Abstract
BACKGROUND
Research into cell-free fetal (cff) nucleic acids has primarily focused on maternal plasma; however, cff DNA and RNA are also detectable in other body fluids such as amniotic fluid (AF). In AF, cff DNA is present in much greater concentrations than in maternal plasma and represents a pure fetal sample uncontaminated by maternal- and trophoblast-derived nucleic acids. The aim of this review was to summarize the current knowledge on cff nucleic acids in AF and to outline future research directions.
METHODS
MEDLINE and PREMEDLINE were searched up to August 2010 for original investigations of cell-free RNA or DNA in AF. Sixteen studies were included in the review.
RESULTS
AF cff DNA represents a physiologically separate pool from cff DNA in maternal plasma. The placenta is not a major source of nucleic acids in AF. It is feasible to isolate cff nucleic acids from small volumes of discarded AF supernatant in sufficient quality and quantity to perform microarray studies and downstream applications such as pathway analysis. This ‘discovery-driven approach’ has resulted in new information on the pathogenesis of Down syndrome and polyhydramnios. There is otherwise a paucity of information relating to the basic biology and clinical applications of cff nucleic acids in AF.
CONCLUSIONS
AF supernatant is a valuable and widely available but under-utilized biological resource. Further studies of cff nucleic acids in AF may lead to new insights into human fetal development and ultimately new approaches to antenatal treatment of human disease.
Keywords: amniotic fluid, cell-free fetal nucleic acids, prenatal diagnosis, gene expression
Introduction
Cell-free fetal (cff) nucleic acids in maternal body fluids are a uniquely dispersed form of fetal genetic material that have become the subject of intense interest in the field of non-invasive prenatal diagnosis. Less than 15 years since their presence in maternal blood was reported (Lo et al., 1997), cff nucleic acids in maternal plasma are now established molecular diagnostic specimens and are routinely used in clinical care for the non-invasive prenatal diagnosis of fetal gender and rhesus D status (Wright and Burton, 2009). Utilizing cff nucleic acids for the non-invasive prenatal diagnosis of trisomy 21 is a more difficult goal due to the formidable technical challenges, but recent advances, such as next generation DNA sequencing, have brought this prospect closer to realization in a universally applicable format (Yang et al., 2003; Lo, 2009; Lo and Chiu, 2009).
Amidst the remarkable progress of cff nucleic acids in maternal plasma, other maternal body fluids such as amniotic fluid (AF; Bianchi et al., 2001), cerebrospinal fluid (Angert et al., 2004) and urine (Koide et al., 2005) have been overlooked. AF specimens represent an opportunity to move the application of cff nucleic acids beyond the prenatal diagnosis of chromosome disorders into broader studies of human development.
What is amniotic fluid?
AF is a complex, dynamic solution that performs multiple functions for the developing fetus at different gestational ages. In the first trimester of pregnancy, AF is primarily formed from maternal plasma that passes through fetal membranes. This free diffusion occurs bi-directionally between the AF and the fetal skin, placenta and umbilical cord. As fetal skin and other organs mature, the composition of AF alters. In the second trimester, AF still has similar levels of primary electrolytes as fetal and maternal blood, but the levels of organic substances are markedly different (Tong et al., 2009). AF provides physical protection of the fetus from mechanical and thermal insults, but also has immune functions, contains growth factors and cytokines and acts as a nutritional source (Underwood et al., 2005). Amniocytes are routinely used to study fetal chromosomes. Cells arising from all three germ layers of the embryo are present in AF, ranging from unspecified progenitors to mature differentiated cells, including those of the renal, heart, lung, liver and hematopoetic cell lineages. Moreover, ∼1% of AF cells have pluripotent properties, making them of particular significance as a novel source of stem cells (Da Sacco et al., 2010).
The amniotic fluid proteome
Research groups that have applied ‘high-dimensional biology’ techniques to AF have primarily relied on proteomic rather than nucleic acid-based approaches (Romero et al., 2006; Tsangaris et al., 2006; Cho et al., 2007; Alam, 2009; Deng et al., 2010; Lee et al., 2010; Romero et al., 2010). Cho et al identified 842 non-redundant proteins in the human AF proteome, including most of the currently known maternal serum biomarkers of pregnancy complications. These authors also analysed the tissue expression profile of AF proteins and found that the top 10 represented organs were kidney, placenta, lung, liver, heart, plasma, brain, testis, pancreas and skeletal muscle in descending order (Cho et al., 2007). However, these results are not consistent with the tissue profiles produced from other proteomic studies (Tsangaris et al., 2006), making conclusions about the relative contributions of fetal organs to the AF proteome difficult. Putative protein markers for premature rupture of membranes, intra-amniotic infection and aneuploidy have also been reported, but these have not yet translated into clinical use (Buhimschi and Buhimschi, 2008; Buhimschi et al., 2008; Anagnostopoulos et al., 2009; Kolialexi et al., 2009).
Nucleic acids in serum and plasma
The major source of cff DNA and mRNA in maternal plasma is the syncytiotrophoblast, which continuously releases cff nucleic acids via apoptosis (Orozco et al., 2006; Tjoa et al., 2006). cff DNA and RNA are detectable in the maternal circulation from the first trimester onwards (Poon et al., 2000; Guibert et al., 2003) and are cleared from the maternal circulation rapidly after delivery, with median half-lives of around 15 min in healthy pregnant women (Lo et al., 1999b; Chiu et al., 2006).
A major technical hurdle in the isolation of cff nucleic acids from plasma is the fact that they only constitute a small proportion of total cell-free nucleic acids, the majority of which is derived from maternal leukocytes. cff DNA in plasma was initially thought to comprise 3–6% of the total circulating cell-free DNA using a real-time PCR assay (Lo et al., 1998), although more recent studies using a second-generation DNA sequencing suggest that the concentration may be as high as 19% (Fan et al., 2008).
Cff nucleic acids in the maternal circulation are stable; this is surprising given the rapid decay that may be expected due to plasma nucleases. The relative stability of cff DNA and RNA is attributed to their association with placenta-derived microparticles that protect them from nuclease degradation (Hasselmann et al., 2001; Gupta et al., 2004; Orozco et al., 2008). Microparticles are a heterogeneous population of small membrane-covered fragments released from cells via several different mechanisms, including apoptosis (programmed cell death) and necrosis (cell death due to primary energy failure). Smaller types of microvesicles that are actively secreted by cells, such as exosomes, are attracting research interest as novel forms of cellular communication (Simpson et al., 2009). They have particular relevance for cell-free nucleic acid biology, as they contain functional forms of RNA that participate in cell-to-cell signalling (Lotvall and Valadi, 2007; Valadi et al., 2007; Luo et al., 2009).
Nucleic acids in amniotic fluid
AF has been relatively understudied due to the risks associated with sample collection and the international research focus on developing non-invasive methods of prenatal diagnosis. However, AF has properties that make it worthy of study for broader applications beyond prenatal diagnosis of aneuploidy. AF contains cff DNA in much greater concentrations than in maternal plasma (Bianchi et al., 2001). Importantly, it represents a relatively pure fetal sample that is largely uncontaminated by maternal- and trophoblast-derived nucleic acids.
To date, most studies of fetal gene expression in AF have used cultured amniocytes. As gene expression is context-dependent and regulated by many factors, including tissue of origin, environmental signals, developmental stage, gender and disease states, these studies do not reflect real-time fetal physiology (Chung et al., 2005; Altug-Teber et al., 2007; Chou et al., 2008). Successful isolation of cff mRNA from AF and its resultant genomic analysis have resulted in the discovery of new information regarding human development in live ongoing pregnancies (Larrabee et al., 2005a; Slonim et al., 2009). Potentially, this will take prenatal diagnosis in new directions and provide fetal medicine specialists with alternative tools to ultrasound examination and maternal serum markers.
The aim of this review was to summarize the current knowledge on cff nucleic acids in AF and to outline possible future research directions.
Methods
Articles were selected for review if they met the following criteria: English-language, original research report on cell-free nucleic acids in AF (human or animal). A search in MEDLINE and PREMEDLINE for articles up to August 2010 was performed using the exploded MeSH search terms: ‘RNA, DNA, nucleic acids, gene expression, OR gene expression profiling’ combined with MeSH term ‘amniotic fluid’ OR ‘amniocentesis’ AND text word ‘cell-free’.
A total of 23 articles were retrieved. One non-English-language study was excluded (Lazar et al., 2007). The titles and abstracts of the remaining 22 articles were reviewed: three review articles that primarily focused on blood (Wataganara and Bianchi, 2004; Bianchi et al., 2006; Maron and Bianchi, 2007) were excluded. A further three studies were excluded, as they did not study cell-free nucleic acids from AF specimens (Crouch et al., 1991; Samura et al., 2003; Miura et al., 2006a). Reference lists from these articles were scanned for any further original research studies, but no further relevant studies were identified (Supplementary data, Fig. S1).
Results
A total of 16 original research studies analysing cff nucleic acids in AF were included in this review. Thirteen studies were on cff DNA and three studies were on cff RNA in AF. All studies were performed on human samples.
cff DNA in amniotic fluid
Presence of cff DNA in AF and extraction methods
Bianchi et al. (2001) first documented that there was ∼100–200-fold more fetal DNA per millilitre of AF compared with maternal plasma. In this study, 38 frozen AF specimens, collected for routine indications at 16–20 weeks gestation, were thawed and centrifuged to remove remaining cells. Extraction was performed with the QIAamp Blood Kit (Qiagen) using the ‘Blood and Body Fluid’ protocol described by the manufacturer. A real-time quantitative PCR analysis was performed for the β-globin gene and for the FCY (DYS1) locus as a basis for detecting male DNA. The investigators were blinded to the karyotype results from the specimens, and in all 38 cases, they were able to correctly predict the fetal sex.
Two further publications refining cff DNA extraction methods were identified, demonstrating improved yields of high-quality cff DNAs from AF supernatant (Lapaire et al., 2006, 2008). With modifications to the original plasma-based protocol, the yield of sequences that amplified glyceraldehyde 3-phosphate dehydrogenase (GAPDH), a housekeeping gene, from 10 ml of AF, increased from 246 to 17 000 GE/ml. Other advantages of the optimized protocol included a reduction in processing time and fewer overall steps to reduce potential contamination. These protocol refinements represented a major step in consistently producing sufficient AF cff DNA for subsequent analysis by comparative genomic hybridization (CGH) microarrays.
The biophysical properties of AF cff DNA
The first data on the biophysical properties of AF cff DNA were published by Lapaire et al. (2007a). DNA was extracted from AF supernatant samples from 39 euploid and 4 aneuploid fetuses. Quantitative real-time PCR amplification of the GAPDH locus, gel electrophoresis and analysis of the DNA fragmentation signature showed significant differences in the concentration of AF cff DNA as a function of gestational age, karyotype and sample storage time. There was a positive correlation between AF cff DNA concentration with gestational age in fresh samples from euploid fetuses, but not in the frozen samples. They also found that the median amount of cff DNA in frozen euploid samples was also significantly lower than that in fresh samples. Storage also reduced the proportion of large DNA fragments in AF, suggesting that freezing at −80°C alone contributed to DNA fragmentation.
There were also observed differences in cff DNA fragment sizes and distributions according to karyotype. The fragmentation signature, which was determined on standard agarose gels, represented differences in the proportion of different sizes of cff DNA fragments. The specific patterns associated with euploid, trisomy 21 and trisomy 13 fetuses, suggested specific kinetic mechanisms unique to each karyotype.
These findings were followed up in a subsequent investigation of AF cff DNA fragmentation patterns in a larger study of samples matched for storage time from women carrying 36 euploid and 29 aneuploid fetuses (Peter et al., 2008). This method allowed for the unique classification of euploid and aneuploid cff DNA samples in AF by their DNA fragmentation signature. In addition, this study showed that archived euploid AF samples gradually lose long cff DNA fragments and that this loss accurately distinguishes them from the fresh samples. These data suggest that archived AF samples consist of large amounts of short cff DNA fragments, which may pass undetected with standard real-time PCR amplification.
In addition to these two studies on the DNA fragment size and distribution, a single study has been published examining the binding of cff DNA in AF to the high-mobility group protein HMGA2 (Winter et al., 2008). HMGA2 is primarily expressed by embryonic and fetal cells and binds to the minor groove of DNA with little sequence specificity. Using chromatin immunoprecipitation, these authors found that the cff DNA in AF was attached to HMGA2. They concluded that cells in AF strongly overexpress HMGA2 according to their fetal origin. The fact that HMGA2 remains attached to cff DNA in AF has potential significance as a method to improve sample yields of cff DNA via immunoprecipitation techniques.
Lack of correlation between AF and serum/plasma cff DNA
A third study on the relative amounts of cff DNA in the fetal and maternal compartments added further information on cff DNA from the celomic cavity (Makrydimas et al., 2008). Lee et al. (2002) examined the quantitative relationship between frozen archived AF and serum cff DNA in Down syndrome pregnancies. They confirmed a higher level of maternal serum cff DNA in Down syndrome pregnancies compared with matched normal controls, but found no such association in AF. This finding suggested that the observed difference in maternal serum cff DNA in trisomy 21 is not a consequence of elevated levels in the amniotic cavity.
Zhong et al. (2006) also examined the relationship between the amount of cff DNA in AF and maternal plasma in 12 euploid pregnancies. They used real-time PCR amplification assays for SRY and RHD to quantify the amount of cff DNA present in maternal plasma and matched AF samples. They confirmed earlier reports that AF contained much larger quantities of cff DNA (median concentration = 3978 copies/ml) than maternal plasma (median concentration = 96.6 copies/ml). However, there was no statistically significant correlation between the amount of cff DNA in AF and maternal plasma.
A third study on the relative amounts of cff DNA in the fetal and maternal compartments added further information on cff DNA from the celomic cavity (Makrydimas et al., 2008). Makrydimas' study on cff DNA in celomic fluid in first-trimester pregnancies undergoing elective termination at 7–9 weeks gestation demonstrated a concentration gradient of cff DNA from the AF (highest), to celomic fluid (lower), to maternal serum (lowest). The authors argued that the size of fetal cff DNA in maternal circulation of 100–300 bp (Chan et al., 2004) made it very unlikely that it could transfer directly through the amniotic membrane to the maternal circulation.
In summary, the results of these three studies show that there is no direct correlation between the amount of cff DNA in AF and maternal plasma, suggesting that AF cff DNA represents a physiologically separate pool from cff DNA in maternal plasma.
Sources of AF cff DNA
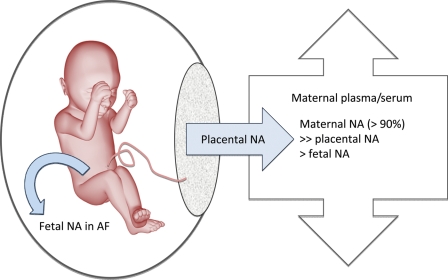
In further developing the concept of the AF pool as being relatively sequestered from the maternal cff DNA pool, Lun et al. (2007) used an epigenetic approach to specifically examine the distribution of placenta-specific cff DNA. They used the tissue-specific epigenetic marker associated with the promoter region of the RAS association family 1A (RASSF1A) gene to determine the relative proportion of placental DNA in AF and plasma. RASSF1A is known to be hypermethylated in the placenta, but hypomethylated in fetal tissues and maternal blood cells (Chan et al., 2006; Chiu et al., 2007). AF and maternal plasma cff DNA samples from 14 second-trimester pregnant women showed that placenta-specific hypermethylated RASSF1A DNA sequences were present in AF, but at a much lower proportion of total RASSF1A DNA compared with maternal plasma. In fact, the proportion of hypermethylated RASSF1A was 30-fold lower in the cff portion of AF than maternal plasma, supporting the concept that the placenta is not a major source of AF cff DNA (Fig. 1).
Figure 1.
Trafficking of cff nucleic acids within fetal and maternal compartments. AF, amniotic fluid; NA, nucleic acids.
Potential clinical applications of AF cff DNA
In tandem with studies investigating the biophysical properties and relationship with plasma cff DNA, several studies explored the clinical applications of AF cff DNA as a method of producing a rapid ‘molecular karyotype’.
Larrabee et al. (2004) performed the first study to utilize DNA from the cell-free fraction of AF for prenatal molecular cytogenetic diagnosis. This feasibility study successfully used cff DNA for CGH microarray analysis with the GenoSensor Array 300. They correctly identified fetal gender and whole chromosome gains or losses, including trisomy 21 and monosomy X, according to the strength of the hybridization signals from chromosome-specific markers. This prenatal ‘molecular karyotype’ using cff DNA has the potential benefit of providing rapid and higher resolution localization of abnormalities in the fetal genome compared with standard metaphase karyotype from cultured amniocytes. However, significant technical challenges were encountered in this preliminary study. Only 17 of 28 AF supernatant samples yielded adequate amounts of high-quality cff DNA (>100 ng DNA) for subsequent analysis.
Another research group investigated the rapid ‘molecular karyotype’ concept with AF cff DNA using a targeted microarray-based CGH panel on which bacterial artificial chromosome clones from human chromosomes 13, 18, 21, X and Y were spotted (Miura et al., 2006b). Array CGH analysis was performed for 13 fetuses with congenital anomalies using cff DNA from their uncultured AF, resulting in successful molecular karyotyping for 12 of 13 fetuses within 5 days. Those fetuses with whole chromosome losses or gains were correctly identified, whereas a false-negative result was obtained for a fetus with a balanced translocation, 45, XY, der(14;21)(q10;q10).
The previously discussed technical advances that increased both the yield and the quality of extracted cff DNA (Lapaire et al., 2006) enabled further investigations of array CGH applications for detecting aneuploidy using smaller volumes of both fresh and frozen AF cff DNA. Lapaire et al. (2007b) extracted cff DNA from 10 ml of residual AF supernatant from 10 samples, including one with a normal karyotype and nine with various aneuploidies. Their results showed that array CGH using AF cff DNA from aneuploid fetuses, when compared with euploid reference AF cff DNA, could detect whole chromosome aneuploidy in eight of nine cases tested, including a case of trisomy 9 mosaicism. Only one case of triploidy was not detected.
Cff mRNA in amniotic fluid
We identified three studies that investigated cff RNA in AF. Larrabee et al. (2005b) performed a filtration study of centrifuged and uncentrifuged AF to determine if AF mRNA was present in a particle-associated form. The authors extracted RNA from uncentrifuged AF and centrifuged AF supernatant and passed it through filters with varying pore sizes of 0.22, 0.45 and 5 µm using the QIAamp Viral RNA Mini Kit (Qiagen). Real-time quantitative reverse transcription PCR amplification was performed to measure GAPDH mRNA concentration in AF. They showed a decrease in GAPDH mRNA concentration in AF samples with cell removal and filtration with decreasing pore size. The greatest decrease in GAPDH mRNA occurred after filtration through a 0.22 μm filter, whereas filtration did not significantly reduce the amount of cff β-globin DNA sequence present. The authors reasoned that the similarity with the data from cff nucleic acids in plasma suggests that there is a universal mechanism of cell-free nucleic acid processing. However, this has not been explored further.
Clinical applications of AF mRNA
Global gene expression analysis of AF was first reported in 2005 when Larrabee et al. (2005a) showed that it was feasible to isolate cff RNA from AF and hybridize it to oligonucleotide microarrays. The four cases were women between 20 and 32 weeks gestation undergoing therapeutic amnioreduction for polyhydramnios associated with twin-to-twin transfusion syndrome (TTTS) or ‘hydrops fetalis’. The RNA was extracted from large volumes of AF (range 57–180 ml), and after amplification, it was labelled and analysed using Affymetrix U133A arrays.
This study demonstrated that gene expression patterns vary among fetuses according to gender, gestational age and disease state. Of the 22 283 probe sets present on the microarray, a median of 20% had significant differences in their levels of expression between the cases and the pooled second-trimester control sample. Of interest was the statistically significant different gene expression observed in the aquaporin genes, a family of water transporters. Fetuses with TTTS had a significantly higher expression of aquaporin-1, suggesting that this gene may play a role in the polyhydramnios associated with this syndrome.
Expression differences according to gestational age in gene families associated with normal fetal development were also examined in this paper. The timing of surfactant gene expression was consistent with known patterns of fetal lung maturation, specifically the appearance of transcripts for surfactant proteins B and C at 17 weeks, their increased expression after 24 weeks, and the appearance of the third-trimester surfactant protein A only after 29 weeks. Numerous keratin gene transcripts were consistently isolated from the 17 week controls, which is not surprising given the large surface area of fetal skin in contact with AF. However, there was an observed decrease in keratin gene transcripts with gestational age, which the authors hypothesized might be due to the developing ‘stratum corneum’ acting as a barrier over the keratin-producing cells in the older fetuses, thereby limiting mRNA release into the AF. Transcripts for salivary and tracheobronchial/gastric mucins also appeared to increase with gestation, in keeping with known epithelial maturation processes.
In addition to these positive findings related to fetal development, there was no evidence that AF mRNA hybridized to placenta-specific gene sequences present on the U133A arrays. This suggests that placenta-derived RNA is absent in AF, despite being detectable in maternal plasma, supporting the concept that AF contains primarily nucleic acids derived from the fetus proper and thus represents a different population of cell-free nucleic acids from that in maternal blood.
More recently, a functional genomic approach has been utilized to study the development of fetuses affected by Down syndrome (Slonim et al., 2009). Slonim et al. characterized gene expression differences in fetuses with either trisomy 21 or normal chromosomes between 16 and 21 weeks gestation. After bioinformatics analysis, genes that differed between the trisomy 21 and euploid samples were identified. After controlling for gestational age and sex, there were 414 probe sets with statistically significant different individual expression levels. Of note, only 5 of these 414 probe sets were physically located on chromosome 21, which casts doubt on the traditional ‘gene dosage’ hypothesis for the pathogenesis of trisomy 21. The five genes on chromosome 21 that were statistically significantly differentially expressed in the Down syndrome fetuses were CLIC6 (a chloride intracellular channel), ITGB2 (integrin, β2), RUNX1 (a transcription factor associated with hematopoiesis) and two open reading frames, C21orf67 and C21orf86.
Further bioinformatics examination using a technique known as gene set enrichment analysis identified only a single chromosomal band, 21q22 (the Down syndrome critical region), with genes that were significantly up-regulated as a group. Synthesizing this gene expression data by using pathway analysis, the functional processes specifically disrupted in Down syndrome fetuses were identified as oxidative stress, ion transport and immune and stress response.
At present, it is unknown whether the cff RNA in AF plays a functional role in the AF. It is more likely that the genes detected originate from tissues in direct contract with AF, and therefore, reflect functional development of those tissues.
Discussion
This literature review identified 16 original studies investigating cff nucleic acids in AF (Table I). This small number is in stark contrast to the numerous primary studies examining cff nucleic acids in maternal plasma, which have been the subject of several recent reviews (Maron and Bianchi, 2007; Lo, 2009; Wright and Burton, 2009).
Table I.
Summary of included studies
| Included studies (in order of citation) | Study population (all mid-trimester AF supernatants unless otherwise stated) | Major findings |
|---|---|---|
| Bianchi et al. (2001) | 38 euploid pregnancies | First report of the presence cff DNA from AF supernatant; fetal DNA >100 times more concentrated than in maternal plasma |
| Lapaire et al. (2006) | 29 euploid pregnancies | Protocol optimization improves yields of cff DNA and reduces processing time |
| Lapaire et al. (2008) | ||
| Lapaire et al. (2007a) | 39 euploid and 4 aneuploid pregnancies | Gestational age, storage time and fetal karyotype influence cff DNA fragmentation patterns |
| Peter et al. (2008) | 36 euploid and 29 aneuploid pregnancies | Unique cff DNA fragmentation signatures identified according to fetal karyotype |
| Winter et al. (2008) | 58 euploid pregnancies | cff DNA is bound to high-mobility group protein HMGA2 |
| Lee et al. (2002) | Matched AF and maternal serum samples from 5 euploid and 6 trisomy 21 pregnancies | No correlation between cff DNA levels in maternal serum and AF |
| Zhong et al. (2006) | 12 matched AF and maternal plasma samples from euploid pregnancies | No correlation between cff DNA levels in maternal plasma and AF |
| Makrydimas et al. (2008) | 11 live pregnancies at 7–9 weeks undergoing surgical termination | Describes concentration gradient of cff DNA in different fetal/maternal compartments: AF (highest) → celomic fluid → maternal serum (lowest) |
| Lun et al. (2007) | 14 euploid pregnancies | Epigenetic approach shows placental DNA sequences are present in much lower proportions in AF compared with maternal plasma |
| Larrabee et al. (2004) | 17 euploid and 4 aneuploid pregnancies | Successful feasibility study of molecular karyotyping using array CGH analysis of AF cff DNA |
| Miura et al. (2006b) | 13 fetuses with congenital anomalies (6 aneuploid, 6 euploid and 1 balanced translocation) | Molecular karyotyping using targeted microarray-based CGH analysis of AF cff DNA |
| Lapaire et al. (2007b) | Nine aneuploid and one euploid fetus | Molecular karyotyping using array CGH to detect aneuploidy |
| Larrabee et al. (2005b) | Six normal pregnancies and one pregnancy with polyhydramnios | Filtration study showing that AF mRNA is particle-associated |
| Larrabee et al. (2005a) | Four polyhydramniotic pregnancies (20–32 weeks) and six normal controls (17 weeks) | Gene expression patterns vary according to gender, gestational age and disease state; placenta-specific gene sequences not detected in AF |
| Slonim et al. (2009) | Seven normal and seven trisomy 21 fetuses | Functional genomic analysis identifies specific pathways disrupted in Down syndrome pregnancies |
The reasons for this disparity are several. The most obvious is the risk of pregnancy loss associated with amniocentesis, which makes the collection of AF for only research indications unfeasible from an ethical viewpoint. Studies are therefore limited to using excess (discarded) fluid remaining from clinically indicated samples. This greatly reduces the numbers and gestational age range of the available samples for study.
The second major reason is that researchers have been justifiably focused on the non-invasive clinical applications of cff DNA for the diagnosis of fetal gender, rhesus genotyping and some single-gene disorders. These have transitioned into clinical care, resulting in reduced invasive testing rates of up to 45% in some populations (Chitty et al., 2007). Eliminating the need for chorionic villus sampling or amniocentesis to accurately diagnose fetal aneuploidy—still the most common indication for an invasive procedure—would revolutionize the prenatal diagnostic sector. There is no doubt that pregnant women would embrace such an advance.
A third reason for the lack of attention paid to the cff nucleic acids in AF is the perceived lack of clinical utility of examining the cell-free portion of the sample when an invasive procedure is indicated. In the majority of cases, all of the clinically relevant diagnostic information can be obtained from the cellular portion of the AF using conventional techniques on cultured amniocytes. Occasionally, other analytes may be measured in the AF, but essentially the supernatant is usually discarded.
There are several conclusions that can be drawn from the results of the studies identified in this literature review. The first is that it is feasible to isolate AF cff nucleic acids from small volumes of AF in sufficient quality for further downstream applications such as microarray analysis. cff DNA in AF is present in a much higher concentration than in maternal plasma and is able to be extracted using commercial kits from fresh or stored clinical specimens (Bianchi et al., 2001; Lapaire et al., 2008). Successive technical refinements over the past decade have also made it possible to extract RNA of sufficient quality and quantity to perform functional genomic analysis (Larrabee et al., 2005a; Slonim et al., 2009).
Second, there is an overall paucity of information relating to the biology of cff nucleic acids in AF. Information regarding differences in fragmentation patterns according to karyotype, gestational age and sample storage time from two studies support the concept that fetal conditions have an impact on the size of cff DNA fragments in the AF, but other than this very little has been published regarding their tissue sources or kinetics (Lapaire et al., 2007a; Peter et al., 2008). What can be concluded from the studies in this review is that the fetus itself is the predominant source of the nucleic acids in the AF, with very little direct mRNA contribution from the placenta, as demonstrated by the microarray (Larrabee et al., 2005a), and epigenetic studies (Lun et al., 2007). Other studies confirm that the pool of cff nucleic acids in the AF is independent from the pool of placenta-derived plasma cff DNA and RNA (Ng et al., 2003; Bischoff et al., 2005; Tjoa et al., 2006). Although it is possible to identify mRNA transcripts present in umbilical cord blood in the maternal circulation (Maron et al., 2007), several studies have established that this trafficking is unidirectional from fetus to mother, making the prospect of contaminating maternal nucleic acids in the amniotic cavity extremely unlikely (Ng et al., 2003; Sekizawa et al., 2003; Maron et al., 2007).
This review also highlights increasing total quantities of cff DNA and differences in mRNA expression as a function of gestational age (Lapaire et al., 2007a). This suggests some similarities with plasma and serum biology. However, specific data on the kinetics, half-life and function of cff nucleic acids in AF are lacking. The issue of kinetics is relevant to AF mRNA, as transcription is a dynamic process that varies rapidly according to the physiological demands on the body, including diurnal rhythms. cff mRNA has a short half-life in maternal plasma of ∼15 min (Chiu et al., 2006), but mRNAs also decay at different rates according to their functional characteristics and sequence attributes (Yang et al., 2003). How the results from other biological models relate to the steady-state population of cff nucleic acids in AF is unknown. This has significance for the interpretation of AF gene expression studies, as to whether they represent a real-time or a delayed snapshot of global fetal transcriptional activity.
Similarly, the relative contributions of various fetal tissues to the cff NA in AF has not been the subject of any research to date. Data from the proteomic studies of AF discussed in the introduction suggest that numerous fetal organs contribute to the protein content of AF, including those systems not in direct physical continuity with amniotic cavity, such as the fetal heart, brain and skeletal muscle. It seems reasonable to assume from these data that the tissue sources of cell-free nucleic acids in AF are similarly multiple, although whether they are in similar proportions to protein expression is uncertain, particularly given that the large number of placental proteins identified in proteomic studies is inconsistent with the low levels of placental mRNA transcripts found in AF. Furthermore, in the proteomic study of Cho et al., all keratin-related entries were removed as contaminants, so that fetal skin was not among the top 10 tissues with the most number of proteins in AF as one might expect (Cho et al., 2007).
Related to both these unanswered questions is the unknown nature of its particle association. The filtration study by Larrabee et al. (2005b) confirmed that filterable and non-filterable forms of cff mRNA exist in AF, supporting the existence of particle-associated forms like those in maternal plasma. However, no studies investigating the types of microparticles associated with NA in AF were identified by this review.
Amniotic fluid supernatant is a valuable biological sample
The final important conclusion from this review is that AF supernatant is a potentially valuable but under-utilized biological sample. Two studies have successfully performed gene expression analyses on cff mRNA in AF using a ‘discovery-driven’, rather than hypothesis-driven, approach (Larrabee et al., 2005a; Slonim et al., 2009). These reports establish that it is possible to identify differentially regulated genes in normal and abnormal fetuses, and employ bioinformatics tools to uncover the pathways and networks involved in the pathogenesis of specific diseases.
In the study comparing fetuses affected by polyhydramnios with normal fetuses, Larrabee et al. (2005a) identified a water transporter gene transcript, aquaporin-1, that is statistically significantly increased by up to 18-fold in recipient twins affected by TTTS. Similarly, Slonim et al. (2009) provided new insights into the pathogenesis of Down syndrome by identifying a role for oxidative stress, ion transport and immune and stress responses in the abnormal development of the trisomy 21 fetus.
Implications for future research
Functional genomics in obstetrics and fetal medicine
Gene expression studies on live human fetuses have been restricted by access to appropriate tissue such as fetal blood. Post-partum gene expression studies using stillborn, neonatal or placental samples are inherently limited in their ability to reflect prenatal events. Until recently, the main applications of RNA-based functional genomics in obstetrics have been in the study of normal parturition (Aguan et al., 2000; Havelock et al., 2005; Hassan et al., 2009; Montenegro et al., 2009) and spontaneous preterm birth (Romero et al., 2006; Enquobahrie et al., 2009), using myometrial, cervical or amniotic membrane specimens. More recently, researchers have applied a global gene expression approach to pre-eclampsia examining various tissues such as maternal blood (Donker et al., 2005; Sun et al., 2009), post-partum placentas (Zhou et al., 2006; Centlow et al., 2008; Enquobahrie et al., 2008; Toft et al., 2008; Sitras et al., 2009; Zhu et al., 2009), umbilical vein epithelial cells (Hoegh et al., 2006b), endometrial epithelial cells (Hoegh et al., 2006a) and first-trimester chorionic villus samples (Founds et al., 2009). Furthermore, mRNA-based microarray analysis has also recently been used to describe the ‘transcriptome of fetal inflammation’ derived from umbilical cord blood (Madsen-Bouterse et al., 2010).
Despite this rapidly growing interest in utilizing functional genomics in obstetrics, very few global gene expression studies have been performed on live ongoing pregnancies for specific abnormalities of fetal development. First-trimester chorionic villus samples have been investigated with microarray analysis to study the mechanisms involved with enlarged nuchal translucencies in euploid fetuses (Farina et al., 2006) and for the prediction of pre-eclampsia (Founds et al., 2009), but these represent placental rather than fetal transcription profiles. An alternative approach in which global gene expression is compared between antenatal, post-natal and neonatal samples to identify significantly up-regulated fetal transcripts in maternal blood has been successfully employed to produce new insights into fetal development in normal term babies (Maron et al., 2007) and to develop potential fetal biomarkers for congenital heart disease (Arcelli et al., 2010). However, beyond the two studies identified in this review (Larrabee et al., 2005a; Slonim et al., 2009), no other reports specifically used AF cff mRNA to advance knowledge regarding fetal development.
AF microparticles and cff mRNA
Another potential direction for future research would be to integrate developments in the biology of microparticles in plasma with those in cff nucleic acids in AF to determine whether cff mRNA has a functional role as a novel form of cell-to-cell genetic signalling within the amniotic cavity. Until recently, cff nucleic acids have been conceived of as by-products of cell turnover and it is unknown if they possess any biological activity in the amniotic cavity.
Exosomes are small vesicles <100 nm diameter of endosomal origin that are released by a wide variety of cells into the extracellular space (Simpson et al., 2009). They contain proteins and RNA derived from the parent cell and are thus able to be characterized according to cell-specific markers. They are relevant to cell-free nucleic acid biology because they contain mRNA and miRNA sequences that are transferrable to other cells, thus leading to the translation of unique proteins in in vitro and in vivo models (Valadi et al., 2007). This suggests that cell-free RNA can play an active role in cell-to-cell genetic exchange, as so-called exosomal shuttle RNA.
Exosomes are secreted by many different cell types and are present in adult and neonatal urine (Pisitkun et al., 2004; Keller et al., 2007). Using the CD24 membrane protein as a marker of renal cell origin, Keller et al. (2007) showed that AF from second-trimester pregnancies contains exosomes that are secreted by the fetal kidney. Their function in the AF is not known, but it is interesting to speculate on whether they have role in fetal and AF homeostasis, particularly as they were shown to contain aquaporin-2. If functional mRNA could be demonstrated in kidney-derived AF exosomes, then this may provide a novel mechanism for the fetal kidneys to directly influence the intra-membranous route of fluid transport in amniotic membranes.
This proposed method of fetal-maternal communication via AF exosomes could extend to other systems than fluid balance. In the only other report on exosomes in AF, Asea et al. (2008) suggest a possible role for exosomes in immune regulation. They detected the presence of heat shock protein-containing exosomes in second-trimester AF specimens. Heat shock protein is known to play a role in microbial-induced and sterile forms of inflammation (Quintana and Cohen, 2005). AF contains decidual natural killer cells. AF exosomes may therefore be a mechanism by which the fetus and the maternal immune system communicate within the uterine cavity.
Neither of these two AF exosome studies has specifically sought to confirm the presence of fetal mRNA in these microparticles. Although the body of literature on exosomes in other body fluids supports the hypothesis that AF exosomes should also contain functional mRNA, it would be an important research priority to confirm this. Delineating the functional significance of these exosomal forms of cff nucleic acids in AF would then open up an entirely new field of research, one in which various fetal organ systems could communicate on a genetic basis with each other, the fetal membranes, placenta and mother.
Conclusions
Cell-free nucleic acids in AF have been relatively neglected in favour of those in maternal plasma. The literature review makes a strong case for researchers in cff nucleic acids to turn their attention to AF because it is a pure fetal sample that provides fetal developmental genetic information not provided by maternal plasma studies. To date, the studies on AF have shown that discarded AF supernatant from clinical specimens is both a feasible and rewarding biological sample for generating hypotheses for further research. Functional genomic analysis allows us to advance applications of cff nucleic acids beyond the prenatal diagnosis of aneuploidy to broader studies on human fetal development. Ultimately, the information derived may lead to new insights into normal development, fetal disease processes, novel biomarkers for diagnosis and new approaches to antenatal treatment of human disease.
Supplementary data
Supplementary data are available at http://humupd.oxfordjournals.org/.
Authors' roles
L.H.: Study design, literature search, data interpretation and manuscript preparation. D.W.B.: Study design and critical review of manuscript.
Funding
L.H. is the recipient of the Albert S. McKern scholarship from the University of Sydney and the Fotheringham Fellowship from the Royal Australian and New Zealand College of Obstetricians and Gynaecologists. D.W.B. receives grant support from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (HD 42053-07).
Supplementary Material
References
- Aguan K, Carvajal JA, Thompson LP, Weiner CP. Application of a functional genomics approach to identify differentially expressed genes in human myometrium during pregnancy and labour. Mol Hum Reprod. 2000;6:1141–1145. doi: 10.1093/molehr/6.12.1141. [DOI] [PubMed] [Google Scholar]
- Alam M. Proteomics-based approach for identification and purification of human phosphate binding apolipoprotein from amniotic fluid. Genet Mol Res. 2009;8:929–937. doi: 10.4238/vol8-3gmr620. [DOI] [PubMed] [Google Scholar]
- Altug-Teber O, Bonin M, Walter M, Mau-Holzmann UA, Dufke A, Stappert H, Tekesin I, Heilbronner H, Nieselt K, Riess O. Specific transcriptional changes in human fetuses with autosomal trisomies. Cytogenet Genome Res. 2007;119:171–184. doi: 10.1159/000112058. [DOI] [PubMed] [Google Scholar]
- Anagnostopoulos AK, Kolialexi A, Mavrou A, Vougas K, Papantoniou N, Antsaklis A, Kanavakis E, Fountoulakis M, Tsangaris GT. Proteomic analysis of amniotic fluid in pregnancies with Klinefelter syndrome fetuses. J Proteomics. 2009;73:943–950. doi: 10.1016/j.jprot.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Angert RM, Leshane ES, Yarnell RW, Johnson KL, Bianchi DW. Cell-free fetal DNA in the cerebrospinal fluid of women during the peripartum period. Am J Obstet Gynecol. 2004;190:1087–1090. doi: 10.1016/j.ajog.2003.10.562. [DOI] [PubMed] [Google Scholar]
- Arcelli D, Farina A, Cappuzzello C, Bresin A, De Sanctis P, Perolo A, Prandstraller D, Valentini D, Zucchini C, Priori S, et al. Identification of circulating placental mRNA in maternal blood of pregnancies affected with fetal congenital heart diseases at the second trimester of pregnancy: implications for early molecular screening. Prenat Diagn. 2010;30:229–234. doi: 10.1002/pd.2443. [DOI] [PubMed] [Google Scholar]
- Asea A, Jean-Pierre C, Kaur P, Rao P, Linhares IM, Skupski D, Witkin SS. Heat shock protein-containing exosomes in mid-trimester amniotic fluids. J Reprod Immunol. 2008;79:12–17. doi: 10.1016/j.jri.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Bianchi DW, LeShane ES, Cowan JM. Large amounts of cell-free fetal DNA are present in amniotic fluid. Clin Chem. 2001;47:1867–1869. [PubMed] [Google Scholar]
- Bianchi DW, Wataganara T, Lapaire O, Tjoa ML, Maron JL, Larrabee PB, Johnson KL. Fetal nucleic acids in maternal body fluids: an update. Ann N Y Acad Sci. 2006;1075:63–73. doi: 10.1196/annals.1368.008. [DOI] [PubMed] [Google Scholar]
- Bischoff FZ, Lewis DE, Simpson JL. Cell-free fetal DNA in maternal blood: kinetics, source and structure. Hum Reprod Update. 2005;11:59–67. doi: 10.1093/humupd/dmh053. [DOI] [PubMed] [Google Scholar]
- Buhimschi IA, Buhimschi CS. Proteomics of the amniotic fluid in assessment of the placenta. Relevance for preterm birth. Placenta. 2008;29(Suppl A):S95–101. doi: 10.1016/j.placenta.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhimschi IA, Zhao G, Rosenberg VA, Abdel-Razeq S, Thung S, Buhimschi CS. Multidimensional proteomics analysis of amniotic fluid to provide insight into the mechanisms of idiopathic preterm birth. PLoS ONE. 2008;3:e2049. doi: 10.1371/journal.pone.0002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centlow M, Carninci P, Nemeth K, Mezey E, Brownstein M, Hansson SR. Placental expression profiling in preeclampsia: local overproduction of hemoglobin may drive pathological changes. Fertil Steril. 2008;90:1834–1843. doi: 10.1016/j.fertnstert.2007.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KC, Zhang J, Hui AB, Wong N, Lau TK, Leung TN, Lo KW, Huang DW, Lo YM. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem. 2004;50:88–92. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
- Chan KC, Ding C, Gerovassili A, Yeung SW, Chiu RW, Leung TN, Lau TK, Chim SS, Chung GT, Nicolaides KH, et al. Hypermethylated RASSF1A in maternal plasma: a universal fetal DNA marker that improves the reliability of noninvasive prenatal diagnosis. Clin Chem. 2006;52:2211–2218. doi: 10.1373/clinchem.2006.074997. [DOI] [PubMed] [Google Scholar]
- Chitty L, Mistry B, Hogg J, Meaney C, Thomasson L, Norbury G, Daniels G, Finning K, Martin P. Prospective register of outcomes of free-fetal DNA testing (PROOF)—results of the first year's audit. Newsletter of the British Society for Human Genetics. 2007;37:8–9. [Google Scholar]
- Chiu RW, Lui WB, Cheung MC, Kumta N, Farina A, Banzola I, Grotti S, Rizzo N, Haines CJ, Lo YM. Time profile of appearance and disappearance of circulating placenta-derived mRNA in maternal plasma. Clin Chem. 2006;52:313–316. doi: 10.1373/clinchem.2005.059691. [DOI] [PubMed] [Google Scholar]
- Chiu RW, Chim SS, Wong IH, Wong CS, Lee WS, To KF, Tong JH, Yuen RK, Shum AS, Chan JK, et al. Hypermethylation of RASSF1A in human and rhesus placentas. Am J Pathol. 2007;170:941–950. doi: 10.2353/ajpath.2007.060641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho CK, Shan SJ, Winsor EJ, Diamandis EP. Proteomics analysis of human amniotic fluid. Mol Cell Proteomics. 2007;6:1406–1415. doi: 10.1074/mcp.M700090-MCP200. [DOI] [PubMed] [Google Scholar]
- Chou CY, Liu LY, Chen CY, Tsai CH, Hwa HL, Chang LY, Lin YS, Hsieh FJ. Gene expression variation increase in trisomy 21 tissues. Mamm Genome. 2008;19:398–405. doi: 10.1007/s00335-008-9121-1. [DOI] [PubMed] [Google Scholar]
- Chung IH, Lee SH, Lee KW, Park SH, Cha KY, Kim NS, Yoo HS, Kim YS, Lee S. Gene expression analysis of cultured amniotic fluid cell with Down syndrome by DNA microarray. J Korean Med Sci. 2005;20:82–87. doi: 10.3346/jkms.2005.20.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch E, Rust K, Marienchek W, Parghi D, Chang D, Persson A. Developmental expression of pulmonary surfactant protein D (SP-D) Am J Respir Cell Mol Biol. 1991;5:13–18. doi: 10.1165/ajrcmb/5.1.13. [DOI] [PubMed] [Google Scholar]
- Da Sacco S, Sedrakyan S, Boldrin F, Giuliani S, Parnigotto P, Habibian R, Warburton D, De Filippo RE, Perin L. Human amniotic fluid as a potential new source of organ specific precursor cells for future regenerative medicine applications. J Urol. 2010;183:1193–1200. doi: 10.1016/j.juro.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng B, Dong Z, Liu Y, Wang C, Liu J, Wang C, Qu X. Effects of pretreatment protocols on human amniotic fluid protein profiling with SELDI-TOF MS using protein chips and magnetic beads. Clinica Chimica Acta. 2010;411:1051–1057. doi: 10.1016/j.cca.2010.03.036. [DOI] [PubMed] [Google Scholar]
- Donker RB, Asgeirsdottir SA, Gerbens F, van Pampus MG, Kallenberg CG, te Meerman GJ, Aarnoudse JG, Molema G. Plasma factors in severe early-onset preeclampsia do not substantially alter endothelial gene expression in vitro. J Soc Gynecol Investig. 2005;12:98–106. doi: 10.1016/j.jsgi.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Enquobahrie DA, Meller M, Rice K, Psaty BM, Siscovick DS, Williams MA. Differential placental gene expression in preeclampsia. Am J Obstet Gynecol. 2008;199:566.e1–11. doi: 10.1016/j.ajog.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquobahrie DA, Williams MA, Qiu C, Muhie SY, Slentz-Kesler K, Ge Z, Sorenson T. Early pregnancy peripheral blood gene expression and risk of preterm delivery: a nested case control study. BMC Pregnancy Childbirth. 2009;9:56. doi: 10.1186/1471-2393-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina A, Volinia S, Arcelli D, Francioso F, Desanctis P, Zucchini C, Pilu G, Carinci P, Morano D, Pittalis MC, et al. Evidence of genetic underexpression in chorionic villi samples of euploid fetuses with increased nuchal translucency at 10–11 weeks' gestation. Prenat Diagn. 2006;26:128–133. doi: 10.1002/pd.1373. [DOI] [PubMed] [Google Scholar]
- Founds SA, Conley YP, Lyons-Weiler JF, Jeyabalan A, Hogge WA, Conrad KP. Altered global gene expression in first trimester placentas of women destined to develop preeclampsia. Placenta. 2009;30:15–24. doi: 10.1016/j.placenta.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibert J, Benachi A, Grebille AG, Ernault P, Zorn JR, Costa JM. Kinetics of SRY gene appearance in maternal serum: detection by real time PCR in early pregnancy after assisted reproductive technique. Hum Reprod. 2003;18:1733–1736. doi: 10.1093/humrep/deg320. [DOI] [PubMed] [Google Scholar]
- Gupta AK, Holzgreve W, Huppertz B, Malek A, Schneider H, Hahn S. Detection of fetal DNA and RNA in placenta-derived syncytiotrophoblast microparticles generated in vitro. Clin Chem. 2004;50:2187–2190. doi: 10.1373/clinchem.2004.040196. [DOI] [PubMed] [Google Scholar]
- Hassan SS, Romero R, Tarca AL, Nhan-Chang CL, Vaisbuch E, Erez O, Mittal P, Kusanovic JP, Mazaki-Tovi S, Yeo L, et al. The transcriptome of cervical ripening in human pregnancy before the onset of labor at term: identification of novel molecular functions involved in this process. J Matern Fetal Neonatal Med. 2009;22:1183–1193. doi: 10.3109/14767050903353216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmann DO, Rappl G, Tilgen W, Reinhold U. Extracellular tyrosinase mRNA within apoptotic bodies is protected from degradation in human serum. Clin Chem. 2001;47:1488–1489. [PubMed] [Google Scholar]
- Havelock JC, Keller P, Muleba N, Mayhew BA, Casey BM, Rainey WE, Word RA. Human myometrial gene expression before and during parturition. Biol Reprod. 2005;72:707–719. doi: 10.1095/biolreprod.104.032979. [DOI] [PubMed] [Google Scholar]
- Hoegh AM, Islin H, Moller C, Sorensen S, Hviid TV. Identification of differences in gene expression in primary cell cultures of human endometrial epithelial cells and trophoblast cells following their interaction. J Reprod Immunol. 2006a;70:1–19. doi: 10.1016/j.jri.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Hoegh AM, Tannetta D, Sargent I, Borup R, Nielsen FC, Redman C, Sorensen S, Hviid TV. Effect of syncytiotrophoblast microvillous membrane treatment on gene expression in human umbilical vein endothelial cells. BJOG. 2006b;113:1270–1279. doi: 10.1111/j.1471-0528.2006.01061.x. [DOI] [PubMed] [Google Scholar]
- Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, Hager HD, Abdel-Bakky MS, Gutwein P, Altevogt P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72:1095–1102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- Koide K, Sekizawa A, Iwasaki M, Matsuoka R, Honma S, Farina A, Saito H, Okai T. Fragmentation of cell-free fetal DNA in plasma and urine of pregnant women. Prenat Diagn. 2005;25:604–607. doi: 10.1002/pd.1213. [DOI] [PubMed] [Google Scholar]
- Kolialexi A, Anagnostopoulos AK, Mavrou A, Tsangaris GT. Application of proteomics for diagnosis of fetal aneuploidies and pregnancy complications. J Proteomics. 2009;72:731–739. doi: 10.1016/j.jprot.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Lapaire O, Stroh H, Peter I, Cowan JM, Tantravahi U, O'Brien B, Bianchi DW, Johnson KL. Larger columns and change of lysis buffer increase the yield of cell-free DNA extracted from amniotic fluid. Clin Chem. 2006;52:156–157. doi: 10.1373/clinchem.2005.058420. [DOI] [PubMed] [Google Scholar]
- Lapaire O, Bianchi DW, Peter I, O'Brien B, Stroh H, Cowan JM, Tantravahi U, Johnson KL. Cell-free fetal DNA in amniotic fluid: unique fragmentation signatures in euploid and aneuploid fetuses. Clin Chem. 2007a;53:405–411. doi: 10.1373/clinchem.2006.076083. [DOI] [PubMed] [Google Scholar]
- Lapaire O, Lu XY, Johnson KL, Jarrah Z, Stroh H, Cowan JM, Tantravahi U, Bianchi DW. Array-CGH analysis of cell-free fetal DNA in 10 ml of amniotic fluid supernatant. Prenat Diagn. 2007b;27:616–621. doi: 10.1002/pd.1752. [DOI] [PubMed] [Google Scholar]
- Lapaire O, Johnson KL, Bianchi DW. Method for extraction of high-quantity and -quality cell-free DNA from amniotic fluid. Methods Mol Biol. 2008;444:303–309. doi: 10.1007/978-1-59745-066-9_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrabee PB, Johnson KL, Pestova E, Lucas M, Wilber K, LeShane ES, Tantravahi U, Cowan JM, Bianchi DW. Microarray analysis of cell-free fetal DNA in amniotic fluid: a prenatal molecular karyotype. Am J Hum Genet. 2004;75:485–491. doi: 10.1086/423288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrabee PB, Johnson KL, Lai C, Ordovas J, Cowan JM, Tantravahi U, Bianchi DW. Global gene expression analysis of the living human fetus using cell-free messenger RNA in amniotic fluid. J Am Med Assoc. 2005a;293:836–842. doi: 10.1001/jama.293.7.836. [DOI] [PubMed] [Google Scholar]
- Larrabee PB, Johnson KL, Peter I, Bianchi DW. Presence of filterable and nonfilterable cell-free mRNA in amniotic fluid. Clin Chem. 2005b;51:1024–1026. doi: 10.1373/clinchem.2004.047670. [DOI] [PubMed] [Google Scholar]
- Lazar L, Nagy B, Ban Z, Nagy GR, Beke A, Papp Z. [Non invasive detection of fetal Rh using real-time PCR method] Orv Hetil. 2007;148:497–500. doi: 10.1556/OH.2007.27904. [DOI] [PubMed] [Google Scholar]
- Lee T, LeShane ES, Messerlian GM, Canick JA, Farina A, Heber WW, Bianchi DW. Down syndrome and cell-free fetal DNA in archived maternal serum. Am J Obstet Gynecol. 2002;187:1217–1221. doi: 10.1067/mob.2002.127462. [DOI] [PubMed] [Google Scholar]
- Lee J, Park JS, Norwitz ER, Kim BJ, Park CW, Jun JK, Syn HC. Identification and characterization of proteins in amniotic fluid that are differentially expressed before and after antenatal corticosteroid administration. Am J Obstet Gynecol. 2010;202:e1–10. doi: 10.1016/j.ajog.2010.01.056. [DOI] [PubMed] [Google Scholar]
- Lo YM. Noninvasive prenatal detection of fetal chromosomal aneuploidies by maternal plasma nucleic acid analysis: a review of the current state of the art. BJOG. 2009;116:152–157. doi: 10.1111/j.1471-0528.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- Lo YM, Chiu RW. Next-generation sequencing of plasma/serum DNA: an emerging research and molecular diagnostic tool. Clin Chem. 2009;55:607–608. doi: 10.1373/clinchem.2009.123661. [DOI] [PubMed] [Google Scholar]
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YM, Lau TK, Zhang J, Leung TN, Chang AM, Hjelm NM, Elmes RS, Bianchi DW. Increased fetal DNA concentrations in the plasma of pregnant women carrying fetuses with trisomy 21. Clin Chem. 1999a;45:1747–1751. [PubMed] [Google Scholar]
- Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999b;64:218–224. doi: 10.1086/302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotvall J, Valadi H. Cell to cell signalling via exosomes through esRNA. Cell Adh Migr. 2007;1:156–158. doi: 10.4161/cam.1.3.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun FM, Chiu RW, Leung TY, Leung TN, Lau TK, Lo YM. Epigenetic analysis of RASSF1A gene in cell-free DNA in amniotic fluid. Clin Chem. 2007;53:796–798. doi: 10.1373/clinchem.2006.084350. [DOI] [PubMed] [Google Scholar]
- Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A, et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- Madsen-Bouterse SA, Romero R, Tarca AL, Kusanovic JP, Espinoza J, Kim CJ, Kim JS, Edwin SS, Gomez R, Draghici S. The transcriptome of the fetal inflammatory response syndrome. Am J Reprod Immunol. 2010;63:73–92. doi: 10.1111/j.1600-0897.2009.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrydimas G, Gerovassili A, Sotiriadis A, Kavvadias A, Nicolaides KH. Cell-free fetal DNA in celomic fluid. Ultrasound Obstet Gynecol. 2008;32:594–595. doi: 10.1002/uog.6117. [DOI] [PubMed] [Google Scholar]
- Maron JL, Bianchi DW. Prenatal diagnosis using cell-free nucleic acids in maternal body fluids: a decade of progress. Am J Med Genet C Semin Med Genet. 2007;145C:5–17. doi: 10.1002/ajmg.c.30115. [DOI] [PubMed] [Google Scholar]
- Maron JL, Johnson KL, Slonim D, Lai CQ, Ramoni M, Alterovitz G, Jarrah Z, Yang Z, Bianchi DW. Gene expression analysis in pregnant women and their infants identifies unique fetal biomarkers that circulate in maternal blood. J Clin Invest. 2007;117:3007–3019. doi: 10.1172/JCI29959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Yoshiura K, Miura S, Yamasaki K, Nakayama D, Ishimaru T, Wagstaff J, Niikawa N, Masuzaki H. Cell-free DNA is more sensitive than cell-free mRNA as a marker for evaluation of fetal-maternal hemorrhage. Clin Chem. 2006a;52:2121–2123. doi: 10.1373/clinchem.2006.075697. [DOI] [PubMed] [Google Scholar]
- Miura S, Miura K, Masuzaki H, Miyake N, Yoshiura K, Sosonkina N, Harada N, Shimokawa O, Nakayama D, Yoshimura S, et al. Microarray comparative genomic hybridization (CGH)-based prenatal diagnosis for chromosome abnormalities using cell-free fetal DNA in amniotic fluid. J Hum Genet. 2006b;51:412–417. doi: 10.1007/s10038-006-0376-7. [DOI] [PubMed] [Google Scholar]
- Montenegro D, Romero R, Kim SS, Tarca AL, Draghici S, Kusanovic JP, Kim JS, Lee DC, Erez O, Gotsch F, et al. Expression patterns of microRNAs in the chorioamniotic membranes: a role for microRNAs in human pregnancy and parturition. J Pathol. 2009;217:113–121. doi: 10.1002/path.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EK, Tsui NB, Lau TK, Leung TN, Chiu RW, Panesar NS, Lit LC, Chan KW, Lo YM. mRNA of placental origin is readily detectable in maternal plasma. Proc Natl Acad Sci USA. 2003;100:4748–4753. doi: 10.1073/pnas.0637450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco AF, Bischoff FZ, Horne C, Popek E, Simpson JL, Lewis DE. Hypoxia-induced membrane-bound apoptotic DNA particles: potential mechanism of fetal DNA in maternal plasma. Ann N Y Acad Sci. 2006;1075:57–62. doi: 10.1196/annals.1368.007. [DOI] [PubMed] [Google Scholar]
- Orozco AF, Jorgez CJ, Horne C, Marquez-Do DA, Chapman MR, Rodgers JR, Bischoff FZ, Lewis DE. Membrane protected apoptotic trophoblast microparticles contain nucleic acids: relevance to preeclampsia. Am J Pathol. 2008;173:1595–1608. doi: 10.2353/ajpath.2008.080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter I, Tighiouart H, Lapaire O, Johnson KL, Bianchi DW, Terrin N. Cell-free DNA fragmentation patterns in amniotic fluid identify genetic abnormalities and changes due to storage. Diagn Mol Pathol. 2008;17:185–190. doi: 10.1097/PDM.0b013e31815bcdb6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon LL, Leung TN, Lau TK, Lo YM. Presence of fetal RNA in maternal plasma. Clin Chem. 2000;46:1832–1834. [PubMed] [Google Scholar]
- Quintana FJ, Cohen IR. Heat shock proteins as endogenous adjuvants in sterile and septic inflammation. J Immunol. 2005;175:2777–2782. doi: 10.4049/jimmunol.175.5.2777. [DOI] [PubMed] [Google Scholar]
- Romero R, Espinoza J, Gotsch F, Kusanovic JP, Friel LA, Erez O, Mazaki-Tovi S, Than NG, Hassan S, Tromp G. The use of high-dimensional biology (genomics, transcriptomics, proteomics, and metabolomics) to understand the preterm parturition syndrome. BJOG. 2006;113:118–135. doi: 10.1111/j.1471-0528.2006.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Kusanovic JP, Gotsch F, Erez O, Vaisbuch E, Mazaki-Tovi S, Moser A, Tam S, Leszyk J, Master SR, et al. Isobaric labeling and tandem mass spectrometry: a novel approach for profiling and quantifying proteins differentially expressed in amniotic fluid in preterm labor with and without intra-amniotic infection/ inflammation. J Matern Fetal Neonatal Med. 2010;23:261–280. doi: 10.3109/14767050903067386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samura O, Miharu N, Hyodo M, Honda H, Ohashi Y, Honda N, Hara T, Ohama K. Cell-free fetal DNA in maternal circulation after amniocentesis. Clin Chem. 2003;49:1193–1195. doi: 10.1373/49.7.1193. [DOI] [PubMed] [Google Scholar]
- Sekizawa A, Yokokawa K, Sugito Y, Iwasaki M, Yukimoto Y, Ichizuka K, Saito H, Okai T. Evaluation of bidirectional transfer of plasma DNA through placenta. Hum Genet. 2003;113:307–310. doi: 10.1007/s00439-003-0987-4. [DOI] [PubMed] [Google Scholar]
- Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- Sitras V, Paulssen RH, Gronaas H, Leirvik J, Hanssen TA, Vartun A, Acharya G. Differential placental gene expression in severe preeclampsia. Placenta. 2009;30:424–433. doi: 10.1016/j.placenta.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Slonim DK, Koide K, Johnson KL, Tantravahi U, Cowan JM, Jarrah Z, Bianchi DW. Functional genomic analysis of amniotic fluid cell-free mRNA suggests that oxidative stress is significant in Down syndrome fetuses. Proc Natl Acad Sci USA. 2009;106:9425–9429. doi: 10.1073/pnas.0903909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CJ, Zhang L, Zhang WY. Gene expression profiling of maternal blood in early onset severe preeclampsia: identification of novel biomarkers. J Perinat Med. 2009;37:609–616. doi: 10.1515/JPM.2009.103. [DOI] [PubMed] [Google Scholar]
- Tjoa ML, Cindrova-Davies T, Spasic-Boskovic O, Bianchi DW, Burton GJ. Trophoblastic oxidative stress and the release of cell-free feto-placental DNA. Am J Pathol. 2006;169:400–404. doi: 10.2353/ajpath.2006.060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft JH, Lian IA, Tarca AL, Erez O, Espinoza J, Eide IP, Bjorge L, Draghici S, Romero R, Austgulen R. Whole-genome microarray and targeted analysis of angiogenesis-regulating gene expression (ENG, FLT1, VEGF, PlGF) in placentas from pre-eclamptic and small-for-gestational-age pregnancies. J Matern Fetal Neonatal Med. 2008;21:267–273. doi: 10.1080/14767050801924118. [DOI] [PubMed] [Google Scholar]
- Tong XL, Wang L, Gao TB, Qin YG, Qi YQ, Xu YP. Potential function of amniotic fluid in fetal development—novel insights by comparing the composition of human amniotic fluid with umbilical cord and maternal serum at mid and late gestation. J Chin Med Assoc. 2009;72:368–373. doi: 10.1016/S1726-4901(09)70389-2. [DOI] [PubMed] [Google Scholar]
- Tsangaris GT, Kolialexi A, Karamessinis PM, Anagnostopoulos AK, Antsaklis A, Fountoulakis M, Mavrou A. The normal human amniotic fluid supernatant proteome. In Vivo. 2006;20:479–490. [PubMed] [Google Scholar]
- Underwood MA, Gilbert WM, Sherman MP. Amniotic fluid: not just fetal urine anymore. J Perinatol. 2005;5:341–348. doi: 10.1038/sj.jp.7211290. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Wataganara T, Bianchi DW. Fetal cell-free nucleic acids in the maternal circulation: new clinical applications. Ann N Y Acad Sci. 2004;1022:90–99. doi: 10.1196/annals.1318.015. [DOI] [PubMed] [Google Scholar]
- Winter N, Neumann A, Bullerdiek J. Cell-free DNA in amniotic fluid remains to be attached to HMGA2-implications for noninvasive prenatal diagnosis. Prenat Diagn. 2008;28:1126–1130. doi: 10.1002/pd.2140. [DOI] [PubMed] [Google Scholar]
- Wright CF, Burton H. The use of cell-free fetal nucleic acids in maternal blood for non-invasive prenatal diagnosis. Hum Reprod Update. 2009;15:139–151. doi: 10.1093/humupd/dmn047. [DOI] [PubMed] [Google Scholar]
- Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE., Jr Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Holzgreve W, Tercanli S, Wenzel F, Hahn S. Cell-free foetal DNA in maternal plasma does not appear to be derived from the rich pool of cell-free foetal DNA in amniotic fluid. Arch Gynecol Obstet. 2006;273:221–226. doi: 10.1007/s00404-005-0068-0. [DOI] [PubMed] [Google Scholar]
- Zhou R, Zhu Q, Wang Y, Ren Y, Zhang L, Zhou Y. Genomewide oligonucleotide microarray analysis on placentae of pre-eclamptic pregnancies. Gynecol Obstet Invest. 2006;62:108–114. doi: 10.1159/000092857. [DOI] [PubMed] [Google Scholar]
- Zhu XM, Han T, Sargent IL, Yin GW, Yao YQ. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 2009;200:661.e1-7. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.