Abstract
A major constituent in the deposit in Alzheimer’s disease (AD) patient brain is the aggregates/fibrils of amyloid-β (Aβ) peptides containing 39–43 amino acids. The total Aβ levels and the concentration ratio between the most abundant Aβ(1–40) peptide and the more aggregation-prone Aβ(1–42) in body fluids (e.g., cerebrospinal fluid or CSF) have been suggested as possible criteria for early diagnosis of AD. By immobilizing capture antibodies specific to the two peptides in separate fluidic channels, surface plasmon resonance (SPR) has been used to quantify Aβ(1–40) and Aβ(1–42) present in CSF samples collected from AD patients and healthy donors. With signal amplification by streptavidin conjugated to an antibody that is selective to the common N-terminus of the Aβ peptides, concentrations as low as 20 pM can be readily measured. The range of Aβ peptide concentrations measurable by this method spans four orders of magnitude. The ability of regenerating the sensor surface for repeated measurements not only improves the reproducibility, but also enhances the sample throughput. Our data reveal that the ratio of Aβ(1–40) concentration versus Aβ(1–42) concentration in CSF samples from AD patients is almost twice as high as that from healthy persons. In contrast to the commonly used enzyme-linked immunosorbent assay (ELISA), SPR obviates the need of a more expensive and less stable enzyme conjugate and the use of carcinogenic substrate for the signal detection, and allows the binding events to be monitored in real time.
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder whose defining features include neuritic (senile) plaques in brain of AD patients.1, 2 A major constituent of the neuritic plaques is the amyloid-β (Aβ) peptides containing 39–43 amino acid residues.3, 4 These Aβ peptides were segments proteolytically cleaved from the amyloid precursor protein (APP).5 Both postmortem analyses of the senile plaques of AD patient brain extracts and in vitro Aβ peptide aggregation studies have firmly established that Aβ(1–42), the 42-residue-long peptide, has a greater tendency to misfold and aggregate than the much more abundant Aβ(1–40). Although the role of Aβ peptides in AD pathogenesis is not well understood, on the basis of the analogy to systemic amyloidosis (deposition of β-sheet-pleated insoluble aggregates/fibrils),1 Aβ peptides are widely believed to be an important biomarker and drug target for AD research and therapy.6–8 A range of methods have been developed to detect Aβ monomers and its aggregates in brain extract,9 including the Aβ-derived diffusible ligands (ADDLs)5 that are currently believed to be the most pernicious among the various Aβ aggregates. However, most of these methods are only applicable to postmortem analyses. For early diagnosis, it is highly desirable to develop sensitive and selective methods9, 10 that are amenable to the detection of Aβ and other AD biomarkers in body fluids (e.g., plasma, urine, and cerebrospinal fluid or CSF). Polyacrylamide gel electrophoresis (PAGE), immunoprecipitation, mass spectrometry, fluorescent staining, and enzyme-linked immunosorbent assay (ELISA) have been employed to detect Aβ species from body fluids and cell media.9, 10 Among them, ELISA has the best sensitivity, selectivity, and versatility.11–14 However, a typical sandwiched ELISA requires 1–2 days, the use of a relatively expensive enzyme-linked antibody, and the need of carcinogenic substrates for the chemiluminescent detection step.15
In recent years, various spectroscopic and electrochemical methods have been developed for detecting monomers, intermediates, and aggregates/fibrils of Aβ16–18 and related amyloidogenic proteins (e.g., α-synuclein19–21). In particular, surface plasmon resonance (SPR)22–24 has also been shown as a promising technique.25–28 SPR is based on the detection of changes in mass concentration of an analyte (also known as prey or target molecule) that is bound to a capture (also known as bait or probe) molecule preimmobilized onto a sensor chip. For analyte concentration determination and kinetic studies of biomolecular interactions, the attractive features of SPR include the high sensitivity, label-free and real-time measurements, relatively simple procedure, and low sample consumption.22–24 Homola and coworkers employed SPR to detect the 17β–hydroxysteroid dehydrogenase type 10 (17β–HSD10) enzyme and a peptidic analog in artificial CSF solutions.27 The interaction between 17β–HSD10 and Aβ has been suggested as a possible cause for mitochondrial dysfunction in AD.11 Lee et al. reported the use of gold nanoparticle-antibody complex for amplified SPR detection of synthetic Aβ(1–40) peptide in buffer solution.28 However, in that work unmodified gold sensors were used for immobilization of the capture antibody and selectivity and the extent of non-specific adsorption inherent in the method were not examined. The work performed by Van Duyne’s group is particularly noteworthy and appears to be most clinically relevant.25, 26 Using an array of Ag nanotriangles fabricated with nanosphere-lithography, 29 localized SPR effect was demonstrated to be attractive in enhancing the detection sensitivity of the detection of ADDL (soluble Aβ oligomers). Although CSF samples from only one AD patient and one control were measured, the feasibility studies clearly highlight that SPR is a powerful technique for clinical assay of AD biomarkers. To our knowledge, SPR has not been explored for sensitive and simultaneous quantification of concentrations of Aβ(1–40) and Aβ(1–42) monomers in human CSF. On the basis that more Aβ(1–42) molecules than their Aβ(1–40) counterparts have aggregated and deposited in AD brain, the concentration ratio between Aβ(1–40) and Aβ(1–42) will be elevated in CSF samples withdrawn from AD patients when compared to that from healthy persons. Therefore, the variation of such concentration ratios has been suggested as a criterion for early diagnosis of AD.
Previously we employed a dual-channel SPR instrument for simultaneous determination of wild-type and mutant p53 (a tumor suppressor protein30) present at low to sub-nanomolar levels in cancer cell lysates.31 The difference in the wild-type and total p53 concentrations reveals the extent of p53 mutation, which is indicative of cancer development. In this work, we utilized such a dual channel SPR for determining the ratio of Aβ(1–40) versus Aβ(1–42) concentrations by immobilizing antibody that specifically recognizes the respective peptide onto the two fluidic channels. Since Aβ peptides (4330 and 4514 Daltons for Aβ(1–40) and Aβ(1–42), respectively) are smaller in size than p53 (53000 Daltons) and their concentrations in CSF are at low (sub-nanomolar) concentrations,12, 32, 33 a signal amplification scheme was devised by using a conjugate preformed between streptavidin and a biotinylated monoclonal antibody that can bind to the common N-terminus of the Aβ(1–40) and Aβ(1–42) peptides. Upon evaluating the analytical figures of merit of this method, its viability for clinical assays was demonstrated by analyzing multiple sets of CSF samples from AD patients and healthy donors for their Aβ(1–40)/Aβ(1–42) concentration ratios.
Experimental Section
Chemicals and Materials
N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), ethanolamine hydrochloride, streptavidin (SA), triethylene glycol mono-11-mercaptoundecyl ether (HSC11PEG3-OH), NaOH, KH2PO4, and K2HPO4 were acquired from Sigma (St. Louis, MO). Hexaethylene glycol mono-11-mercaptoundecyl acid (HSC11PEG6-COOH) was purchased from Sensopath Technologies (Bozeman, MT). Aβ(1–40) and Aβ(1–42) peptides were purchased from American Peptide Inc. (Sunnyvale, CA). Monoclonal antibodies (mAbs) of Aβ(1–40) (clone 11A50-B10) and Aβ(1–42) (clone 12F4) specific to the C-termini of Aβ(1–40) and Aβ(1–42), respectively, and an mAb capable of binding to the common N-terminus of these two peptides (clone 6E10) were obtained from Covance Inc. (Dedham, MA). Other reagents were all of analytical purity and used as received. All stock solutions were prepared daily with deionized water treated with a water purification system (Simplicity 185, Millipore Corp, Billerica, MA).
Instruments
The FI-SPR measurements were conducted with a BI-SPR 3000 system (Biosensing Instrument Inc, Tempe, AZ). Phosphate-buffered saline (PBS buffer, pH 7.4, 10 mM phosphate/10 mM NaCl/0.01% (V/V) Tween 20) was degassed via vacuum pumping for 30 min and used as the carrier solution. The samples were preloaded into a 200-µL sample loop on a six-port valve and then delivered to the flow cell with an internal volume of ca. 1.0 μL by a syringe pump (Model KDS260, Kd Scientific, Holliston, MA). The instrument is capable of cutting off the dispersed front and tail ends of injected sample plugs prior to introducing samples into the SPR sensing areas.
Procedures
Solution Preparation
HSC11PEG6-COOH and HSC11PEG3-OH were dissolved in anhydrous ethanol. Samples of Aβ(1–40), Aβ(1–42) and antibodies were diluted with PBS buffer. The Aβ stock solutions (0.5 mM) were prepared daily as in our previous studies.34 Briefly, lyophilized Aβ samples were dissolved in 10 mM NaOH solution in which the aggregation of Aβ is known to be effectively inhibited. Upon sonication for 1 min, the solution was centrifuged at 13,000 rpm for 30 min to remove any insoluble particles, and the supernatants were pipetted out and diluted by PBS buffer for the experiments. The stock peptide concentrations were determined from the UV-vis spectra using the extinction coefficient at 276 nm (ε = 1410 M−1cm−1). Ethanolamine was dissolved in water.
SPR Sensor Surface Modification
Au films coated onto BK7 glass slides were purchased from Biosensing Instrument Inc., and annealed in a hydrogen flame to eliminate surface contaminants. The PEG (polyethylene glycol)-covered SPR chip was formed by immersing the cleaned substrate in a mixed solution of HSC11PEG6-COOH and HSC11PEG3-OH in the dark for 48 h. The final concentrations of HSC11PEG6-COOH and HSC11PEG3-OH were maintained at 0.2 and 1.8 mM, respectively. Upon formation of the PEG self-assembled monolayers (SAMs), the chips were removed from the solution, rinsed with absolute ethanol and deionized water, and then dried with nitrogen. For SA immobilization, 100 µL aliquots of the EDC/NHS mixture were cast onto the chip surface for 10 min. EDC/NHS solution was prepared by mixing 0.4 M EDC with 0.1 M NHS in water right before the PEG film activation step. These activated chips were then washed with water and soaked in an SA solution for 0.5 h. An SA sensor chip was preformed by cross-linking SA molecules onto a mixed SAM composed of carboxyl- and hydroxyl-terminated PEG molecules via the standard amine coupling reaction. The mixed PEG SAM has been shown to be effective in eliminating nonspecific adsorption but allowing controllable attachment of sensing molecules.35 This was followed by casting 1.0 M ethanolamine onto the chips to block the unreacted sites. The resultant chip was mounted onto the SPR instrument for measurements. After a stable baseline had been obtained, in the parallel flow configuration, the biotinylated mAbs for Aβ(1–40) and Aβ(1–42) were injected separately into the two different fluidic channels. Once the mAb plugs had entered the channels, the solution flows were stopped to allow solution contact with the sensor surface for 1–2 h.
CSF Samples
The Huntington Hospital Institutional Review Board for Human Research approved the study protocol and informed consent forms. Consecutive controls and AD patients from the Pasadena area were recruited through advertising. Signed informed consent was obtained, supplemented by consent from the durable power of attorney for all AD participants. Criteria for diagnosis of clinically probable AD were met, using the national criteria.36 Controls were subject to a full history and physical examination that excluded any active, untreated disorder. CSF samples were collected between 1:00 and 6:00 pm by lumbar puncture using a 22-gauge Quincke-type needle, aliquoted and stored at −80° C until thawed for SPR assay.
Simultaneous SPR detection of Aβ(1–40) and Aβ(1–42) concentrations
In a serial flow configuration, buffer solutions containing synthetic Aβ(1–40) and Aβ(1–42) or CSF samples were injected into the fluidic channels covered with the respective antibody of Aβ(1–40) or Aβ(1–42). To achieve low detection level and to conserve sample, a slow flow rate of 5 μL/min, unless otherwise stated, was used. For signal amplification, the conjugate preformed between biotinylated Aβ(1–16) mAb and streptavidin was injected into the SPR cell to react with the Aβ(1–40) and Aβ(1–42) peptides that had been captured by their respective antibodies. Duration of the conjugate injection was 900 s at 10 µL/min. After each measurement, the surface was regenerated via one or more injections of 10 mM NaOH.
Results and Discussion
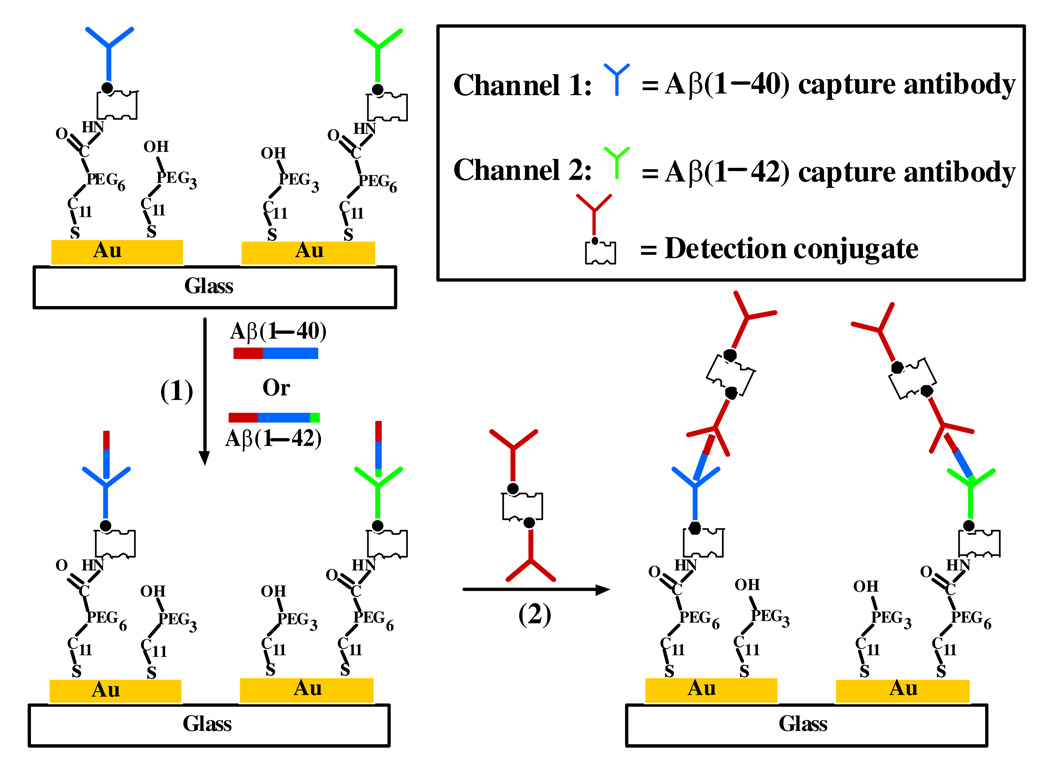
A schematic representation of the simultaneous SPR determination of Aβ(1–40) and Aβ(1–42), amplified by using a conjugate of streptavidin-biotinylated Aβ(1–16) mAb (referred to as the detection conjugate hereafter), is illustrated in Figure 1. The biotinylated Aβ(1–40) and Aβ(1–42) antibodies (referred to as the capture antibodies henceforth), respectively depicted in blue and green (Figure 1), were individually immobilized onto the sensor regions covered by the two fluidic channels. If the sample contains relatively high concentrations (5.00–150.00 nM) of Aβ(1–40) and/or Aβ(1–42), the specific binding between a capture antibody and its cognate Aβ peptide enables the direct and label-free determination of the corresponding Aβ concentration (first step in Figure 1). If the Aβ peptide concentration is too low to give an appreciable SPR signal, injection of detection conjugate can be made (second step). The SPR signal is known to be proportional to the amount of mass deposited on the sensor surface. Aβ(1–16) mAb (shown in red) on the detection conjugate can recognize the N-terminus of both Aβ(1–40) and Aβ(1–42), resulting in amplified SPR signals. Such an amplification stems from the much greater molecular weights of SA (52800 Daltons) and Aβ(1–16) mAb (150000 Daltons) than those of the Aβ(1–40) and Aβ(1–42) peptides (4330 and 4514 Daltons, respectively). Because of the low (sub-nanomolar) Aβ concentrations in CSF, we found that signal amplification by the detection conjugate, capable of determining Aβ peptide concentrations between 0.02 and 5.00 nM (vide infra), was necessary for the real-sample measurements.
Figure 1.
Schematic diagram showing the simultaneous SPR detection of Aβ(1–40) and Aβ(1–42). One fluidic channel is covered with the capture antibody (shown in blue) for Aβ(1–40) whose hydrophobic domain (residues 17–40) is depicted in blue and hydrophilic (residues 1–16) in red, while the other channel was precoated with the capture antibody (shown in green) for Aβ(1–42) whose additional residues (41–42) are shown in green. Injection of Aβ samples results in the attachment of Aβ(1–40) or Aβ(1–42) to the respective channel (Step 1), and injection of the detection conjugate that can recognize the common hydrophilic domain of Aβ(1–40) and Aβ(1–42) leads to signal amplification (Step 2). For simplicity, the two fluidic channels are represented by two Au stripes and the Aβ peptides are represented by sticks.
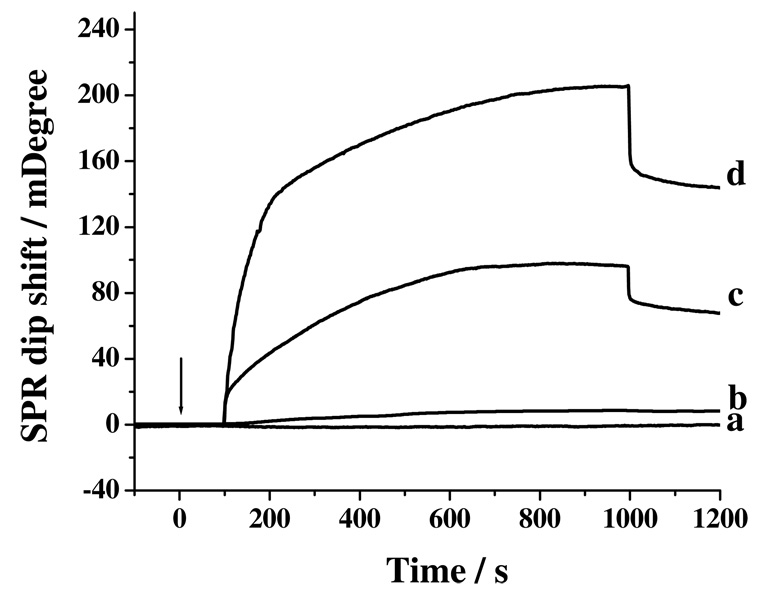
A direct injection of 1.00 nM Aβ(1–40) into a channel covered with its capture antibody did not produce a detectable signal (curve a in Figure 2). However, directly injecting a sample containing 50.00 nM Aβ(1–40) resulted in a signal change by 7.51 mDeg (equivalent to 75.1 resonance units in curve b). Interestingly, the channel that originally produced curve a gave rise to a much more pronounced baseline shift after 30.00 nM Aβ(1–16) mAb had been injected (curve c). A net change of 69.58 mDeg was observed at ca. 1200 s in the sensorgram (or 200 s after replacing the injected sample with the carrier solution). The signal amplification is originated from attachment of Aβ(1–16) mAb whose molecular weight is greater than that of Aβ(1–40). The transient increase and decrease at the beginning and end of the injection peaks in curves c are attributed to the slight refractive index differences between the Aβ(1–16) mAb and the carrier solutions. Notice that the net change in the baseline of curve d is 2.06 times of that of curve c and recall that SA is a much smaller molecule than Aβ(1–16) mAb. Thus the difference in the changes between curves c and d is largely contributed by the mAb molecules and suggests that, on an average, two Aβ(1–16) mAb molecules are bound to one SA molecule, as alluded by the schematics shown in Figure 1. This is conceivable, given the steric hindrance imposed by Aβ(1–16) mAb, that not all of the four sites in the tetravalent SA are occupied by the Aβ(1–16) mAb molecule. We also found that injection of gold nanoparticles capped with Aβ(1–16) mAb molecules resulted in much greater nonspecific adsorption at the PEG SAM.
Figure 2.
SPR sensorgrams after injections of (a) 1.00 and (b) 50.00 nM of Aβ(1–40) into precoated channel with Aβ(1–40) capture antibody, (c) curve (a) + 30.00 nM of Aβ(1–16) mAb, and (d) curve (a) + 30.00 nM of the detection conjugate. The arrow indicates the time when the injections were made and the flow rate was 10 μL/min. The delays in all the sensorgrams between the times when injections were made and signals were observed are times needed for the injected sample plugs to travel from the injection valve to the SPR flow cell.
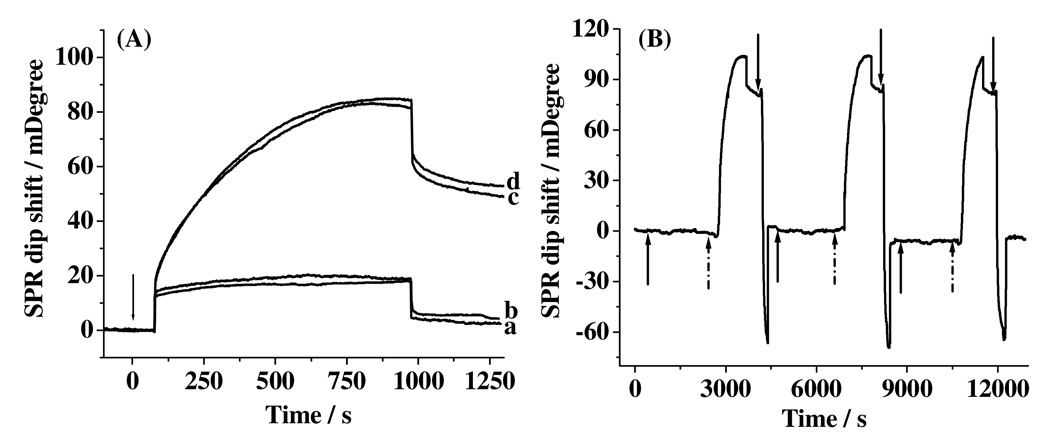
We first examined the selectivity of and efficacy for eliminating nonspecific adsorption by the capture antibody-covered surface by injecting the conjugate into fluidic channels covered with different molecules. Shown in Figure 3A are four overlaid sensorgrams acquired from surfaces covered with Aβ(1–40) capture antibody (curve a), Aβ(1–40) capture antibody to which 0.50 nM Aβ(1–42) had been exposed (curve b), Aβ(1–42) capture antibody with which 0.20 nM Aβ(1–42) was allowed to react (curve c), and Aβ(1–40) capture antibody that had been in contact with a 0.20 nM Aβ(1–40) solution (curve d). When no peptides were captured, little SPR dip shifts appeared in the sensorgrams (~ 3.02 mDeg in curve a and 5.27 mDeg in curve b). When a given Aβ peptide has been captured by its cognate capture antibody, injections of the detection conjugate produced large SPR dip shifts (77.23 and 83.51 mDeg in curves c and d, respectively). These data suggest that the method is quite selective and the rather small nonspecific adsorption is indicative of the efficacy of the mixed PEG SAM in reducing nonspecific adsorption.37–40
Figure 3.
SPR sensorgrams after injections of 30.00 nM detection conjugate to Aβ(1–40) capture antibody (a) before and (b) after exposed to 0.50 nM Aβ(1–42), (c) Aβ(1–42) capture antibody that had been exposed to 0.20 nM Aβ(1–42), and (d) same as (a) but with exposure to 0.20 nM Aβ(1–40). The arrow indicates the time when the injections were made. (B) A sensorgram showing three repeated cycles for the injections of 0.50 nM Aβ(1–40) (signified by the solid upward arrows) and 30.00 nM detection conjugate (starting from the dotted arrows) into an SPR channel covered with the Aβ(1–40) capture antibody and the regeneration of the sensor surface using 10 mM NaOH (starting from the downward arrows).
We found that reproducible signals can be obtained by injecting 10 mM NaOH into the fluidic channels to regenerate the surface (i.e., desorbing both the conjugate and Aβ peptide). After 10 regenerations, the binding signal decreased by less than 8%. Therefore, a single chip can be repeatedly used for multiple samples (cf. Figure 3B), dramatically increasing the sample throughput and decreasing the sample analysis time. Notice that one measurement cycle (continuous injections of Aβ solutions, the detection conjugate, and the NaOH regeneration solution) takes about 1 h, which is shorter than that needed for a typical ELISA (~1 day). More importantly, the binding, signal amplification, and regeneration steps can all be monitored in real time.
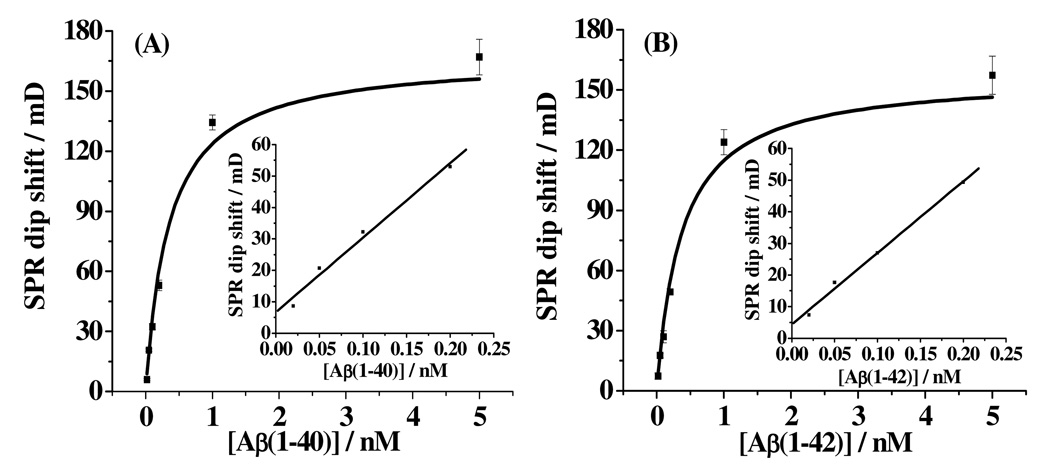
With the selectivity of the method established, we assessed other analytical figures of merit such as reproducibility, sensitivity, dynamic ranges and detection limits. The dependences of the SPR dip shift on the Aβ(1–40) and Aβ(1–42) concentrations are presented in panels A and B of Figure 4, respectively. The aforementioned regeneration of the sensor surface contributes to the good reproducibility of the method, as the relative standard deviations (RSDs), shown as the error bars in Figure 4, are all less than 10%. For both Aβ peptides, the SPR dip shift increases sharply within the concentration range of 0.02–1.00 nM, but begins to level off beyond 1.00 nM. We found that the data can be fitted with the Langmuir isotherm:
| (1) |
where θ is surface coverage and K is binding affinity between the detection conjugate and the Aβ peptide. The K values from the fits were deduced to be 2.51 nM−1 for Aβ(1–40) and 2.31 nM−1 for Aβ(1–42). The highly comparable K values suggest that the detectionconjugate binds to both Aβ(1–40) and Aβ(1–42) with similar affinities. This is conceivable since the Aβ(1–16) mAb on the conjugate binds to the same N-terminus of the two peptides. These values are also in reasonable agreement with the reported nanomolar level KD value between the Aβ(1–40) mAb and Aβ(1–40).41 The nanomolar level KD values deduced from equation (1) suggest that the binding of the mAb to Aβ(1–40) or Aβ(1–42) is rather high. This is not surprising since the mAbs we used in this work are known to be specific only to the monomeric forms of Aβ(1–40) and Aβ(1–42).42 The plateau exhibited by the curves in Figure 4 is indicative of the attainment of the “saturated” coverage by the detection conjugate. The concentration ranges shown in Figure 4 are more than adequate to encompass the Aβ concentrations in both healthy people and AD patients.12, 32, 43–45
Figure 4.
Dependence of SPR dip shifts on concentrations of Aβ(1–40) (A) and Aβ(1–42) (B). The absolute errors were deduced from at least three replicate measurements and are shown as the error bars. The curves are responses predicted by the Langmuir isotherm. Concentrations of Aβ(1–40) and Aβ(1–42) determined were 0.02, 0.05, 0.10, 0.20, 1.00, and 5.00 nM. Insets show the linear portions of the curves when the surface coverage of Aβ(1–40) or Aβ(1–42) is less than 35% of a full monolayer.
We also found that Aβ peptide concentrations that are higher than 5.00 nM can be detected by decreasing the time for the Aβ capture step. When the Aβ concentrations are high, the SPR signal changes can be easily discerned without signal amplification by the detection conjugate. By keeping the experimental condition for the detection step the same but reducing the reaction time to 30 s (instead of 1800 s used for obtaining Figures 2–4), we were able to observe continuous SPR signal changes between 5.00 and 150.00 nM and found that the responses also obey the Langmuir behavior. Thus, our method is amenable for the determination of Aβ peptide concentrations between 0.02 and 150.00 nM, a span of almost four orders of magnitude. That the method is capable of detecting higher Aβ peptide concentrations may offer a useful means for quantifying Aβ in other types of studies (e.g., measuring free Aβ monomers in an incubated Aβ solution for probing the dynamics and mechanism of Aβ aggregation/fibrillation processes under different experimental conditions).46
When the surface coverage is less than ~35% of a full monolayer (between 0.02 and 0.20 nM), the SPR responses were found to be linearly dependent on the concentration (insets of Figure 4). The detection limits, estimated from 3σ of the baseline signals, are 3.3 pM for Aβ(1–40) and 3.5 pM for Aβ(1–42). Although the detection limits are governed by factors such as the sensitivity of the method and the binding affinity between Aβ peptides and the antibodies, based on the fact that ELISA can also detect low or subpicomolar Aβ concentrations (5 to 20 pM12, 13, 47, 48), it appears that more sensitive SPR methods (e.g., localized SPR biosensors using Ag nanotriangles25, 26 or signal amplification schemes with gold nanoparticles at appropriate substrates49, 50) may further reduce the detection limits.
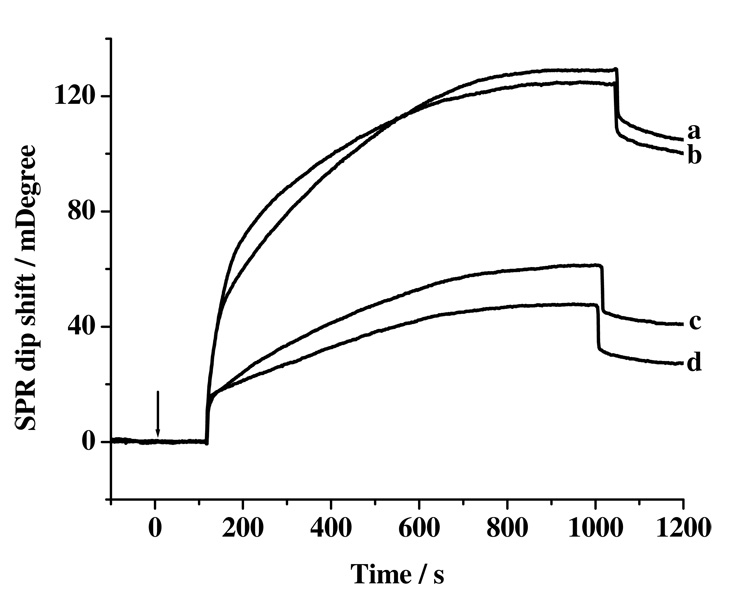
Finally, to demonstrate the viability of the method for clinical assays, simultaneous quantifications of Aβ(1–40) and Aβ(1–42) in CSF samples from five AD patients and five healthy donors were carried out. Two interesting conclusions can be drawn from the results shown in Figure 5 and Table 1. First, the Aβ(1–40) concentration is much higher than its Aβ(1–42) counterpart in the CSF samples collected from both the healthy and patient donors. This is expected since, among the various peptidic segments cleaved from the Aβ precursor protein (APP), the abundance of Aβ(1–40) (~60–70%) is greater than that of Aβ(1–42) (~5–15%).9, 33, 51, 52 Second, the SPR dip shift for Aβ(1–40) from a healthy donor (e.g., 105.89 mDeg from curve a in Figure 5) is only slightly greater (or statistically indifferent) than that from a AD patient (100.31 mDeg from curve b). However, Table 1 shows that the Aβ(1–42) concentration from the AD patients’ CSF samples is considerably lower. This result is plausible since Aβ(1–42), among the different Aβ peptide variants, is known to have the highest propensity to form oligomers and the higher-ordered protofibrils and fibrils. The deposition of these insoluble aggregates in brain reduces the amount of soluble Aβ(1–42) monomers in CSF. Our results are also consistent with findings derived from ELISA measurements of similar samples.12, 32, 45, 48
Figure 5.
SPR sensorgrams depicting simultaneous detection of Aβ(1–40) and Aβ(1–42) concentrations in CSF collected from healthy donors (curves a and c for Aβ(1–40) and Aβ(1–42), respectively) and AD patients (curves b and d for Aβ(1–40) and Aβ(1–42), respectively) with signal amplification by the detection conjugate. The arrow indicates the time when the injections of the detection conjugate were made.
Table 1.
Clinical parameters and Aβ(1–40) and Aβ(1–42) concentrations in CSF
| Sample | [Aβ(1–40)] | [Aβ(1–42)] | [Aβ(1–40) ]/[Aβ(1–42)] |
|---|---|---|---|
| Healthy controls (n = 5) 3 males, 2 females Ages of 78 yrs (3.2)* |
1.25 ± 0.31 nM | 0.31 ± 0.12 nM | 3.91 ± 1.05 |
| AD patients (n =5) 3 males, 2 females Ages of 76.4 yrs (4.2)* |
1.13 ± 0.35 nM | 0.16 ± 0.07 nM | 6.89 ± 1.55 |
Mean age (standard error of the mean) and matched (Mann-Whitney test: p = 0.402)
Conclusion
Simultaneous SPR quantification of Aβ(1–40) and Aβ(1–42) peptide concentrations has been accomplished using the respective antibodies preimmobilized onto two separate fluidic channels. With signal amplification by a conjugate formed between streptavidin and an N-terminus-specific monoclonal antibody, concentrations as low as 20 pM of the Aβ peptides can be readily measured. The range of concentration measurable by the SPR method spans almost four orders of magnitude, making it versatile for measuring samples from different sources and/or systems. The method developed herein is also quite reproducible (%RSD < 10%) and selective. The amenability of the method to clinical assays has been demonstrated by determining the variation of the concentration ratio between Aβ(1–40) and Aβ(1–42) in CSF samples collected from AD patients and healthy persons. Our method retains the powerful features inherent in ELISA (e.g., quantitative, sensitive and selective), but offers three additional advantages: faster analysis time, obviation of an enzyme-linked antibody for signal detection (amplification), and regeneration of the sensor surface for replicate measurements. Obviation of an enzyme reduces the overall assay cost and avoids the use of carcinogenic substrate for chemiluminescent detection. The renewable chips not only enhance the sample throughput (multiple samples assayed with a single chip), but also allow reproducible measurements to be made. Although not explored in this work, the use of multichannel SPR instrument is expected to provide an even greater sample throughput. The proof-of-concept experiment demonstrates that SPR can potentially serve as a viable alternative for facile and sensitive clinical analyses of important biomarkers related to neurodegenerative diseases.
Acknowledgments
We thank Mr. Shengmu Xiao for his technical help. Partial support of this work by the National Natural Science Foundation of China (Nos. 20975114 and 20775093 to JW) and an NINDS grant (No. SC1NS070155-01 to FZ), an NSF-RUI grant (No. 0555224 to FZ) and the NIH-RIMI Program at California State University, Los Angeles (P20-MD001824-01 to FZ) is gratefully acknowledged. MGH thanks HMRI and the Anne and Jerry Dunlevy Foundation for support.
References
- 1.Hardy J, Selkoe DJ. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Proc. Natl. Acad. Sci. U. S. A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haass C, Schlossmacher MG, Hung AY, Vigopelfrey C, Mellon A, Ostaszewski BL, Lieberburg I, Koo EH, Schenk D, Teplow DB, Selkoe DJ. Nature. 1992;1992:322–325. doi: 10.1038/359322a0. [DOI] [PubMed] [Google Scholar]
- 4.Shoji M, Golde TE, Ghiso J, Cheung TT, Estus S, Shaffer LM, Cai XD, McKay DM, Tintner R, Frangione B, Younkin SG. Science. 1992;258:126–129. doi: 10.1126/science.1439760. [DOI] [PubMed] [Google Scholar]
- 5.Thinakaran G, Koo EH. In: APP Biology, Processing and Function. Sisodia SS, Tanzi RE, editors. New York: Springer; 2007. [Google Scholar]
- 6.Andreasen N, Zetterberg H. Curr. Med. Chem. 2008;15:766–771. doi: 10.2174/092986708783955572. [DOI] [PubMed] [Google Scholar]
- 7.Blennow K, Hampel H, Weiner M, Zetterberg H. Nat. Rev. Neurol. 2010;6:131–144. doi: 10.1038/nrneurol.2010.4. [DOI] [PubMed] [Google Scholar]
- 8.Hansson O, Zetterberg H, Buchhave P, Andreasson U, Londos E, Minthon L, Blennow K. Dement. Geriatr. Cogn. Disord. 2007;23:316–320. doi: 10.1159/000100926. [DOI] [PubMed] [Google Scholar]
- 9.Golde TE, Eckman CB, Younkin SG. Biochim. Biophys. Acta. 2000;1502:172–187. doi: 10.1016/s0925-4439(00)00043-0. [DOI] [PubMed] [Google Scholar]
- 10.Munishkina LA, Fink AL. Biochim. Biophys. Acta. 2007;1768:1862–1885. doi: 10.1016/j.bbamem.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Gravina SA, Ho LB, Eckman CB, Long KE, Otvos L, Younkin LH, Suzuki N, Younkin SG. J. Biol. Chem. 1995;270:7013–7016. doi: 10.1074/jbc.270.13.7013. [DOI] [PubMed] [Google Scholar]
- 12.Mehta PD, Pirttila T, Patrick BA, Barshatzky M, Mehta SP. Neurosci. Lett. 2001;304:102–106. doi: 10.1016/s0304-3940(01)01754-2. [DOI] [PubMed] [Google Scholar]
- 13.Seubert P, Vigopelfrey C, Esch F, Lee M, Dovey H, Davis D, Sinha S, Schlossmacher M, Whaley J, Swindlehurst C, McCormack R, Wolfert R, Selkoe D, Lieberburg I, Schenk D. Nature. 1992;359:325–327. doi: 10.1038/359325a0. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki N, Cheung TT, Cai XD, Odaka A, Otvos L, Eckman C, Golde TE, Younkin SG. Science. 1994;264:1336–1340. doi: 10.1126/science.8191290. [DOI] [PubMed] [Google Scholar]
- 15.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. New York: W. H. Freeman; 2004. [Google Scholar]
- 16.Makin OS, Serpell LC. FEBS J. 2005;272:5950–5961. doi: 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]
- 17.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vestergaard M, Kerman K, Saito M, Nagatani N, Takamura Y, Tamiya E. J. Am. Chem. Soc. 2005;127:11892–11893. doi: 10.1021/ja052522q. [DOI] [PubMed] [Google Scholar]
- 19.Ferreon ACM, Moran CR, Ferreon JC, Deniz AA. Angew. Chem. Int. Ed. 2010;49:3469–3472. doi: 10.1002/anie.201000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masarik M, Stobiecka A, Kizek R, Jelen F, Pechan Z, Hoyer W, Jovin TM, Subramaniam V, Palecek E. Electroanalysis. 2004;16:1172–1181. [Google Scholar]
- 21.Palecek E, Ostatna V, Masarik M, Bertoncini CW, Jovin TM. Analyst. 2008;133:76–84. doi: 10.1039/b712812f. [DOI] [PubMed] [Google Scholar]
- 22.Hanken DG, Jordan CE, Frey BL, Corn RM. In: Electroanalytical Chemistry: A Series of Advances. Bard AJ, Rubenstein I, editors. Vol. 20. New York: Marcel Dekker; 1998. pp. 141–225. [Google Scholar]
- 23.Homola J. Surface Plasmon Resonance Based Sensors. Berlin: Springer; 2006. [Google Scholar]
- 24.Phillips KS, Cheng Q. Anal. Bioanal. Chem. 2007;387:1831–1840. doi: 10.1007/s00216-006-1052-7. [DOI] [PubMed] [Google Scholar]
- 25.Haes AJ, Chang L, Klein WL, Van Duyne RP. J. Am. Chem. Soc. 2005;127:2264–2271. doi: 10.1021/ja044087q. [DOI] [PubMed] [Google Scholar]
- 26.Haes AJ, Hall WP, Chang L, Klein WL, Van Duyne RP. Nano Lett. 2004;4:1029–1034. [Google Scholar]
- 27.Hegnerová K, Bockova M, Vaisocherova H, Kristofikova Z, Ricny J, Ripova D, Homola J. Sens. Actuators B. 2009;139:69–73. [Google Scholar]
- 28.Lee JH, Kang DY, Lee T, Kim SU, Oh BK, Choi JW. J. Nanosci. Nanotechnol. 2009;9:7155–7160. doi: 10.1166/jnn.2009.1613. [DOI] [PubMed] [Google Scholar]
- 29.Haynes CL, Van Duyne RP. J. Phys. Chem. B. 2001;105:5599–5611. [Google Scholar]
- 30.Levine AJ, Finlay CA, Hinds PW. Cell. 2004;S116:S67–S69. doi: 10.1016/s0092-8674(04)00036-4. [DOI] [PubMed] [Google Scholar]
- 31.Wang YC, Zhu X, Wu MH, Xia N, Wang JX, Zhou FM. Anal. Chem. 2009;81:8441–8446. doi: 10.1021/ac9014269. [DOI] [PubMed] [Google Scholar]
- 32.Tamaoka A, Sawamura N, Fukushima T, Shoji S, Matsubara E, Shoji M, Hirai S, Furiya Y, Endoh R, Mori H. J. Neurol. Sci. 1997;148:41–45. doi: 10.1016/s0022-510x(96)00314-0. [DOI] [PubMed] [Google Scholar]
- 33.Wang R, Sweeney D, Gandy SE, Sisodia SS. J. Biol. Chem. 1996;271:31894–31902. doi: 10.1074/jbc.271.50.31894. [DOI] [PubMed] [Google Scholar]
- 34.Jiang DL, Men LJ, Wang JX, Zhang Y, Chickenyen S, Wang YS, Zhou FM. Biochemistry. 2007;46:9270–9282. doi: 10.1021/bi700508n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sigal GB, Bamdad C, Barberis A, Strominger J, Whitesides GM. Anal. Chem. 1996;68:490–497. doi: 10.1021/ac9504023. [DOI] [PubMed] [Google Scholar]
- 36.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 37.Das J, Huh CH, Kwon K, Park S, Jon S, Kim K, Yang H. Langmuir. 2009;25:235–241. doi: 10.1021/la802531d. [DOI] [PubMed] [Google Scholar]
- 38.Herrwerth S, Rosendahl T, Feng C, Fick J, Eck W, Himmelhaus M, Dahint R, Grunze M. Langmuir. 2003;19:1880–1887. [Google Scholar]
- 39.Muñoz EA, Yu HN, Hallock J, Edens RE, Linhardt RJ. Anal. Biochem. 2005;343:176–178. doi: 10.1016/j.ab.2005.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piehler J, Brecht A, Valiokas R, Liedberg B, Gauglitz G. Biosens. Bioelectron. 2000;15:473–481. doi: 10.1016/s0956-5663(00)00104-4. [DOI] [PubMed] [Google Scholar]
- 41.Ramakrishnan M, Kandimalla KK, Wengenack TM, Howell KG, Poduslo JF. Biochemistry. 2009;48:10405–10415. doi: 10.1021/bi900523q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Agnaf OMA, Mahil DS, Patel BP, Austen BM. Biochem. Biophys. Res. Commun. 2000;273:1003–1007. doi: 10.1006/bbrc.2000.3051. [DOI] [PubMed] [Google Scholar]
- 43.Galasko D, Chang L, Motter R, Clark CM, Kaye J, Knopman D, Thomas R, Kholodenko D, Schenk D, Lieberburg I, Miller B, Green R, Basherad R, Kertiles L, Boss MA, Seubert P. Arch. Neurol. 1998;55:938–945. doi: 10.1001/archneur.55.7.937. [DOI] [PubMed] [Google Scholar]
- 44.Motter R, Vigopelfrey C, Kholodenko D, Barbour R, Johnsonwood K, Galasko D, Chang L, Miller B, Clark C, Green R, Olson D, Southwick P, Wolfert R, Munroe B, Lieberburg I, Seubert P, Schenk D. Ann. Neurol. 1995;38:643–648. doi: 10.1002/ana.410380413. [DOI] [PubMed] [Google Scholar]
- 45.Shoji M, Matsubara E, Kanai M, Watanabe M, Nakamura T, Tomidokoro Y, Shizuka M, Wakabayashi K, Igeta Y, Ikeda Y, Mizushima K, Amari M, Ishiguro K, Kawarabayashi T, Harigaya Y, Okamoto K, Hirai S. J. Neurol. Sci. 1998;158:134–140. doi: 10.1016/s0022-510x(98)00122-1. [DOI] [PubMed] [Google Scholar]
- 46.Murphy RM. Biochim. Biophys. Acta. 2007;1768:1923–1934. doi: 10.1016/j.bbamem.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 47.Jensen M, Schroder J, Blomberg M, Engvall B, Pantel J, Ida N, Basun H, Wahlund LO, Werle E, Jauss M, Beyreuther K, Lannfelt L, Hartmann T. Ann. Neurol. 1999;45:504–511. doi: 10.1002/1531-8249(199904)45:4<504::aid-ana12>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 48.Mehta PD, Pirttila T, Mehta SP, Sersen EA, Aisen PS, Wisniewski HM. Arch. Neurol. 2000;57:100–105. doi: 10.1001/archneur.57.1.100. [DOI] [PubMed] [Google Scholar]
- 49.He L, Musick MD, Nicewarner SR, Salinas FG, Benkovic SJ, Natan MJ, Keating CD. J. Am. Chem. Soc. 2000;122:9071–9077. [Google Scholar]
- 50.Yao X, Li X, Toledo F, Zurita-Lopez C, Gutova M, Momand J, Zhou FM. Anal. Biochem. 2006;354:220–228. doi: 10.1016/j.ab.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 51.Selkoe DJ. Nature. 1999;399:A23–A31. doi: 10.1038/399a023. [DOI] [PubMed] [Google Scholar]
- 52.Sinha S, Lieberburg I. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11049–11053. doi: 10.1073/pnas.96.20.11049. [DOI] [PMC free article] [PubMed] [Google Scholar]