Abstract
It is hard to imagine a more extreme contrast than that between the dilute solutions used for in vitro studies of protein folding and the crowded, compartmentalized, sticky, spatially inhomogeneous interior of a cell. This review highlights recent research exploring protein folding in the cell with a focus on issues that are generally not relevant to in vitro studies of protein folding, such as macromolecular crowding, hindered diffusion, co-translational folding, molecular chaperones, and evolutionary pressures. The technical obstacles that must be overcome to characterize protein folding in the cell are driving methodological advances, and we draw attention to several examples, such as fluorescence imaging of folding in cells and genetic screens for in-cell stability.
Introduction
Chris Anfinsen launched the field of protein folding by showing that ribonuclease (specifically, bovine pancreatic ribonuclease A) could refold to an active enzyme after reductive denaturation. Naturally, ribonuclease became emblematic of the fundamental tenet of protein folding—that the primary sequence of a protein specifies an energy landscape and a successful route to the native state at the global energy minimum. Yet ribonuclease folds in vivo during a complex journey through the secretory pathway of the cell. Notably, in its biological folding process, ribonuclease confronts milieux that are densely crowded with macromolecules; it samples the microenvironments of the ribosome tunnel, the translocon, and the ER lumen; it has the opportunity to fold from its N- to C-terminus; and it is not left on its own, but instead is accompanied by lumenal chaperones that facilitate its folding and post-translational modification. As this journey abundantly illustrates, protein folding in the cell confronts many issues that are nonexistent in high dilution refolding experiments. It is thus not surprising that an increasing research effort is being applied to issues and processes involved in cellular protein folding.
This review presents research in the expanding area of protein folding in the cell. We mainly confine our discussion to publications on cellular protein folding that have appeared in the last two years and to areas that have not been recently reviewed. We first describe a number of issues, such as macromolecular crowding and co-translational folding, that arise when considering folding in the cell but are absent in vitro (Figure 1). Next, we describe innovative methods to address these issues, particularly in intact cells where the risks of reductionism are minimized. Lastly, we describe some new papers that highlight the complex biological pressures on protein folding in the cell and how these influence protein evolution. Throughout, space constraints have required us to selectively cite the literature in this exciting area. We apologize to any colleagues whose relevant work is not mentioned. In turn we hope that we provide the interested reader with a sense of the major questions touching in vivo folding and examples of provocative recent papers that address these questions.
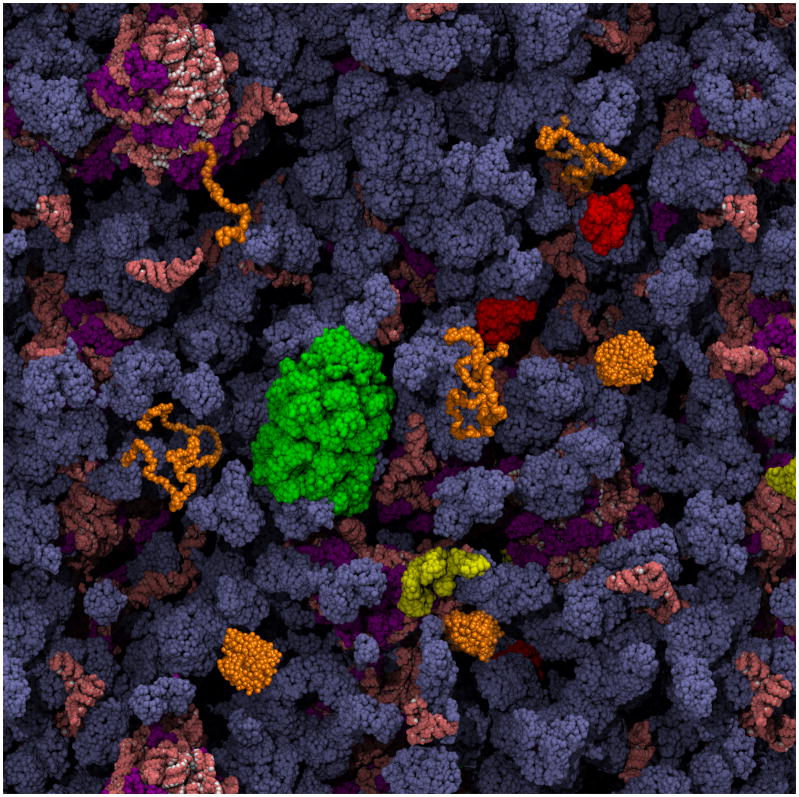
Figure 1. Schematic depiction of a protein folding reaction in the cytoplasm of an E. coli cell, showing vividly how different the environment is from dilute in vitro refolding experiments.
The cytoplasmic components are present at their known concentrations. As discussed in this review, features of particular importance to the folding of a protein of interest (in orange) are: the striking extent of volume exclusion due to macromolecular crowding, the presence of molecular chaperones that interact with nascent and incompletely folded proteins (GroEL in green, DnaK in red, and trigger factor in yellow), and the possibility of co-translational folding upon emergence of the polypeptide chain from the ribosome (ribosomal proteins are purple; all RNA is salmon). The cytoplasm image is courtesy of A. Elcock.
Macromolecular crowding
A striking difference between most in vitro folding experiments and the cellular environment is the high concentration of macromolecules, which severely limits the cellular volume accessible to a polypeptide chain. Like many issues related to folding in the cell, determining the effects of crowding on folding presents major technical challenges to both computational and experimental studies; moreover, crowding is generally accompanied by other effects including altered diffusion and weak interactions.
The effects of macromolecular crowding have been discussed in an extensive 2008 review by Zhou, Rivas and Minton [1], and recent computational work in this area was critically reviewed by Adrian Elcock [2]. In general, macromolecular crowding is predicted [3] and found experimentally [4,5] to favor compaction, shifting denatured and intermediate ensembles in a folding reaction away from more extended states. The net effect of crowding on native state stability is as yet not entirely clear [1,6], but recent experimental results suggest that any change in stability is modest [4,7]. McGuffee and Elcock recently reported results of an impressive Brownian dynamics simulation of the E. coli cytoplasm showing that steric and electrostatic potentials alone did not recapitulate experimental results on either protein stability in the cell or diffusion rates, but instead they observed much better fits when they included short-range attractive hydrophobic interactions [8]. Additionally, their calculations showed that the impact of crowding depends on the properties of a given protein and the size differential between it and the surrounding macromolecules, as was also concluded by Christiansen et al. [7].
Macromolecular crowding also affects the viscosity of the cellular environment and solvent viscosity has been invoked as an important factor in determining folding rates and mechanisms (see for example [9]). In a recent study, Dhar et al. used computational and experimental approaches to study the effects of a model crowder, Ficoll, on activity and folding of phosphoglycerate kinase (PGK), a 412-aa protein with two domains connected by a flexible hinge [10]. The protein became strikingly more active in the presence of Ficoll, apparently because crowding decreased the inter-domain separation. Also, the relaxation rate from temperature jump unfolding experiments showed a maximum at 100 g/L Ficoll. The authors interpreted this result as arising from opposing effects of crowding and viscosity [10].
Hindered mobility and sticky neighbors
Several recent studies show that the cellular environment affects macromolecular motion and provide insight into how. For example, two recent papers have used fluorescence recovery after photobleaching to monitor translational diffusion of GFP constructs in E. coli cells [11,12]. Using fusion constructs consisting of GFP and native E. coli proteins, Kumar et al. found unexpectedly low translational diffusion rates and argued that macromolecular crowding and macromolecular networks in the cytoplasm lead to molecular sieving even for small, 27 kDa, proteins [12]. In contrast, for proteins of less than ~115 kDa Nenninger and coworkers argued that intermolecular interactions were the main contributor to reduced diffusion [11]. Intriguingly, Pielak and co-workers observed significantly slower rotational diffusion for protein solutes when another protein rather than an inert molecule such as Ficoll or Dextran was used as a crowding agent, arguing for greater effects on rotational than translational diffusion from weak interactions [13,14]. The importance of weak interactions mirrors results from McGuffee and Elcock's computational modeling of the E. coli cytoplasm [8]. However, a recent study concludes that hydrodynamic effects reproduce experimental size dependence of protein mobility in cells better than non-specific weak interactions [15]. Despite the numerous studies into motion in the cell, the impact of this altered mobility on folding is as yet uncharted territory.
Vectorial synthesis and roles of mRNA and ribosomes in folding
Newly synthesized polypeptide chains emerge from the ribosome vectorially, allowing their N-terminal portions to sample conformational space before the chain is completely synthesized. Additionally, the earliest environments encountered by a nascent chain are the ribosome tunnel and ribosome-associated chaperones. There have been excellent recent reviews on issues related to co-translational folding including one in this volume [16–19], and we will not duplicate their coverage.
A single domain stabilized by many long-range contacts is not expected to fold until the entire chain is complete, and recent studies of ribosome-bound nascent chains (RNCs) have confirmed this expectation for an SH3 domain by NMR [20] and GFP by observing chromophore maturation [21]. In fact, the NMR studies on SH3 RNCs reveal little or no compaction until the entire chain has exited the ribosomal tunnel [20]. By comparison, RNCs of the larger GFP may populate a more compact state prior to full translation [21].
The interrelationship of translation rate and folding has been discussed in a number of recent reviews [22–25]. Intuitively, slowing translation might allow more time for proper folding, and indeed in a recent study mutant ribosomes with reduced translation rates increased the soluble expression of eukaryotic proteins in E. coli [26]. The messenger RNA sequence can also affect the translation rate either through the use of rare codons [22,25,27–29] or by RNA folding [29,30]. Either of these factors may be changed by synonymous mutations, where mRNA sequences are altered without affecting the encoded amino acid. Increasing the translation rate of the multidomain E. coli protein SufI by synonymously exchanging rare codon clusters for common codons was found to decrease co-translational folding and the production of mature, folded protein [31]. Intriguingly, the mutation that leads to deletion of F508 (ΔF508) in the cystic fibrosis transmembrane conductance regulator (CFTR), the most common mutation linked to cystic fibrosis, also changes the preceding codon for Ile507 [32]. Alteration of the local mRNA structure in the mutant retards translation and presumably impairs folding, increasing co-translational ubiquitination and leading to protein degradation [32,33]. Restoration of the original Ile507 codon in the ΔF508 background significantly increases the amount of mature CFTR in the plasma membrane, demonstrating the potential impact of synonymous mutations on in vivo protein folding and maturation [32,33].
As described in recent reviews [16,17,19,34], the nascent chain can also be influenced by the ribosomal exit tunnel and, for proteins targeted to the bacterial periplasm or the eukaryotic ER, by the environment of the translocon. Specific sequences are known to stall ribosomes [35], and exciting recent cryo-EM structures from the Beckmann laboratory reveal the nature of their interactions with the tunnel [36,37] The impact of chain conformations within the tunnel on folding and targeting has been described for the E. coli EspP protein [38], and elegant studies by the Deutsch laboratory [39,40] as well as by Johnson, Skach and collaborators [34,41] have demonstrated that membrane protein structure formation can initiate co-translationally within both the ribosome tunnel and the translocon.
Molecular chaperones remodel the in-vivo folding energy landscape
The ability of molecular chaperones to interact with nascent or incompletely folded chains so as to favor successful folding and disfavor aggregation is well established. Yet the impact of chaperones on the folding mechanisms and stabilities of their clients is less clear, despite expanding literature on the functions and substrate-omes of several chaperones (e.g., [42,43]).
Consider the case of arguably the best-studied chaperone, the E. coli chaperonin GroEL and its partner GroES: Based on their own and others’ experimental results, Horwich and co-workers argue that GroEL/ES is a passive ‘Anfinsen cage’ protecting its substrate from aggregation during folding but otherwise having little effect on its folding landscape [44,45]. Conversely, interactions between the substrate and the walls of the GroEL cavity may actively expand and compress substrate proteins [46,47]. Direct examination of complexes between GroEL and substrates show binding of several GroEl subunits to a single substrate with some distortion in the overall chaperonin conformation [48,49]. This image is consistent with models emerging from recent FRET studies of labeled GroEL substrates [50,51], which implicate a hierarchy of hydrophobically-mediated chaperonin-substrate binding events in remodeling the substrate folding mechanism.
Spatial organization, membranes and compartmentalization
Cellular interiors are highly anisotropic with elaborate and physiologically critical architectures. This subcellular organization plays a major role in folding at several levels: For example, the native structures of membrane proteins are tuned to the diverse microenvironments and two-dimensional character of membranes. Additionally, membranes are barriers, requiring proteins made on one side but destined to perform their functions extracytoplasmically to be translocated across the membrane, in most cases unfolded. For secretory proteins, passage through cellular compartments is highly choreographed with an assembly line of modifying enzymes, chaperones and transport mechanisms. Compartmentalization also opens up the possibility of chemical gradients, e.g. pH or oxidizing potential. Taken together, the spatial organization and compartmentalization of the cellular environment enable folding to occur in temporally optimized steps.
Bacterial proteins destined for the periplasm or the cell exterior can be secreted across membranes either folded (e.g., by the Tat system) or unfolded (e.g., by the Sec system) [52]. Recent work by Ignatova and coworkers reveals that for both E. coli and B. subtilis transcriptomes, proteins targeted to Tat are more likely to have slowly translated regions, which presumably encourage co-translational folding and subsequent translocation [31]. The autotransporter bacterial virulence factors transport their unfolded passenger domain via a channel formed, at least in part, by the C terminal porin domain, and extracellular folding of the C-terminal portion of the passenger domain drives translocation of the remainder of this domain across the membrane [53,54]. This coupling of folding and membrane translocation represents a distinct mechanistic difference between in vitro and in vivo folding.
Proteins that translocate through the Sec channel in bacteria and eukaryotes are greeted by an array of chaperones and modifying enzymes that alter the folding energy landscape. In addition, they move from an ATP-rich, reducing environment to one that is ATP-poor and oxidizing as they enter either the bacterial periplasm, mitochondrial intermembrane space (IMS) or the eukaryotic ER. We refer interested readers to recent reviews of protein folding in the ER [55], in the mitochondrial IMS [56,57], and in the periplasm [58,59]. Here, we discuss only a few highlights of the recent literature in this active research area.
Exposure of nascent chains to the oxidizing environment of the periplasm or ER lumen enables step-wise disulfide bond formation, fixing the topology of secretory proteins. Not surprisingly, the timing and specificity of disulfide bond formation is integral to their in vivo folding, and this issue has been widely studied in eukaryotic proteins [55]. Recently, Kadorkura and Beckwith provided an analogous picture for folding in the bacterial periplasm by trapping folding intermediates of alkaline phosphatase (PhoA) with incomplete disulfide bond formation [60]. They showed that protein disulfide isomerase-facilitated disulfide bond formation in PhoA occurs sequentially as the Cys residues emerge from the translocon [60]. Tapley et al. recently identified another example of coupling folding with the environmental gradient between cytoplasm and periplasm in their discovery of a pH-triggered periplasmic chaperone system [61].
Many proteins that fold in the ER lumen are large with complicated topologies including multiple domains, disulfides, and glyosylation sites, which play roles in specialized in vivo folding mechanisms. A recent study on the influenza membrane glycoprotein neuraminidase [62] found that its interactions with the lectin chaperones calnexin and calreticulin, which bind in a glycosyl-specific manner, help ensure proper folding and oligomerization. In another exciting report, a novel mechanism for the coordinated folding and assembly of heavy and light chains of IgG antibodies was recently unveiled [63,64]. IgG maturation requires that the intrinsically disordered CH1 heavy chain domain undergo isomerization of a specific trans X-Pro bond to cis, disulfide bond formation, and then binding-induced folding upon association with the CL domain of the light chain [63,64]. The unfolded, reduced CH1 domain has a high affinity for the ER-resident Hsp70 chaperone BiP, and so spends its time waiting for the partner light chain in complex with BiP.
Promising new methods for study of in vivo protein folding
The Holy Grail in studies of protein folding in the cell is to directly observe a protein of interest (POI) in intact cells and to characterize its folding, both thermodynamically and kinetically, in situ. Not surprisingly, this has proven exceedingly difficult. Several years ago, Ghaemmaghami and Oas took advantage of E. coli’s urea tolerance to perform in-cell urea titrations of the λ repressor headpiece utilizing a novel hydrogen exchange/mass spectrometry method to assess stability [65]. The resulting analysis showed little change in stability relative to dilute solution, although placing the bacteria in hyperosmotic medium markedly increased stability. Subsequently, our lab combined this in-cell urea titration approach with an in-cell fluorescence folding-reporter system to monitor protein stability [66,67]. We also observed little change in stability but a substantial change in the urea dependence of the apparent equilibrium constant (m-value), not explained by our subsequent in vitro study of the effect of crowding [4]. While promising, these early studies underlined the desirability of approaches to probe in-cell stability that do not rely on harsh denaturants.
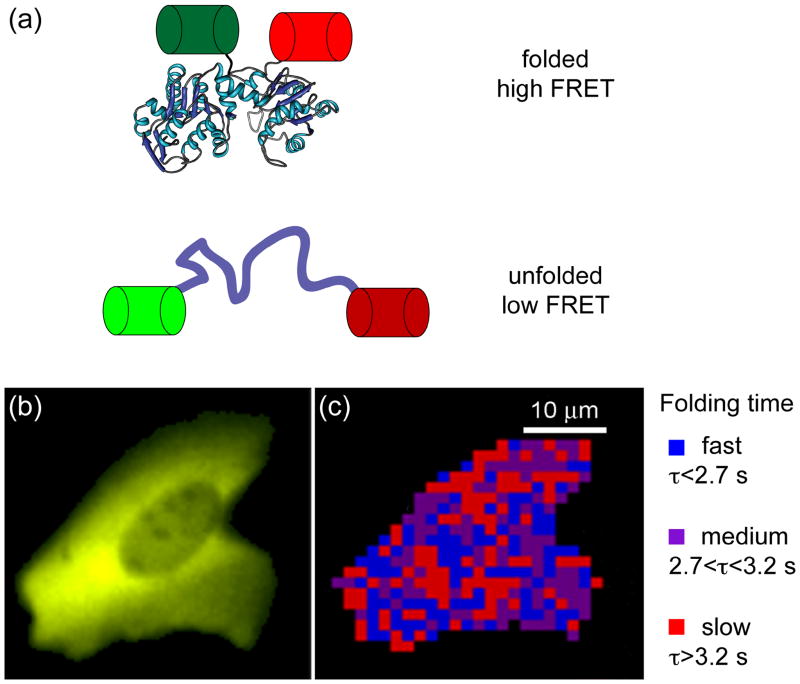
Exciting recent work from the Gruebele lab combines temperature-jump perturbation methods with fast relaxation imaging (FreI) to interrogate the in vivo folding landscape of a POI, here a temperature-sensitive variant of phosphoglycerate kinase (tsPGK) [68]. tsPGK was fused at each terminus with a fluorescent protein to form a folding-sensitive FRET reporter system (Figure 2a), similar to earlier work by Philipps et al. [69]. Using localized short laser-initiated temperature jumps to trigger tsPGK unfolding, these researchers measured protein folding kinetics in real time in living cells (Figure 2b) [68]. Tantalizingly, FreI experiments show that diffusion is slow in the cytoplasm and that folding kinetics are significantly different in different regions of the cell (Figure 2c) [68].
Figure 2. Monitoring protein folding kinetics in a living cell using fast relaxation imaging (FReI) [68].
(a) The folding sensor was created by sandwiching a temperature-sensitive POI (here, PGK) between two fluorescent proteins, GFP and mCherry, allowing folding to be monitored by FRET. (b) Image of the fluorescence arising from expression of the fusion protein in a human cell. (c) Map of the refolding dynamics of the fusion protein in the cell shown in (b). The time dependence of the FRET signal following a temperature jump is spatially mapped. Figures 2b and c are courtesy of M. Gruebele.
In-cell NMR, recently reviewed by Pielak [70] and by Ito and Selenko [71], is a potentially powerful approach to study proteins in vivo and to gain insight into their stability and folding mechanisms. Unfortunately, many folded proteins fail to show measurable NMR spectra in the cellular environment, most likely due to hindered rotational diffusion. Despite the inherent obstacles, the Shirakawa group has had impressive success applying NMR to small proteins in eukaryotic cells [72], including interrogating stability by hydrogen exchange. They found significant enhancement in exchange kinetics for ubiquitin inside HeLa cells. Interestingly, a mutant ubiquitin that should bind less well to partner proteins had lower hydrogen exchange rates in-cell than did wild-type ubiquitin, arguing that protein-protein interactions may modulate protein stability in the cell.
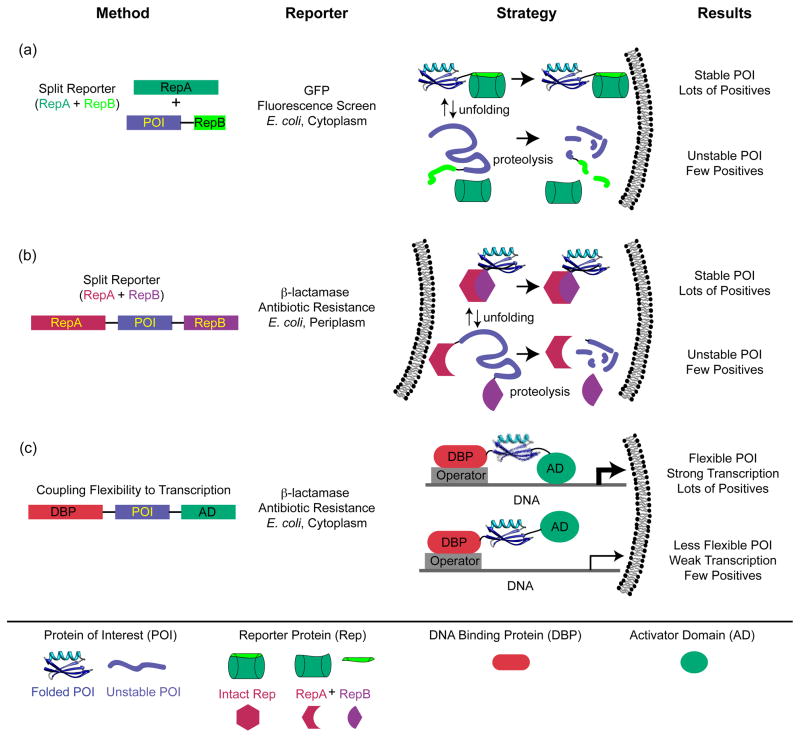
Clever use of split reporters, in which folding of the POI is coupled to successful binding and folding of two pieces of a reporter protein (Figure 3), has shed light on in vivo folding properties. This strategy was introduced to screen for soluble, and by inference stable, proteins in earlier work by Thomas and coworkers using a genetic marker [73] and by Waldo and coworkers using recombination of GFP [74] (Figure 3a). This approach has been exploited with Im7 as the POI and β-lactamase as the split reporter (Figure 3b) [75], and developed into more general screens for successful folders in the bacterial periplasm [76] and in yeast expression systems [77]. In the Im7 work, Foit et al. correlated resistance to β-lactam antibiotics with the stability of Im7 variants in the periplasm and selected for variants with enhanced stability [75]. Intriguingly, many stabilizing mutations altered functional residues that would normally be involved in interactions between Im7 and its binding partner colicin E7. These results point to the conflicting pressures on protein sequences placed by multiple evolutionary constraints, and the trade-offs in folding properties required to meet in vivo requirements for function (see next section).
Figure 3. Selection and screening strategies designed to assess protein stability in vivo.
(a) Construct designed such that GFP fluorescence reports on protein solubility, which is correlated to stability [74]. The gene for the POI is C-terminally fused to a β strand from GFP, and a GFP construct missing this strand is co-expressed with the fusion. GFP fluorescence reports on the successful docking of the missing strand onto the truncated GFP. In turn, unstable (or insoluble) POIs with their attached GFP strands are degraded (or inaccessible to recombine with the truncated GFP). (b) Screen designed such that successful periplasmic expression of β-lactamase requires a stably folded POI and hence recombination of two β-lactamase fragments that flank the POI [75]. Here, the level of functional periplasmic β-lactamase, which relies on stability of the POI, is read out as resistance to increasing concentrations of β–lactam antibiotic [75]. As in (a), this correlation is presumably based on the enhanced proteolytic susceptibility of the fusion protein when the POI is unstable. (c) Construct designed such that flexibility of the POI controls transcription of the β-lactamase gene [78,79]. When the POI is flanked by the N-terminal DNA-binding domain of bacteriophage λ, which binds to the λ operator, and the RNA polymerase α subunit, which activates transcription of the β-lactamase gene, flexible POIs lead to greater transcriptional activation and higher antibiotic resistance.
These approaches to in-cell stability use a single flanking reporter and are clearly powerful as selections for enhanced stability. However, they are end-point assays based on the proteolytic lability of the fusion construct when the POI is unstable and therefore cannot readily yield an estimate of folding free energy. A related in vivo screen flanks the POI between a DNA-binding domain and a transcriptional activation domain [78] (Figure 3c). Only when the POI is sufficiently flexible can the flanking domains effectively activate transcription of a reporter gene. The authors tested streptococcal protein G B1 variants with melting temperatures over a wide range, from 38 to 100 °C. The resulting range of expression levels was remarkably well correlated to the Tm values of the test protein (higher Tm, lower expression)--reliably enough to select for stabilizing mutations in a directed evolution experiment [79]. In this intriguing stability reporter system, the read-out may be more directly related to the thermodynamic stability of the POI.
Only the fittest survive: evolutionary implications for folding in the cell
How the competition for fitness at the organismal level imposes evolutionary constraints on protein molecular properties is complex. Attaining the native functional state is biologically essential both because a protein must perform its cellular function(s) and because misfolded or incompletely folded proteins can be deleterious [80]. However, the optimal molecular properties to benefit the organism are not simply stability and activity, and even these may be in conflict, as illustrated in the Im7 work described above [75]. Nature is, in a real sense, carrying out a selection experiment with an array of selective pressures, many of which we have yet to identify.
The relationship between protein folding and evolvability has recently been reviewed by Tokuriki and Tawfik [81], and a stimulating earlier review appeared in 2008 [82]. Thus, we mention only a few particularly relevant papers that speak to the biological pressures on protein folding in the cell.
How do in-vivo folding properties evolve? A key challenge is how an organism copes with ‘weak-link’ protein variants as it moves towards more successfully evolved versions. One strategy exploits chaperones as buffers, enabling the organism to cope with mutated proteins that have impaired folding properties [81,83,84]. In a recent study, Shamoo and coworkers explicitly designed a weak-link challenge and asked how it was overcome: They substituted the normal (and essential) gene for adenylate kinase in the thermophile Geobacillus stearothermophilus by the analogous gene from the mesophile B. subtilis and grew the resulting bacterial population at high temperatures to force competition between mutants [85,86]. Success in the population was achieved by a combination of mutations that led to enhanced activity and improved folding properties [85,86].
While the evolution of a single protein is informative, computational analyses of sequence databases provide a more global view of evolutionary trends. For example, Vendruscolo and collaborators have used physicochemical analysis of proteins to predict cellular properties of proteins such as expression levels [87]. More recently, these researchers found that aggregation propensity and folding propensity tend to be anti-correlated [88].
Stability, function, and evolvability are only part of the equation: In thought-provoking work on p53 from Alan Fersht’s lab, an additional factor emerges in biological requirements on protein folding: It appears that the amount of unfolded protein in the cell may be under a selective pressure because of the constraints on degradation and consequent regulation of the pool of protein available for its transcriptional regulatory activity [89].
Concluding thoughts
Elucidating how protein expression, folding, function, misfolding and aggregation, chaperone interactions, and degradation are balanced dynamically will require aggressive, multidisciplinary, and creative research in the future. Most importantly, a holistic view will be required as we seek to understand protein folding in the cell and to gain and integrate both biophysical and physiological insights. Some aspects of in vivo folding have thus far been the domain of particular fields: for example, how proteins fold as they traverse the secretory pathway in eukaryotes has been a topic of study in cell biology. Also, with few exceptions and for obvious reasons, computational modeling has been applied most extensively to isolated, small proteins. We urge that the challenges of folding in the cell be tackled by teams of investigators, capable of developing the needed arsenal of novel and powerful methods and simultaneously retaining a keen sense of the biological realities imposed on this already complex chemical reaction: the successful formation of a native protein capable of insuring the fitness of the host organism.
Acknowledgments
We thank Adrian Elcock and Martin Gruebele for generously providing figures, Rob Smock for help with Figure 1, and Dan Hebert for critical reading of the manuscript. Funding from the National Institutes of Health (grants OD000945, GM027616, and GM094848) is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elcock AH. Models of macromolecular crowding effects and the need for quantitative comparisons with experiment. Curr Opin Struct Biol. 2010;20:196–206. doi: 10.1016/j.sbi.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsao D, Dokholyan NV. Macromolecular crowding induces polypeptide compaction and decreases folding cooperativity. Phys Chem Chem Phys. 2010;12:3491–3500. doi: 10.1039/b924236h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong J, Gierasch LM. Macromolecular crowding remodels the energy landscape of a protein by favoring a more compact unfolded state. J Am Chem Soc. 2010;132:10445–10452. doi: 10.1021/ja103166y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, He H, Li S. Effect of Ficoll 70 on thermal stability and structure of creatine kinase. Biochemistry. 2010;75:648–654. doi: 10.1134/s0006297910050160. [DOI] [PubMed] [Google Scholar]
- 6.Qin S, Zhou HX. Atomistic modeling of macromolecular crowding predicts modest increases in protein folding and binding stability. Biophys J. 2009;97:12–19. doi: 10.1016/j.bpj.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christiansen A, Wang Q, Samiotakis A, Cheung MS, Wittung-Stafshede P. Factors defining effects of macromolecular crowding on protein stability: an in vitro/in silico case study using cytochrome c. Biochemistry. 2010;49:6519–6530. doi: 10.1021/bi100578x. [DOI] [PubMed] [Google Scholar]
- ●●8.McGuffee SR, Elcock AH. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. This paper reports a bold computational simulation of the E. coli cytoplasm using Brownian dynamics methods. The authors vary the nature of the potentials between macromolecules to determine the best fit to known diffusion, stability, association, and aggregation properties of proteins in the bacterial cytoplasm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagen SJ. Solvent viscosity and friction in protein folding dynamics. Curr Protein Pept Sci. 2010;11:385–395. doi: 10.2174/138920310791330596. [DOI] [PubMed] [Google Scholar]
- 10.Dhar A, Samiotakis A, Ebbinghaus S, Nienhaus L, Homouz D, Gruebele M, Cheung M. Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding. Proc Natl Acad Sci USA. 2010;107:17586–17591. doi: 10.1073/pnas.1006760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nenninger A, Mastroianni G, Mullineaux CW. Size dependence of protein diffusion in the cytoplasm of Escherichia coli. J Bacteriol. 2010;192:4535–4540. doi: 10.1128/JB.00284-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar M, Mommer MS, Sourjik V. Mobility of cytoplasmic, membrane, and DNA-binding proteins in Escherichia coli. Biophys J. 2010;98:552–559. doi: 10.1016/j.bpj.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Wang Y, Pielak GJ. Translational and rotational diffusion of a small globular protein under crowded conditions. J Phys Chem B. 2009;113:13390–13392. doi: 10.1021/jp907744m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Li C, Pielak GJ. Effects of proteins on protein diffusion. J Am Chem Soc. 2010;132:9392–9397. doi: 10.1021/ja102296k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●15.Ando T, Skolnick J. Crowding and hydrodynamic interactions likely dominate in vivo macromolecular motion. Proc Natl Acad Sci USA. 2010;107:18457–18462. doi: 10.1073/pnas.1011354107. Like McGuffee and Elcock (ref. 8), these authors carried out Brownian dynamics simulations of the E. coli cytoplasm, but Ando and Skolnick concluded that hydrodynamic interactions were the major influence on the observed reduced motion of GFP inside cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 17.Cabrita LD, Dobson CM, Christodoulou J. Protein folding on the ribosome. Curr Opin Struct Biol. 2010;20:33–45. doi: 10.1016/j.sbi.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Weinreis SA, Ellis JP, Cavagnero S. Dynamic fluorescence depolarization: A powerful tool to explore protein folding on the ribosome. Methods. 2010;52:57–73. doi: 10.1016/j.ymeth.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ignatova Z. Co-translational folding of a nascent chain. [DOI] [PubMed] [Google Scholar]
- 20.Eichmann C, Preissler S, Riek R, Deuerling E. Cotranslational structure acquisition of nascent polypeptides monitored by NMR spectroscopy. Proc Natl Acad Sci USA. 2010;107:9111–9116. doi: 10.1073/pnas.0914300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●21.Ugrinov KG, Clark PL. Cotranslational folding increases GFP folding yield. Biophys J. 2010;98:1312–1320. doi: 10.1016/j.bpj.2009.12.4291. Ugrinov and Clark isolated poly-ribosomes translating GFP nascent chains from E. coli cells and demonstrated that full-length GFP chains released from ribosomes fold faster than full-length chains isolated from inclusion bodies. Importantly, this work allowed a direct comparison of two different in vivo GFP species, nascent and misfolded chains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czech A, Fedyunin I, Zhang G, Ignatova Z. Silent mutations in sight: covariations in tRNA abundance as a key to unravel consequences of silent mutations. Mol BioSyst. 2010;6:1767–1772. doi: 10.1039/c004796c. [DOI] [PubMed] [Google Scholar]
- 23.Fredrick K, Ibba M. How the sequence of a gene can tune its translation. Cell. 2010;141:227–229. doi: 10.1016/j.cell.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Wilke CO, Drummond DA. Signatures of protein biophysics in coding sequence evolution. Curr Opin Struct Biol. 2010;20:385–389. doi: 10.1016/j.sbi.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●26.Siller E, Dezwaan DC, Anderson JF, Freeman BC, Barral JM. Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J Mol Biol. 2010;396:1310–1318. doi: 10.1016/j.jmb.2009.12.042. These authors demonstrated by a clever experimental strategy that decreasing E. coli translation rates to approximate the folding rate in eukaryotic cells increases the yield of correctly folded, soluble eukaryotic proteins in E. coli expression systems even for proteins prone to inclusion body formation. [DOI] [PubMed] [Google Scholar]
- 27.Clarke TF, Clark PL. Increased incidence of rare codon clusters at 5′ and 3′ gene termini: implications for function. BMC Genomics. 2010;11:118. doi: 10.1186/1471-2164-11-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 29.Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci USA. 2010;107:3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●31.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. Rare codon clusters identified by bioinformatic methods were found to be more common in long proteins and proteins that are translocated via the Tat system. For the multidomain E. coli protein Suf1 such clusters caused translational pausing, and the associated co-translational folding allowed sequential structure formation. [DOI] [PubMed] [Google Scholar]
- ●32.Bartoszewski RA, Jablonsky M, Bartoszewska S, Stevenson L, Dai Q, Kappes J, Collawn JF, Bebok Z. A synonymous single nucleotide polymorphism in ΔF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J Biol Chem. 2010;285:28741–28748. doi: 10.1074/jbc.M110.154575. This work is an important demonstration that synonymous mutations that alter mRNA structure and thus slow translation can exacerbate the effects of non-synonymous mutations, which caust defects by changing the amino acid sequence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunt R, Sauna ZE, Ambudkar SV, Gottesman MM, Kimchi-Sarfaty C. Silent (synonymous) SNPs: should we care about them? Methods Mol Biol. 2009;578:23–39. doi: 10.1007/978-1-60327-411-1_2. [DOI] [PubMed] [Google Scholar]
- 34.Skach WR. Cellular mechanisms of membrane protein folding. Nat Struct Mol Biol. 2009;16:606–612. doi: 10.1038/nsmb.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 36.Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, Berninghausen O, Sattler M, Wilson DN, Beckmann R. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol Cell. 2010;40:138–146. doi: 10.1016/j.molcel.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 37.Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache J, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson JH, Woolhead CA, Bernstein HD. The conformation of a nascent polypeptide inside the ribosome tunnel affects protein targeting and protein folding. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07325.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosolapov A, Deutsch C. Tertiary interactions within the ribosomal exit tunnel. Nat Struct Mol Biol. 2009;16:405–411. doi: 10.1038/nsmb.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●40.Tu LW, Deutsch C. A folding zone in the ribosomal exit tunnel for Kv1.3 helix formation. J Mol Biol. 2010;396:1346–1360. doi: 10.1016/j.jmb.2009.12.059. Implementing an important method pioneered in the Deutsch laboratory, the authors used engineered Cys residues and a high molecular weight Cys-reactive maleimide to probe structure formation within the ribosome and translocon. A number of α-helices in the Kv1.3 potassium channel formed in the vestibule of the ribosome tunnel and persisted into the translocon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daniel CJ, Conti B, Johnson AE, Skach WR. Control of translocation through the Sec61 translocon by nascent polypeptide structure within the ribosome. J Biol Chem. 2008;283:20864–20873. doi: 10.1074/jbc.M803517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujiwara K, Ishihama Y, Nakahigashi K, Soga T, Taguchi H. A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 2010;29:1552–1564. doi: 10.1038/emboj.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 44.Apetri AC, Horwich AL. Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc Natl Acad Sci USA. 2008;105:17351–17355. doi: 10.1073/pnas.0809794105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horwich AL, Fenton WA. Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q Rev Biophys. 2009;42:83–116. doi: 10.1017/S0033583509004764. [DOI] [PubMed] [Google Scholar]
- 46.Lin Z, Madan D, Rye HS. GroEL stimulates protein folding through forced unfolding. Nat Struct Mol Biol. 2008;15:303–311. doi: 10.1038/nsmb.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma S, Chakraborty K, Muller BK, Astola N, Tang YC, Lamb DC, Hayer-Hartl M, Hartl FU. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell. 2008;133:142–153. doi: 10.1016/j.cell.2008.01.048. [DOI] [PubMed] [Google Scholar]
- 48.Elad N, Farr GW, Clare DK, Orlova EV, Horwich AL, Saibil HR. Topologies of a substrate protein bound to the chaperonin GroEL. Mol Cell. 2007;26:415–426. doi: 10.1016/j.molcel.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clare DK, Bakkes PJ, van Heerikhuizen H, van der Vies SM, Saibil HR. Chaperonin complex with a newly folded protein encapsulated in the folding chamber. Nature. 2009;457:107–110. doi: 10.1038/nature07479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chakraborty K, Chatila M, Sinha J, Shi Q, Poschner BC, Sikor M, Jiang G, Lamb DC, Hartl FU, Hayer-Hartl M. Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell. 2010;142:112–122. doi: 10.1016/j.cell.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 51.Hofmann H, Hillger F, Pfeil SH, Hoffmann A, Streich D, Haenni D, Nettels D, Lipman EA, Schuler B. Single-molecule spectroscopy of protein folding in a chaperonin cage. Proc Natl Acad Sci USA. 2010;107:11793–11798. doi: 10.1073/pnas.1002356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Natale P, Brüser T, Driessen AJ. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane--distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778:1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- ●53.Junker M, Besingi RN, Clark PL. Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol Microbiol. 2009;71:1323–1332. doi: 10.1111/j.1365-2958.2009.06607.x. [DOI] [PubMed] [Google Scholar]
- ●54.Peterson JH, Tian P, Ieva R, Dautin N, Bernstein HD. Secretion of a bacterial virulence factor is driven by the folding of a C-terminal segment. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1009491107. in press. The paper by Junker et al. proposes model for directional (C-terminus first), coupled translocation and folding for the bacterial outer membrane protein autotransporter virulence factor, pertactin, from Bordatella pertussis. Using a homologous E. coli autotransporter virulence factor, EspP, Peterson et al. draw similar conclusions and more specifically identify the region of EspP, the C-terminal 17-kDa of its so-called passenger domain, the folding of which provides the driving force for translocation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hebert DN, Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 56.Riemer J, Bulleid N, Herrmann JM. Disulfide formation in the ER and mitochondria: Two solutions to a common process. Science. 2009;324:1284–1287. doi: 10.1126/science.1170653. [DOI] [PubMed] [Google Scholar]
- 57.Sideris DP, Tokatlidis K. Oxidative protein folding in the mitochondrial intermembrane space. Antioxid Redox Signal. 2010;13:1189–1204. doi: 10.1089/ars.2010.3157. [DOI] [PubMed] [Google Scholar]
- 58.Inaba K. Disulfide bond formation system in Escherichia coli. J Biochem. 2009;146:591–597. doi: 10.1093/jb/mvp102. [DOI] [PubMed] [Google Scholar]
- 59.Kadokura H, Beckwith J. Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid Redox Signal. 2010;13:1231–1246. doi: 10.1089/ars.2010.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●60.Kadokura H, Beckwith J. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell. 2009;138:1164–1173. doi: 10.1016/j.cell.2009.07.030. Kadokura and Beckwith devised a strategy to trap folding intermediates in the secreted protein PhoA and used it to demonstrate that the E. coli protein disulfide isomerase, DsbA, preferentially interacts with specific Cys residues catalyzing disulfide bond formation sequentially during co-translational translocation from the cytoplasm to the periplasm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●61.Tapley TL, Franzmann TM, Chakraborty S, Jakob U, Bardwell JC. Protein refolding by pH-triggered chaperone binding and release. Proc Natl Acad Sci USA. 2010;107:1071–1076. doi: 10.1073/pnas.0911610107. The authors characterize a novel periplasmic stress chaperone in E. coli, HdeA, which relies on pH changes to bind and release its substrates, and in so doing facilitates their folding and rescues them from aggregation at low pH in an ATP-independent mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang N, Glidden EJ, Murphy SR, Pearse BR, Hebert DN. The cotranslational maturation program for the type II membrane glycoprotein influenza neuraminidase. J Biol Chem. 2008;283:33826–33837. doi: 10.1074/jbc.M806897200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●63.Feige MJ, Groscurth S, Marcinowski M, Shimizu Y, Kessler H, Hendershot LM, Buchner J. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol Cell. 2009;34:569–579. doi: 10.1016/j.molcel.2009.04.028. This paper relates the steps in immunoglobulin maturation in B-cells to the folding properties of the IgG CH1 heavy chain domain, showing how its proper assembly with a light chain is timed to avoid aggregation or wasted material. The natively disordered heavy chain domain binds to the ER-resident Hsp70, BiP, such that a pool of CH1 can be maintained in a folding-competent state until its partner light chain is available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feige MJ, Hendershot LM, Buchner J. How antibodies fold. Trends Biochem Sci. 2010;35:189–198. doi: 10.1016/j.tibs.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghaemmaghami S, Oas TG. Quantitative protein stability measurement in vivo. Nat Struct Biol. 2001;8:879–882. doi: 10.1038/nsb1001-879. [DOI] [PubMed] [Google Scholar]
- 66.Ignatova Z, Gierasch LM. Monitoring protein stability and aggregation in vivo by real-time fluorescent labeling. Proc Natl Acad Sci USA. 2004;101:523–528. doi: 10.1073/pnas.0304533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ignatova Z, Krishnan B, Bombardier JP, Marcelino AM, Hong J, Gierasch LM. From the test tube to the cell: exploring the folding and aggregation of a β-clam protein. Biopolymers. 2007;88:157–163. doi: 10.1002/bip.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●68.Ebbinghaus S, Dhar A, McDonald JD, Gruebele M. Protein folding stability and dynamics imaged in a living cell. Nat Meth. 2010;7:319–323. doi: 10.1038/nmeth.1435. In this landmark paper, the authors use a temperature-sensitive protein labeled with a FRET pair to create a folding sensor in living human cells. They then apply a novel imaging method to map the rate of recovery of equilibrium following temperature jumps. The rates of folding/unfolding are surprisingly spatially and temporally heterogenous even in a single cell. [DOI] [PubMed] [Google Scholar]
- 69.Philipps B, Hennecke J, Glockshuber R. FRET-based in vivo screening for protein folding and increased protein stability. J Mol Biol. 2003;327:239–249. doi: 10.1016/s0022-2836(03)00077-9. [DOI] [PubMed] [Google Scholar]
- 70.Pielak GJ, Li C, Miklos AC, Schlesinger AP, Slade KM, Wang GF, Zigoneanu IG. Protein nuclear magnetic resonance under physiological conditions. Biochemistry. 2009;48:226–234. doi: 10.1021/bi8018948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ito Y, Selenko P. Cellular structural biology. Curr Opin Struct Biol. 2010 doi: 10.1016/j.sbi.2010.07.006. In Press. [DOI] [PubMed] [Google Scholar]
- 72.Inomata K, Ohno A, Tochio H, Isogai S, Tenno T, Nakase I, Takeuchi T, Futaki S, Ito Y, Hiroaki H, et al. High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature. 2009;458:106–109. doi: 10.1038/nature07839. [DOI] [PubMed] [Google Scholar]
- 73.Stidham RD, Wigley WC, Hunt JF, Thomas PJ. Assessment of protein folding/solubility in live cells. Methods Mol Biol. 2003;205:155–169. doi: 10.1385/1-59259-301-1:155. [DOI] [PubMed] [Google Scholar]
- 74.Cabantous S, Rogers Y, Terwilliger TC, Waldo GS. New molecular reporters for rapid protein folding assays. PloS One. 2008;3:e2387. doi: 10.1371/journal.pone.0002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●75.Foit L, Morgan GJ, Kern MJ, Steimer LR, von Hacht AA, Titchmarsh J, Warriner SL, Radford SE, Bardwell JC. Optimizing protein stability in vivo. Mol Cell. 2009;36:861–871. doi: 10.1016/j.molcel.2009.11.022. This paper reports an ingenious application of a split β-lactamase folding reporter to assess stability of the small protein Im7 in the periplasm of E. coli. The authors then utilize the resulting stability-dependent antibiotic resistance read-out to select Im7 mutants with increased stability. The results illustrate vividly the tension between retention of function and enhancement of stability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mansell T, Linderman S, Fisher A, Delisa M. A rapid protein folding assay for the bacterial periplasm. Protein Sci. 2010;19:1079–1090. doi: 10.1002/pro.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lyngsø C, Kjaerulff S, Müller S, Bratt T, Mortensen U, Dal Degan F. A versatile selection system for folding competent proteins using genetic complementation in a eukaryotic host. Protein Sci. 2010;19:579–592. doi: 10.1002/pro.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barakat NH, Carmody LJ, Love JJ. Exploiting elements of transcriptional machinery to enhance protein stability. J Mol Biol. 2007;366:103–116. doi: 10.1016/j.jmb.2006.10.091. [DOI] [PubMed] [Google Scholar]
- ●79.Barakat NH, Love JJ. Combined use of experimental and computational screens to characterize protein stability. Protein Eng Des Sel. 2010;23:799–807. doi: 10.1093/protein/gzq052. This paper uses a combination of directed evolution and protein design to select for streptococcal protein G B1 domains with enhanced thermostability. The authors test libraries of clones using a selection in which β-lactamase transcription is up-regulated by increased dynamics of the B1 domain. A tight inverse correlation is observed between antibiotic resistance and thermostability of the B1 domain. [DOI] [PubMed] [Google Scholar]
- 80.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 81.Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 82.Zeldovich KB, Shakhnovich EI. Understanding protein evolution: from protein physics to Darwinian selection. Annu Rev Phys Chem. 2008;59:105–127. doi: 10.1146/annurev.physchem.58.032806.104449. [DOI] [PubMed] [Google Scholar]
- 83.Bogumil D, Dagan T. Chaperonin-dependent accelerated substitution rates in prokaryotes. Genome Biol Evol. 2010;2:602–608. doi: 10.1093/gbe/evq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Williams TA, Fares MA. The effect of chaperonin buffering on protein evolution. Genome Biol Evol. 2010;2:609–619. doi: 10.1093/gbe/evq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Counago R, Wilson CJ, Pena MI, Wittung-Stafshede P, Shamoo Y. An adaptive mutation in adenylate kinase that increases organismal fitness is linked to stability-activity trade-offs. Protein Eng Des Sel. 2008;21:19–27. doi: 10.1093/protein/gzm072. [DOI] [PubMed] [Google Scholar]
- ●86.Peña MI, Davlieva M, Bennett MR, Olson JS, Shamoo Y. Evolutionary fates within a microbial population highlight an essential role for protein folding during natural selection. Mol Syst Biol. 2010;6:387. doi: 10.1038/msb.2010.43. The authors used a ‘weak-link’ approach and competition in a microbial population to identify adenylate kinase mutants that improve the evolutionary fate of the host organism. A mesophilic version of this essential enzyme was incorporated into a thermophile to select for mutations that enhance the fitness of the protein via increased activity and/or stability. By testing in vitro properties of the variant enzymes they find both are important for molecular adaptation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vendruscolo M, Tartaglia GG. Towards quantitative predictions in cell biology using chemical properties of proteins. Mol Biosyst. 2008;4:1170–1175. doi: 10.1039/b805710a. [DOI] [PubMed] [Google Scholar]
- 88.Tartaglia GG, Vendruscolo M. Proteome-Level relationship between folding and aggregation propensities of proteins. J Mol Biol. 2010 doi: 10.1016/j.jmb.2010.08.013. In Press. [DOI] [PubMed] [Google Scholar]
- 89.Mayer S, Rudiger S, Ang HC, Joerger AC, Fersht AR. Correlation of levels of folded recombinant p53 in Escherichia coli with thermodynamic stability in vitro. J Mol Biol. 2007;372:268–276. doi: 10.1016/j.jmb.2007.06.044. [DOI] [PubMed] [Google Scholar]