Abstract
The nucleotide-binding and oligomerization, leucine-rich repeat (NLR) family of proteins sense microbial infections and activate the inflammasome, a multi-protein complex that promotes microbial clearance. Kaposi's sarcoma-associated herpesvirus (KSHV) is linked to several human malignancies. We report that KSHV Orf63 is a viral homolog of human NLRP1. Orf63 blocked NLRP1-dependent innate immune responses, including caspase-1 activation and processing of interleukin (IL)-1β and IL-18. KSHV Orf63 interacted with NLRP1, NLRP3, and NOD2. Inhibition of Orf63 expression resulted in increased expression of IL-1β during the KSHV lifecycle. Furthermore, inhibition of NLRP1 was necessary for efficient reactivation and generation of progeny virus. The viral homolog subverts the function of cellular NLRs, which suggests that modulation of NLR-mediated innate immunity is important for the life-long persistence of herpesviruses.
Keywords: Inflammasome, NLRP1, NOD2, NLRP3, KSHV, Innate Immunity
Kaposi's sarcoma-associated herpesvirus (KSHV/HHV8) is the etiological agent of several human cancers including Kaposi's sarcoma (KS), primary effusion lymphoma (PEL) and multicentric Castleman's disease (MCD) (1, 2). The ability of KSHV to evade host innate immunity is essential for productive infection, latency and life-long persistence (3).
Several families of pattern recognition receptors (PRRs) have been described: Toll-like receptors (TLRs), nucleotide-binding and oligomerization, leucine-rich repeat (NLR) proteins, retinoic acid inducible gene (RIG)-I-like receptors (RLRs) and C-type lectin receptors (CLRs) (4, 5). Upon recognition of pathogen-associated molecular patterns (PAMPS), PRRs signal immune cell activation. TLRs play an important role in the lifecycle of KSHV (6, 7), but whether NLRs also do is unknown. Over twenty NLR family members have been identified in humans, and polymorphisms in several NLRs are linked to various autoinflammatory diseases (8–12). Activation of a subset of NLRs by PAMPs causes the formation of large multimeric complexes termed inflammasomes, which are composed of oligomers of a specific NLR, procaspase-1 and apoptotic-associated speck-like (ASC) adaptor protein (13). Inflammasome formation results in the proteolytic processing of proinflammatory cytokines IL-1β and IL-18 by active caspase-1. Excessive IL-1β and IL-18 production in response to pathogen infection is associated with pyroptosis, an inflammatory process involving caspase-1-mediated cell death.
KSHV Orf63 is an uncharacterized tegument protein. Basic Local Alignment Search Tool for proteins (BLASTP) of NLRP1 and KSHV Orf63 revealed that these proteins are homologous (E value=0.0002) and that Orf63 showed significant similarity to the LRR domain of NLRP1 (fig. S1A). A ClustalW2 alignment of the two proteins also showed homology of the nucleotide-binding domain (NBD) of NLRP1 (fig. S1A) and full-length NLRP1 (fig. S1B) with KSHV Orf63. However, KSHV Orf63 does not contain the effector caspase activation and recruitment domain (CARD) or pyrin domain (PYD) of NLRP1, which are required for its activation, suggesting that Orf63 may function as an inhibitor of NLRP1.
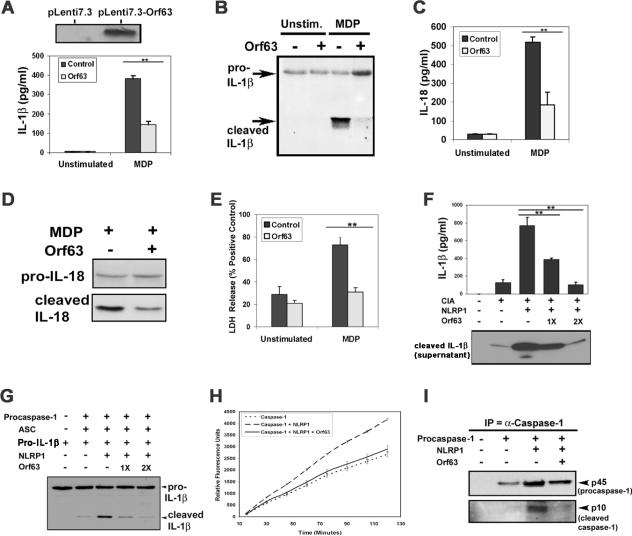
To investigate whether Orf63 inhibits NLRP1 activity, THP-1 cells stably expressing Orf63 and control cells were primed with lipopolysaccharide (LPS) to upregulate IL-1β transcription, followed by stimulation of NLRP1 with the bacteria cell wall constituent, muramyldipeptide (MDP). We found that Orf63 expression significantly inhibited MDP-induced IL-1β and IL-18 production compared to control cells (Fig. 1A–D and fig. S2A). Furthermore no changes in the inflammasome-independent cytokine, tumor necrosis factor (TNF)-α, were observed, confirming the specificity of Orf63 inhibition of the NLRP1 inflammasome (fig. S2B).
Fig. 1. Orf63 is a viral homolog and inhibitor of NLRP1.
(A–D) Stable Orf63 expression in THP-1 cells is detected by immunoblot (top panel in A). THP-1-Control and THP-1-Orf63 cells were either treated with vehicle control or primed with 5ng/ml LPS for 1 hour, followed by stimulation with 10μg/ml MDP for 6 hours. Supernatants were harvested and analyzed for IL-1β by enzyme linked immunosorbent assay (ELISA) and values were normalized to extracellular lactate dehydrogenase (LDH) released (A) or analyzed for pro-IL-1β and cleaved IL-1β by immunoblot (B). Supernatants were also analyzed for IL-18 by ELISA and normalized to extracellular LDH (C) and by immunoblot (D). (E) THP-1-Control or THP-1-Orf63 cells were treated as described above and extracellular LDH was calculated relative to positive control. (F) Procaspase-1, pro-IL-1β, ASC (together denoted as CIA) and NLRP1 expression plasmids were transfected into 293T cells with Orf63 or vector control. Cell extracts and supernatants were harvested 24 hours later and subjected to an IL-1β ELISA and immunoblot for cleaved IL-1β (G) 293T cells were transfected with the indicated expression constructs followed by immunoblot for IL-1β expression. (H) NLRP1 and procaspase-1 were transfected into 293T cells with Orf63 or vector control for 24 hours. Caspase-1 enzymatic activity was determined by incubating lysates with caspase-1 substrate, WEHD-7-amino-4-trifluoromethyl coumarin (AFC). Fluorescence emission was measured every 15 minutes for 2 hours. (I) Caspase-1 was immunoprecipitated from 293T cells transfected with the indicated expression plasmids followed by immunoblot. ** indicates statistical significance P ≤ 0.05 by two-tailed student's t-test. Data are representative of a minimum of three experimental replicates.
Inflammasome activation leads to production of proinflammatory cytokines and eventual cell death. In cells expressing Orf63, NLRP1-dependent cell death (as measured by lactate dehydrogenase (LDH) activity) was significantly inhibited compared to vector alone (Fig. 1E), demonstrating that Orf63 protects cells from NLRP1-dependent cell death.
To further examine the role of Orf63 in blocking NLRP1, we transfected 293T cells with Orf63 and the NLRP1 inflammasome components ASC, procaspase-1 and pro-IL-1β (14). Reconstitution of the NLRP1 inflammasome resulted in an increase in IL-1β secretion, which was inhibited by expression of Orf63 in a dose-dependent fashion (Fig. 1F and G). In contrast, another KSHV viral protein, replication and transcription activator (RTA) was unable to inhibit IL-1β secretion (fig. S2C). We also investigated the ability of Orf63 to inhibit caspase-1 enzymatic activity. Transfection of procaspase-1 and NLRP1 into 293T cells resulted in increased caspase-1 specific activity that was inhibited by co-expression of Orf63 (Fig. 1H). Furthermore, NLRP1-induced proteolytic processing of procaspase-1 to activated caspase-1 was also inhibited (Fig. 1I).
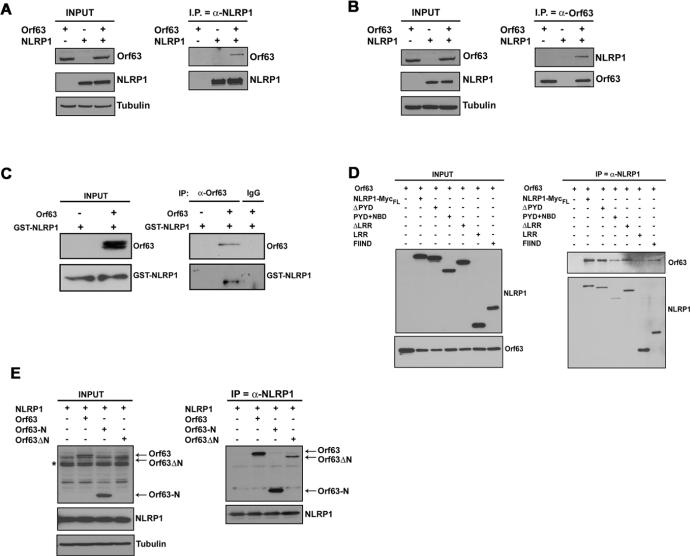
Next, we investigated whether Orf63 could interact with NLRP1. Orf63 co-immunoprecipitated with NLRP1, and vice versa, in 293T cells co-transfected with plasmids to express these proteins (Fig. 2A and B). Endogenous NLRP1 was also shown to interact with Orf63 when Orf63 was expressed in THP-1 cells (fig. S2D). Thus, Orf63 is capable of interacting with NLRP1 and/or in a complex with components of the NLRP1 inflammasome. To investigate the latter scenario, we tested the ability of Orf63 to interact with ASC or caspase-1. We found that Orf63 did not interact with either ASC or caspase-1 in the absence of NLRP1 (fig. S3). Next, Orf63 was immunoprecipitated from a mixture of purified Orf63 made in bacteria and purified NLRP1 made in insect cells, and we found that NLRP1 protein co-immunoprecipitated with Orf63, suggesting that Orf63 directly interacts with NLRP1 in the absence of other proteins (Fig. 2C).
Fig. 2. Orf63 interacts with NLRP1.
(A) 293T cells were transfected with expression plasmids for NLRP1, Orf63 or both plasmids and 48 hours later, NLRP1 was immunoprecipitated followed by immunoblotting for Orf63. (B) A reverse immunoprecipitation was performed. 293T cells were transfected with expression plasmids for NLRP1, Orf63 or both plasmids and 48 hours later, Orf63 was immunoprecipitated followed by immunoblotting for NLRP1. (C) Co-immunoprecipitation of purified Orf63 and NLRP1-GST proteins followed by immunoblotting for Orf63-FLAG and GST-NLRP1. (D) 293T cells were transfected with Orf63 and full-length or mutant NLRP1 expression plasmids followed by immunoprecipitation of Orf63 and immunoblotting for NLRP1. (E) 293T cells were transfected with Orf63-N and Orf63ΔN mutants and full-length NLRP1 expression plasmids for 48 hours. Immunoprecipitations were performed for NLRP1 followed by immunoblots for Orf63-N and Orf63ΔN mutants. Asterisk indicates non-specific band in input lanes. Data are representative of a minimum of three experimental replicates.
To identify the structural elements of NLRP1 that were required for interaction with Orf63, we tested several domain mutants of NLRP1 (15). KSHV Orf63 interacted with the NBD, LRR and FIIND (function to find) domains of NLRP1, whereas no interaction was observed with the PYD and CARD effector domains of NLRP1 (Fig. 2D and fig. S4A–D). The LRR domain of NLRP1 is thought to negatively regulate its activation by folding back onto the NBD domains (14, 16). Interaction of Orf63 with NLRP1 suggests that Orf63 may be functioning similarly to the NLRP1 LRR to block activity. Sequence alignments indicated that Orf63 most closely aligned with the LRR and NBD domains of NLRP1 (fig. S1A). Hence, we created mutants of Orf63; Orf63-N, which contains most of the region similar to NBD but lacks the region that aligns with the LRR, and Orf63ΔN, which contains the entire LRR and only a part of the NBD (fig. S5A), and tested the ability of these mutants to bind NLRP1 by co-immunoprecipitation assays. Either domain was sufficient for interacting with NLRP1 (Fig. 2E) similar to previous reports on NLR proteins (17, 18). Furthermore, in NLRP1 inflammasome reconstitution assays, both Orf63 mutants were capable of inhibiting NLRP1 activity (fig. S5B).
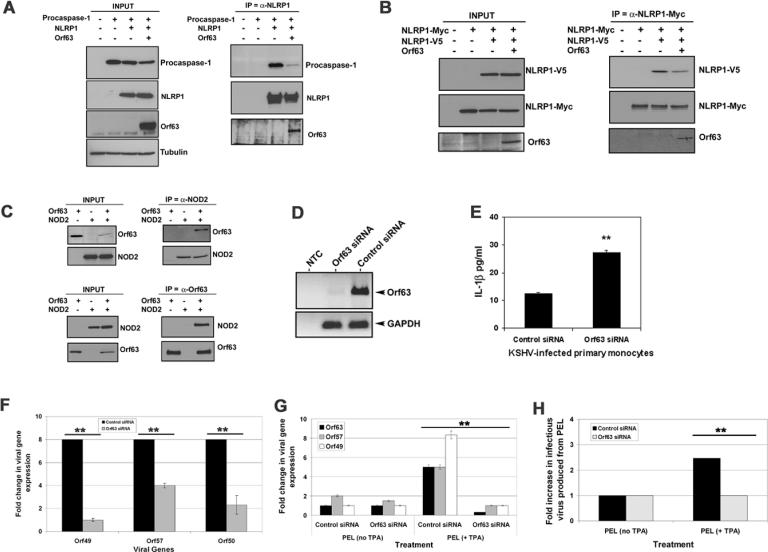
The NLRP1 inflammasome is comprised of multiple proteins complexed with NLRP1, caspase-1 and ASC (13). Our data demonstrate that Orf63 can inhibit NLRP1 function and can also interact with NLRP1. To determine the mechanism of NLRP1 inhibition, we tested the ability of Orf63 to bind NLRP1 and to inhibit the formation of the NLRP1 inflammasome. The presence of Orf63 inhibited the interaction of NLRP1 with procaspase-1, but not its interaction with ASC (Fig. 3A and fig. S6A and B). NLRP1 oligomerization has been shown to be required for activation (14). We confirmed that NLRP1 self-associates and found that Orf63 blocked this self-association (Fig. 3B). Gel fractionation analysis under native conditions revealed that the presence of Orf63 caused more NLRP1 to fractionate in the lower molecular weight fractions, where Orf63 is also expressed (fig. S6C). This provides further support that Orf63 inhibits NLRP1 oligomerization. Taken together, our findings indicate that Orf63 hinders inflammasome formation by both preventing NLRP1 oligomerization and inhibiting the association of NLRP1 with procaspase-1.
Fig. 3. Orf63 inhibits NLRP1 inflammasome formation and is necessary for IL-1β inhibition during viral infection.
(A) Indicated expression plasmids were transfected into 293T cells. NLRP1 was immunoprecipitated 48 hours later and procaspase-1 interactions with NLRP1 were determined by immunoblot. (B) 293T cells were transfected with indicated expression plasmids for 48 hours. NLRP1-myc was immunoprecipitated with anti-myc-antibody and interactions with NLRP1-V5 detected by immunoblot. (C) 293T cells were transfected with indicated expression plasmids followed by immunoprecipitation of either Orf63 or NOD2 48 hours later. Interaction of Orf63 with NOD2 was detected by immunoblot (D) KSHV-infected primary human monocytes were transfected with siRNAs against Orf63 or non-targeting controls. 48 hours later, Orf63 and GAPDH transcription was analyzed by PCR. (E) Control and Orf63 siRNA monocytes were analyzed for IL-1β expression by ELISA 48 hours after transfection with Orf63 siRNA or non-targeting control siRNA. (F) Transcription of lytic viral genes, Orf49, Orf50 and Orf57, in KSHV-infected monocytes was analyzed by qPCR 48 hours after knock-down of Orf63. (G) BCBL-1 PEL cells were nucleofected with siRNAs against Orf63 or a non-targeting control. 24 hours later, the cells were treated with 25ng/ml TPA. 96 hours post-TPA treatment, Orf49, Orf57 and Orf63 transcription was analyzed by qPCR. (H) BCBL-1 PEL cells were treated as in panel G, and 96 hours later, supernatants were used to infect naïve Vero cells. 72 hours later, KSHV viral load in the infected Vero cells was determined by qPCR for Orf49. ** indicates statistical significance P ≤ 0.05 by two-tailed student's t-test. Data are representative of a minimum of three experimental replicates.
NLRP1 can also interact with NOD2 and this interaction enhances its activity in terms of inflammasome function (19). Reciprocal immunoprecipitations revealed that Orf63 interacts with NOD2 (Fig. 3C). Similar to NLRP1, the interaction of Orf63 with NOD2 required the NBD domain (fig. S7A). In contrast, Orf63 did not interact with NOD1 (fig. S7B).
NLRP1 activation and subsequent IL-1β and IL-18 secretion is detrimental to viral infection. We hypothesized that inhibition of Orf63 would result in increased proinflammatory cytokine production during KSHV infection. KSHV has been shown to infect human monocytes (7), therefore we isolated primary human monocytes from blood from healthy donors (fig. S8A and B) and infected them with KSHV in the presence and absence of siRNAs against Orf63 (Fig. 3D). We detected increased IL-1β expression and decreased viral gene expression in KSHV-infected monocytes transfected with siRNAs targeting Orf63 compared to non-targeting control (Fig. 3E and F, respectively).
We next determined the effect of Orf63 during viral reactivation. KSHV-293T cells containing endogenous NLRP1 were transfected with a plasmid encoding RTA to induce KSHV reactivation and lytic gene expression (20). Knockdown of Orf63 resulted in a statistically significant increase in IL-1β expression compared to non-targeting control (fig. S9A and B). Reconstitution of cells with the NLRP1 inflammasome inhibited expression of KSHV reactivation as measured by expression of the vIL-6 lytic protein (fig. S9C); however, when only the LRR domain of NLRP1, which is an inhibitor of NLR activity (14, 16, 21), was co-transfected with the inflammasome components, vIL-6 expression was restored (fig. S9C).
Similarly, when NLRP1 expression was knocked-down in KSHV-infected latent BCBL-1 primary effusion lymphoma (PEL) cells (fig. S10A), we detected an increase in viral genomes and infectious virus, which was enhanced after induction of lytic replication (fig. S10B and C). To confirm that Orf63 is necessary for KSHV lytic gene expression, we inhibited Orf63 expression in BCBL-1 PEL cells using Orf63 siRNA. Orf63 siRNA transfected PEL showed a significant loss of Orf63 expression (Fig. 3G). Further, Orf63 knockdown resulted in the loss of lytic gene expression as measured by loss of Orf49 and Orf57 viral lytic transcript expression (Fig. 3G). After knockdown of Orf63 in reactivated PEL, supernatant was transferred to naïve Vero cells and viral infectivity on Vero cells was quantitated by real-time PCR. We found that Orf63 knockdown resulted in a block of infectious virus produced during PEL reactivation (Fig. 3H). Taken together, our data show that Orf63 can block NLRP1 activation and the production of IL-1β. This function of Orf63 appears to be critical for viral gene expression and viral genome replication during KSHV primary infection as well as KSHV reactivation from latency.
BLASTP alignments revealed homology of Orf63 to the NBD and LRR domains of NLRP1, which are conserved motifs among the NLR family members. To investigate whether Orf63 could also inhibit the activation of other NLR family members, we stimulated THP-1-Orf63 or control THP-1 cells with several NLRP3 agonists (12). Orf63 inhibited IL-1β and IL-18 cleavage and production after treatment with the NLRP3 agonists, ATP and Alum (Fig. 4A–D, fig. S11 and fig. S12A–D). Orf63 also protected cells from NLRP3-dependent cell death in response to these agonists as measured by a LDH release assay (Fig. 4E and fig. S12E). Orf63 co-immunoprecipitated with NLRP3 when both proteins were exogenously expressed in 293T cells (Fig. 4F). Furthermore, 35S-methionine-labelled NLRP1 and NLRP3 bound to purified GST-Orf63 protein (Fig. 4G and H).
Fig. 4. Orf63 inhibits the NLRP3 inflammasome.
(A–D) THP-1-control or THP-1-Orf63 cells were mock treated or primed with 5ng/ml LPS for 1 hour followed by stimulation with 2.5mM ATP for 6 hours. Supernatants were harvested and analyzed by ELISA for IL-1β and normalized to extracellular LDH (A) or for pro-IL-1β and cleaved IL-1β by immunoblot (B). ATP-stimulated cells were also examined for IL-18 expression by ELISA and values were normalized to extracellular LDH (C) and pro-IL-18 and cleaved IL-18 by immunoblot (D). (E) THP-1-control or THP-1-Orf63 cells were treated with ATP as described above and subjected to an LDH release assay. (F) 293T cells were transfected with NLRP3 and Orf63 expression plasmids for 48 hours. NLRP3 was immunoprecipitated followed by immunoblotting for Orf63. (G) GST and GST-Orf63 was purified and subjected to SDS-PAGE and Coomassie staining. (H) GST or GST-Orf63 protein was incubated with glutathione beads and 35S-methionine-labeled NLRP1, NLRP3 or luciferase protein as previously described (29). Complexes were washed several times and subjected to SDS-PAGE. The gel was dried and the bound radiolabeled proteins were detected by a phosphorimager. The first three lanes represent 20% of the input of each of the three labeled proteins added to the GST and GST-Orf63 binding reactions. ** indicates statistical significance P ≤ 0.05 by two-tailed student's t-test. Data are representative of a minimum of three experimental replicates.
KSHV exists in diverse cell types including monocytes, B cells, epithelial, and endothelial cells (22–24). NLRP1 is ubiquitously expressed compared to NLRP3, which is more restricted in its expression (25, 26). Herpesviruses establish life-long latency and must encode proteins that function in the context of all types of cellular events including adverse circumstances. Given KSHV's broad tropism, it seems advantageous for KSHV to encode an inhibitor of the inflammasome.
The NLR nomenclature requires that in order to be classified as a NLR, the protein must contain a NBD and a LRR domain, which are two domains that are evolutionarily conserved in all NLRs (27). Poxviruses encode a pyrin-only containing protein named M13L that inhibits IL-1β and IL-18 secretion (28) through a different mechanism since M13L lacks both a NBD and a LRR domain. KSHV Orf63 does show homology to both the NBD and LRR domains of NLRs and is a homolog of NLRP1. We also found that another herpesvirus encodes a viral homolog of NLRP1 (Fig. S13). This suggests that the targeting of NLR proteins might play a very important role in the herpesvirus lifecycle.
KSHV encodes a viral homolog of cellular NLRP1 without the CARD and PYD effector domains of its cellular counterpart. Although Orf63 did not demonstrate significant similarity to NLRP3, it blocked NLRP3 activity, suggesting that Orf63 is capable of broad inhibition of NLR inflammasome responses. Thus, during the course of evolution with its human host, KSHV has usurped and modified a cellular NLR gene to inhibit the host inflammasome response.
One-sentence summary.
This is the first report of molecular mimicry of a NLR protein by a human virus and we show that KSHV Orf63, a viral homolog of human NLRP1, blocks NLR-dependent innate immune responses, including caspase-1 activation and processing of IL-1β and IL-18.
Supplementary Material
Acknowledgments
We thank S. Krall for assistance with tissue culture, Y. Matsuzawa for plasmid preparation, and D. Dittmer for proofreading. This work was supported by NIH grants DE018281, CA096500, American Heart Association grant 0640041N, and a Burroughs Wellcome Fund grant to BD, and NIH grants AI057157, AI077437, CA156330 and UCRF to JPT and AI56324 and AI91967 to JCR. SG was supported in part by NIH training grants T32-AI007419 and T32-AI007001, BKD is supported by the Crohn's & Colitis Foundation of America and 5R21CA131645, and JAW was supported in part by F32-AI78735. BD is a Leukemia & Lymphoma Society Scholar and Burroughs Wellcome Fund Investigator in Infectious Disease. The NLRP1 and mutant NLRP1 expression plasmids as well as purified GST-NLRP1 protein were obtained from Dr. John Reed and require a material transfer agreement. The NLRP3, NOD1, NOD2, mutant NOD2, ASC, procaspase1, and pro-IL-1β expression plasmids were obtained from Dr. Jenny Ting and also require a material transfer agreement. Authors (BD, JPT and SMG) have filed a provisional patent on the KSHV Orf63 protein and/or any peptides derived from this protein in treatment for human disease.
Footnotes
Supporting Online Material www.sciencemag.org Materials and Methods Figs. S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, S12, S13
REFERENCES & NOTES
- 1.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. N Engl J Med. 1995;332:1186. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 2.Chang Y, et al. Science. 1994;266:1865. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 3.Coscoy L. Nat Rev Immunol. 2007;7:391. doi: 10.1038/nri2076. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. Nat. 2010;11:373. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 5.Meylan E, Tschopp J, Karin M. Nature. 2006;442:39. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 6.Gregory SM, et al. Proc Natl Acad Sci U S A. 2009;106:11725. doi: 10.1073/pnas.0905316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West J, Damania B. J Virol. 2008;82:5440. doi: 10.1128/JVI.02590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boschan C, et al. Am J Med Genet A. 2006;140:883. doi: 10.1002/ajmg.a.31148. [DOI] [PubMed] [Google Scholar]
- 9.Jin Y, et al. N Engl J Med. 2007;356:1216. [Google Scholar]
- 10.Lequerre T, et al. Rheumatology (Oxford) 2007;46:709. doi: 10.1093/rheumatology/kel399. [DOI] [PubMed] [Google Scholar]
- 11.Masters SL, Simon A, Aksentijevich I, Kastner DL. Annu Rev Immunol. 2009;27:621. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroder K, Tschopp J. Cell. 2010;140:821. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 13.Martinon F, Burns K, Tschopp J. Mol Cell. 2002;10:417. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 14.Faustin B, et al. Mol Cell. 2007;25:713. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 15.Bruey JM, et al. Cell. 2007;129:45. doi: 10.1016/j.cell.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 16.Tanabe T, et al. Embo J. 2004;23:1587. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damiano JS, Oliveira V, Welsh K, Reed JC. Biochem J. 2004;381:213. doi: 10.1042/BJ20031506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hake SB, et al. Mol Cell Biol. 2000;20:7716. doi: 10.1128/mcb.20.20.7716-7725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu LC, et al. Proc Natl Acad Sci U S A. 2008;105:7803. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun R, et al. Proc Natl Acad Sci U S A. 1998;95:10866. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poyet JL, et al. J Biol Chem. 2001 Jul 27;276:28309. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 22.Blasig C, et al. J Virol. 1997;71:7963. doi: 10.1128/jvi.71.10.7963-7968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monini P, et al. Blood. 1999;93:4044. [PubMed] [Google Scholar]
- 24.Pauk J, et al. N Engl J Med. 2000;343:1369. doi: 10.1056/NEJM200011093431904. [DOI] [PubMed] [Google Scholar]
- 25.Hlaing T, et al. J Biol Chem. 2001;276:9230. doi: 10.1074/jbc.M009853200. [DOI] [PubMed] [Google Scholar]
- 26.Manji GA, et al. J Biol Chem. 2002;277:11570. doi: 10.1074/jbc.M112208200. [DOI] [PubMed] [Google Scholar]
- 27.Ting JP, et al. Immunity. 2008;28:285. [Google Scholar]
- 28.Johnston JB, et al. Immunity. 2005;23:587. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Damania B, Alwine JC. Genes Dev. 1996 Jun 1;10:1369. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.