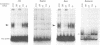Abstract
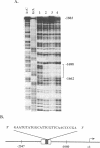
Fibronectin (FN) is a widely distributed extracellular matrix protein that is essential for cell adhesion in a variety of biological processes such as wound healing, tissue development and remodeling and oncogenic transformation. Appropriate FN levels are obtained by induction or repression of the FN gene in response to specific factors or circumstances in vivo. In order to identify regulatory regions involved in tissue-specific expression of FN, we have examined the transcriptional activity of overlapping fragments, within 4 kb upstream of the rat FN gene, following transfection into different cell types. Two regions conferred increases in transcription. The region between -1.08 and -2.6 displayed tissue-specificity and was active in fibroblasts but not hepatoma cells. The second region, between -3.2 and -3.9, was active in both cell types. Further characterization of the -1.08 to -2.6 segment demonstrated that it acts as an enhancer. Exonuclease III deletions of the 3' and 5' ends of the enhancer localized essential sequences between -1.5 and -1.7 and indicate that this fragment acts in concert with other sites between -1.08 and -2.6 to provide maximum enhancer activity. Gel mobility shift assays demonstrated fibroblast-specific binding of nuclear protein(s) to a 65 bp fragment within the essential region and DNase I footprinting localized this binding to a 27 bp sequence. Deletion of the sequence abolished the activity of the 1.5 kb enhancer. These studies show that a novel DNA sequence at -1688 is involved in regulating transcription of the FN gene in fibroblasts.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blatti S. P., Foster D. N., Ranganathan G., Moses H. L., Getz M. J. Induction of fibronectin gene transcription and mRNA is a primary response to growth-factor stimulation of AKR-2B cells. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1119–1123. doi: 10.1073/pnas.85.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowlus C. L., McQuillan J. J., Dean D. C. Characterization of three different elements in the 5'-flanking region of the fibronectin gene which mediate a transcriptional response to cAMP. J Biol Chem. 1991 Jan 15;266(2):1122–1127. [PubMed] [Google Scholar]
- Cognet M., Bergot M. O., Kahn A. cis-acting DNA elements regulating expression of the liver pyruvate kinase gene in hepatocytes and hepatoma cells. Evidence for tissue-specific activators and extinguisher. J Biol Chem. 1991 Apr 25;266(12):7368–7375. [PubMed] [Google Scholar]
- Dean D. C., Bowlus C. L., Bourgeois S. Cloning and analysis of the promotor region of the human fibronectin gene. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1876–1880. doi: 10.1073/pnas.84.7.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D. C. Expression of the fibronectin gene. Am J Respir Cell Mol Biol. 1989 Jul;1(1):5–10. doi: 10.1165/ajrcmb/1.1.5. [DOI] [PubMed] [Google Scholar]
- Dean D. C., McQuillan J. J., Weintraub S. Serum stimulation of fibronectin gene expression appears to result from rapid serum-induced binding of nuclear proteins to a cAMP response element. J Biol Chem. 1990 Feb 25;265(6):3522–3527. [PubMed] [Google Scholar]
- Dean D. C., Newby R. F., Bourgeois S. Regulation of fibronectin biosynthesis by dexamethasone, transforming growth factor beta, and cAMP in human cell lines. J Cell Biol. 1988 Jun;106(6):2159–2170. doi: 10.1083/jcb.106.6.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D. A relational database of transcription factors. Nucleic Acids Res. 1990 Apr 11;18(7):1749–1756. doi: 10.1093/nar/18.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese K., Cox J., Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992 Apr 3;69(1):185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- Godbout R., Ingram R., Tilghman S. M. Multiple regulatory elements in the intergenic region between the alpha-fetoprotein and albumin genes. Mol Cell Biol. 1986 Feb;6(2):477–487. doi: 10.1128/mcb.6.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignotz R. A., Massagué J. Transforming growth factor-beta stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J Biol Chem. 1986 Mar 25;261(9):4337–4345. [PubMed] [Google Scholar]
- Judde J. G., Max E. E. Characterization of the human immunoglobulin kappa gene 3' enhancer: functional importance of three motifs that demonstrate B-cell-specific in vivo footprints. Mol Cell Biol. 1992 Nov;12(11):5206–5216. doi: 10.1128/mcb.12.11.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles B. B., Howe C. C., Aden D. P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980 Jul 25;209(4455):497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- Kornblihtt A. R., Umezawa K., Vibe-Pedersen K., Baralle F. E. Primary structure of human fibronectin: differential splicing may generate at least 10 polypeptides from a single gene. EMBO J. 1985 Jul;4(7):1755–1759. doi: 10.1002/j.1460-2075.1985.tb03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsman D., Bustin M. A signature for the HMG-1 box DNA-binding proteins. Bioessays. 1993 Aug;15(8):539–546. doi: 10.1002/bies.950150807. [DOI] [PubMed] [Google Scholar]
- Luckow B., Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987 Jul 10;15(13):5490–5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao S., Suri P. K., Shu-Ling L., Abraham A., Cook N., Milos P., Zern M. A. Role of the cyclic AMP response element in rat fibronectin gene expression. Hepatology. 1993 May;17(5):882–890. [PubMed] [Google Scholar]
- Müller U., Roberts M. P., Engel D. A., Doerfler W., Shenk T. Induction of transcription factor AP-1 by adenovirus E1A protein and cAMP. Genes Dev. 1989 Dec;3(12A):1991–2002. doi: 10.1101/gad.3.12a.1991. [DOI] [PubMed] [Google Scholar]
- Nakajima T., Nakamura T., Tsunoda S., Nakada S., Oda K. E1A-responsive elements for repression of rat fibronectin gene transcription. Mol Cell Biol. 1992 Jun;12(6):2837–2846. doi: 10.1128/mcb.12.6.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. S., Odermatt E., Schwarzbauer J. E., Hynes R. O. Organization of the fibronectin gene provides evidence for exon shuffling during evolution. EMBO J. 1987 Sep;6(9):2565–2572. doi: 10.1002/j.1460-2075.1987.tb02545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogulis R. J., Freytag S. O. Contribution of specific cis-acting elements to activity of the mouse pro-alpha 2(I) collagen enhancer. J Biol Chem. 1993 Feb 5;268(4):2493–2499. [PubMed] [Google Scholar]
- Polly P., Nicholson R. C. Sequence of the mouse fibronectin-encoding gene promoter region. Gene. 1993 Dec 31;137(2):353–354. doi: 10.1016/0378-1119(93)90036-3. [DOI] [PubMed] [Google Scholar]
- Rossi P., de Crombrugghe B. Identification of a cell-specific transcriptional enhancer in the first intron of the mouse alpha 2 (type I) collagen gene. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5590–5594. doi: 10.1073/pnas.84.16.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Patel R. S., Fonda D., Hynes R. O. Multiple sites of alternative splicing of the rat fibronectin gene transcript. EMBO J. 1987 Sep;6(9):2573–2580. doi: 10.1002/j.1460-2075.1987.tb02547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Tamkun J. W., Lemischka I. R., Hynes R. O. Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell. 1983 Dec;35(2 Pt 1):421–431. doi: 10.1016/0092-8674(83)90175-7. [DOI] [PubMed] [Google Scholar]
- Seed B., Sheen J. Y. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene. 1988 Jul 30;67(2):271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- Shirakawa F., Saito K., Bonagura C. A., Galson D. L., Fenton M. J., Webb A. C., Auron P. E. The human prointerleukin 1 beta gene requires DNA sequences both proximal and distal to the transcription start site for tissue-specific induction. Mol Cell Biol. 1993 Mar;13(3):1332–1344. doi: 10.1128/mcb.13.3.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srebrow A., Muro A. F., Werbajh S., Sharp P. A., Kornblihtt A. R. The CRE-binding factor ATF-2 facilitates the occupation of the CCAAT box in the fibronectin gene promoter. FEBS Lett. 1993 Jul 19;327(1):25–28. doi: 10.1016/0014-5793(93)81031-t. [DOI] [PubMed] [Google Scholar]
- Sztul E., Colombo M., Stahl P., Samanta R. Control of protein traffic between distinct plasma membrane domains. Requirement for a novel 108,000 protein in the fusion of transcytotic vesicles with the apical plasma membrane. J Biol Chem. 1993 Jan 25;268(3):1876–1885. [PubMed] [Google Scholar]
- Tyagi J. S., Hirano H., Merlino G. T., Pastan I. Transcriptional control of the fibronectin gene in chick embryo fibroblasts transformed by Rous sarcoma virus. J Biol Chem. 1983 May 10;258(9):5787–5793. [PubMed] [Google Scholar]
- Wu C. Analysis of hypersensitive sites in chromatin. Methods Enzymol. 1989;170:269–289. doi: 10.1016/0076-6879(89)70052-5. [DOI] [PubMed] [Google Scholar]