurements of mean food intake during day (12 h) and night (12 h) by wild-type (white columns) and sg/sg (black columns) mice; n = 8–11 per group, mean ± SEM. d Oxygen consumption (VO2) during day (12 h), night (12 h) and fasting (6 h) by wild-type (white circles) and sg/sg (black circles) mice, with bar chart showing mean VO2 during day, night and fasting of wild-type (white column) and sg/sg (black column) mice; n = 8–11 per group, mean ± SEM; **p < 0.01 by two-way ANOVA with Bonferroni’s post hoc test (PDF 43 kb)
Quantitative PCR of (a–d) myosin heavy chain mRNA as labelled from quadriceps muscle of male wild-type (white columns) and homozygous sg/sg (black columns) mice, n = 10–12 per group, 3–7 months old, mean ± SEM. e Tnni1 and Tnni2 (f) in quadriceps muscle of wild-type (white columns) and sg/sg (black columns) mice; n = 7–8 per group, mean ± SEM. Relative mRNA expression was normalised against 18S rRNA (PDF 644 kb)
PDF 2,338 kb
PDF 2,414 kb
PDF 1,890 kb
Ingenuity pathway analysis of differentially expressed genes. Differentially expressed genes from the Illumina BeadArray analysis comparing sg/sg with wild-type mice were analysed via Ingenuity pathway analysis to reveal the top ten: (a) functions associated with upregulated genes; (b) functions associated with downregulated genes; (c) pathways associated with upregulated genes; and (d) pathways associated with upregulated genes. p values represent Fisher's exact test with the threshold at p < 0.05 (PDF 1.01 mb)
Functional annotation of genes that were up- or downregulated in sg/sg mice and associated with metabolic disorders. Differentially expressed genes from the Illumina BeadArray analysis comparing sg/sg to wild-type mice were analysed via Ingenuity pathway analysis. Differentially expressed genes with known associations to metabolic disorders are shown. Up arrows, upregulation; down arrows, downregulation (PDF 69 kb)
a Quantitative PCR of Rev-erbα mRNA in quadriceps muscle of wild-type (white column) and sg/sg (black column) mice; n = 7–8 per group, mean ± SEM; **p < 0.01 by t test. b Western blot of REV-ERBα (Affinity BioReagents, Golden, CO, USA, diluted 1:1000; antibody not commercially available) in quadriceps muscle of wild-type (WT) and sg/sg mice (PDF 331 kb)