Abstract
DNA-intercalating molecules can impair DNA replication, DNA repair, and gene transcription. We previously demonstrated that XR5944, a DNA bis-intercalator, specifically blocks binding of estrogen receptor-α (ERα) to the consensus estrogen response element (ERE). The consensus ERE sequence is AGGTCAnnnTGACCT, where nnn is known as the tri-nucleotide spacer. Recent work has shown that the tri-nucleotide spacer can modulate ERα-ERE binding affinity and ligand-mediated transcriptional responses. To further understand the mechanism by which XR5944 inhibits ERα-ERE binding, we tested its ability to interact with consensus EREs with variable tri-nucleotide spacer sequences and with natural but non-consensus ERE sequences using one dimensional nuclear magnetic resonance (1D 1H NMR) titration studies. We found that the tri-nucleotide spacer sequence significantly modulates the binding of XR5944 to EREs. Of the sequences that were tested, EREs with CGG and AGG spacers showed the best binding specificity with XR5944, while those spaced with TTT demonstrated the least specific binding. The binding stoichiometry of XR5944 with EREs was 2:1, which can explain why the spacer influences the drug-DNA interaction; each XR5944 spans four nucleotides (including portions of the spacer) when intercalating with DNA. To validate our NMR results, we conducted functional studies using reporter constructs containing consensus EREs with tri-nucleotide spacers CGG, CTG, and TTT. Results of reporter assays in MCF-7 cells indicated that XR5944 was significantly more potent in inhibiting the activity of CGG- than TTT-spaced EREs, consistent with our NMR results. Taken together, these findings predict that the anti-estrogenic effects of XR5944 will depend not only on ERE half-site composition but also on the tri-nucleotide spacer sequence of EREs located in the promoters of estrogen-responsive genes.
Keywords: Estrogen, XR5944, estrogen response element, tri-nucleotide spacer, nuclear magnetic resonance
1. Introduction
Estrogens are steroid hormones that play critical roles in the initiation, development, and metastasis of breast and uterine cancers [1]. The estrogen (E2) response in breast cancer cells is predominantly mediated by the estrogen receptor-α (ERα), a ligand-activated transcription factor. ERα regulates transcription of target genes through direct binding to its cognate recognition sites, known as estrogen response elements (EREs), or by modulating the activity of other DNA-bound transcription factors at alternative DNA sequences [2]. Endocrine therapy, often effective for ERα-positive breast tumors, impairs the hormone-receptor complex or inhibits E2 production. Unfortunately, a significant fraction (~20–50%) of ERα-positive breast tumors fails to respond [3], or eventually develops resistance, to antiestrogen treatments [4]. Hence, there remains an urgent need for new and effective agents that overcome the resistance to existing endocrine therapies.
We previously showed that XR5944, a DNA bis-intercalator with potent anticancer activity, is capable of inhibiting ERα-mediated transcriptional responses via its ability to block the binding of ERα to the ERE sequence [5]. This blocking activity was predicted to have a certain degree of specificity for EREs based on the structural observation that the preferred DNA-intercalating sequence of XR5944 contains 5′-(TpG):(CpA) sites [6]. Such sites are twice represented in a consensus ERE (AGGTCAnnnTGAGGT). This prediction was supported by our determination that XR5944 did not inhibit transactivation of the Sp1 consensus binding site which contains multiple guanines and cytosines (5′-GGGGCGGGGC-3′) but no 5′-TG-3′ motifs [5].
The consensus ERE is an inverted repeat comprised of two ERE half-sites separated by three bases: AGGTCAnnnTGACCT where nnn is known as the tri-nucleotide spacer [7]. Historically, this spacer was considered to be irrelevant to ERα-DNA binding and receptor-mediated transcriptional response. However, recently we demonstrated that the sequence of the tri-nucleotide spacer is non-random at receptor-bound genomic loci, influences ERα-DNA-binding affinity, and modulates transactivation potential of the receptor-ligand-DNA complex [8, 9]. We found that binding of ERα to the canonical ERE is modulated by the tri-nucleotide spacer sequence such that binding affinity and the estrogen-stimulated transcriptional response is favored by spacer sequences of CTG>GCC>TTT [9]. These studies also demonstrated that the spacer sequence modulates the sensitivity of EREs to repression engendered by the receptor antagonist hydroxytamoxifen. Here, we tested the possibility that the tri-nucleotide spacer plays a similar role in determining the binding characteristics of the XR5944 to consensus and non-consensus ERE sequences. Further, we tested whether the spacer sequence could modulate the inhibitory effects of XR5944 on ERE-mediated gene transcription.
Using 1D 1H NMR titration [6] to study XR5944 interactions with canonical EREs spaced by diverse tri-nucleotide spacers, we show that the spacer sequence affects the binding specificity of XR5944 with the ERE. XR5944 binding specificity correlated with the efficacy of XR5944 to inhibit ERE-mediated transactivation in response to liganded ERα. Together, these findings demonstrate that ERE spacer sequences modulate XR5944-DNA interactions and ERα-mediated transcriptional responses at consensus EREs. These findings have important implications for the prediction of XR5944-responsive EREs in the human genome and may form the basis for the development of promoter-specific DNA intercalators to target subgroups of naturally occurring EREs with common spacer sequences.
2. Materials and methods
2.1 Sample preparation
Sense and complimentary ERE oligonucleotides were synthesized (1 μmol scale) using b-cyanoethylphosphoramidite solid-phase chemistry on an Expedite™ 8909 Nucleic Acid Synthesizer (Applied Biosystem, Inc.) in DMT-on mode. Samples were purified using MicroPure II Columns from BioSearch Technologies (Novato, CA) as previously described [6]. DNA concentrations were determined by UV absorbance at 260 nm. XR5944 was provided by Xenova Ltd. (Slough, UK). 20 mM XR5944 stock solutions were prepared by dissolving the drug in deionized water.
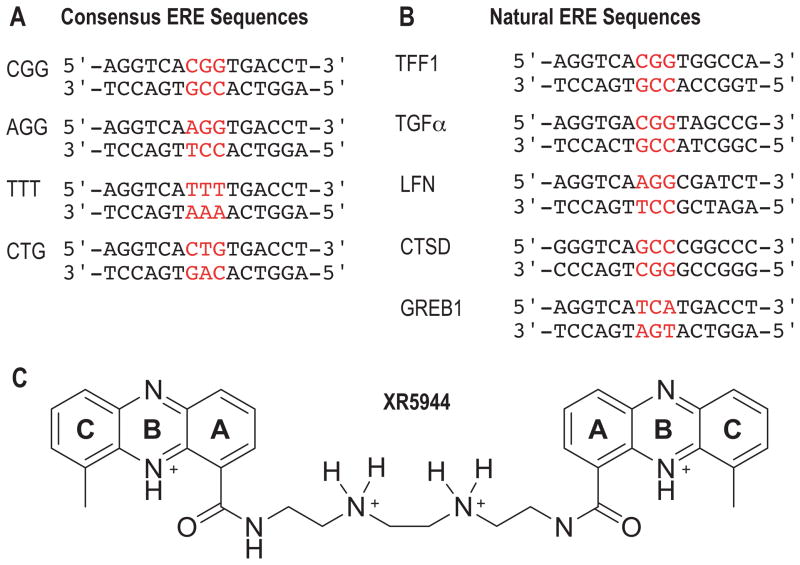
Each ERE duplex DNA is a 15-mer hetero-duplex containing two 6-nt ERE half-sites with a 3-nt spacer (Fig. 1). The DNA samples were prepared by dissolving single-stranded DNA oligonucleotides in 50 mM sodium phosphate buffer at pH 7 in D2O/H2O (10%/90%). Each ERE duplex was prepared by titrating one strand into the solution of another strand. The precise ratio of the two strands was monitored by the imino signals using 1D 1H NMR spectra. At the 1:1 ratio, the two strands form a clean DNA duplex. The final concentrations of DNA oligonucleotides were around 0.5–3 mM. The ERE-XR5944 complexes (with ratio 1:0.5, 1:1, 1:2) were prepared by adding an appropriate amount of drug to DNA samples.
Figure 1.
The ERE sequences tested in this study for interaction with XR5944. (A) Consensus ERE sequences with variable tri-nucleotide spacer sequences and (B) natural ERE sequences. (C) The chemical structure of XR5944 in protonated form.
2.2. 1D 1H NMR Experiments
1D 1H NMR experiments were carried out on a Bruker Avance 600 MHz spectrometer at 5ºC utilizing 50 mM sodium phosphate buffer at pH 7. The NMR experiments for samples in water were performed with WATERGATE or Jump-Return water suppression techniques. High-resolution 1H NMR spectra were acquired with the following acquisition parameters: time domain 32 K; 90º pulse width 11.0 μs; spectral width 16 ppm; relaxation delay 1.0 s, acquisition time 3.2 s. 128 scans were accumulated. Fourth-order polynomial functions were applied for the base-line correction.
2.3. Luciferase reporter assays
Luciferase reporter assays were performed using the Luciferase Assay System (Promega) as previously described [10]. Single-copy ERE regulatory elements were cloned into pGL2-Promoter (Promega) and all constructs were sequence-verified prior to use in reporter assays. The EREs used in this study consisted of the consensus sequence with tri-nucleotide spacers CGG, CTG, and TTT (Fig. 1). Reporter constructs were transfected into ERα-expressing MCF-7 breast cancer cells using the TransIT-LT1 transfection reagent (Mirus) per the manufacturer’s instructions. Cells were changed to estrogen-depleted, phenol-free media consisting of MEM alpha (Gibco) with 10% charcoal/dextran-stripped calf serum, insulin (4 μg/ml, Sigma), penicillin G, streptomycin, and L-glutamine (all Gibco), for 72 hours prior to treatments. Stock concentrations of 2 mM XR5944 were prepared in DMSO and diluted into the assay medium to yield final XR5944 concentrations of ≤100 nM. Solvent controls contained DMSO at 0.005% corresponding to the highest concentration of XR5944 tested and had no effect on the assay results. Cells were transfected with 0.6 μg of plasmid DNA per well of a 12-well plate and were incubated for 4 hours. Cells were then treated with either E2 or XR5944 combined with E2 for 16 hours. After this time period, the cells were collected in 200 μl of Reporter Lysis buffer (Promega) per well. Cotransfection with a β-galactosidase-expressing plasmid (Promega) enabled normalization of transfection efficiency across samples using a β-galactosidase assay kit (Promega) according to the manufacturer’s instructions.
2.4. Statistics
Transfection experiments were performed a minimum of three times, in triplicate, for each reporter construct. SPSS software was used for data analysis, and the data were expressed as mean ± SEM. Experimental results using reporters with different spacer sequences were compared at the indicated concentrations of XR5944 by t test (two tailed) where P < 0.05 was considered statistically significant.
3. Results and Discussion
3.1. The tri-nucleotide spacer sequence between ERE half-sites modulates XR5944 binding to the consensus EREs
We conducted 1D 1H NMR titration studies of the interaction between XR5944 (for molecular structure see Fig. 1C) and consensus ERE (5′-AGGTCA-nnn-TGACCT) containing unique tri-nucleotide spacer sequences. Each ERE duplex was prepared by titrating one strand into the solution of another strand to a final ratio of 1:1, monitored by the imino signals using 1D 1H NMR, until a clean DNA duplex was formed. The tri-nucleotide spacers tested included CGG, AGG, CTG, and TTT (Fig. 1A). The imino protons of guanines (H1) and thymines (H3) of Watson-Crick base pairs, i.e., GC/CG and TA/AT base pairs are located in regions of 12–13 and 13–14 ppm in 1H-NMR spectra, respectively, which are well separated from other protons (bottom spectra, Figs. 2A–D). For each 15-mer consensus ERE sequence, imino proton peaks were detectable for all base pairs except two terminal base pairs (Fig 2 and Fig S1). The imino protons of the two terminal base pairs are not detectable due to their rapid exchange with water because of the end-fraying effect [6, 11]. For example, for the consensus ERE DNA duplex with the CGG spacer (Fig 1A-top), four thymine imino protons were observed for the four non-terminal T:A/A:T base pairs (13.2–13.6 ppm) and nine guanine imino protons were observed for nine G:C/C:G base pairs (12–12.8 ppm)(Fig S1A). Compared to the ERE sequence with the CGG spacer, the consensus ERE sequence with the AGG spacer has an additional A:T base pair in place of a C:G base pair (Fig 1A). Therefore, five thymine imino protons were observed (13.2–13.6 ppm) and eight guanine imino protons were observed (12–12.8 ppm) for the consensus ERE DNA duplex with the AGG spacer (Fig S1B-top).
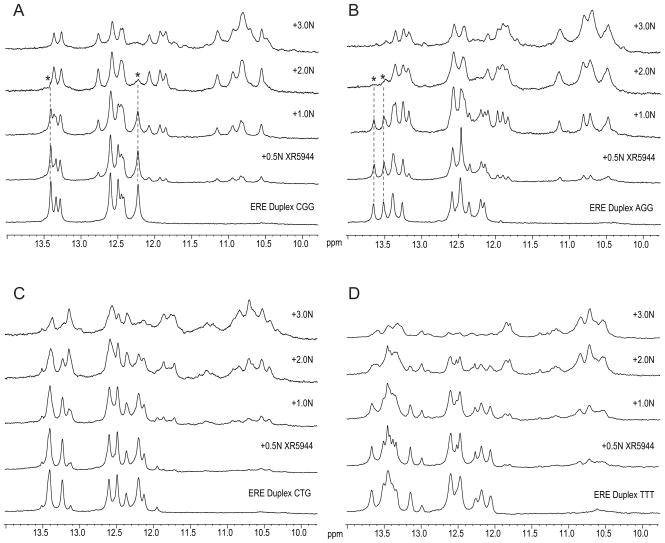
Figure 2.
Binding of XR5944 with consensus ERE sequences containing different tri-nucleotide spacers by NMR. The tri-nucleotide spacers tested were CGG (A), AGG (B), CTG (C), and TTT (D) titrated with XR5944 at drug equivalence from 0 (bottom) to 3 (top). Upon addition of increasing concentrations of XR5944, the imino proton peaks of the free ERE DNAs started to vanish. Concomitantly, a new set of imino proton peaks representing the ERE-drug complexes emerged in a dose dependent fashion peaking at a drug equivalence of 2. While some imino proton peaks from the drug-DNA complex were located in the same region as those of the free DNA (12–13.5 ppm), some of the emerging imino peaks from the drug-DNA complex were upfield-shifted, e.g., those observed at 10–11 ppm. For the DNA sequences to which XR5944 binds with high specificity, the imino protons of the free DNAs almost completely vanished at the drug equivalence of 2, as indicated by asterisks (*) and dashed lines (---) for isolated imino proton peaks of the free DNA with CGG- and AGG-spacers.
The NMR titration results indicated that XR5944 binds the different ERE sequences with variable specificity (Fig. 2). To this end, 1D 1H NMR titration experiments can be considered a useful method to monitor drug-DNA binding properties. The well-separated region of imino protons of a duplex DNA provides a direct and unambiguous detection of drug binding interactions [6]. In our 1D 1H NMR titration experiments, upon XR5944 binding, the imino peaks of the free ERE DNA vanished in a dose-dependent fashion while a new set of imino peaks reflecting the drug-DNA complex emerged (top four spectra of Fig 2A–D). Of those spacer sequences tested, the CGG spacer showed the greatest binding specificity. As seen in Fig. 2A, upon adding XR5944 (i.e., at 0.5 N to 3.0 N), a new set of imino proton peaks with sharp line-widths started to emerge, whereas the imino proton peaks of the free DNA started to vanish. The observation of two sets of imino peaks, one from the free DNA and another from the drug-complexed DNA, indicates that XR5944 binds the ERE in a slow-exchange binding mode on the NMR time scale. The slow-exchange binding mode suggests a drug binding to DNA with high affinity, while the occurrence of a single set of sharp NMR peaks from the drug-DNA complex indicates a specific binding site (i.e. preferred drug intercalation at specific base pair sites)[6]. The binding stoichiometry of XR5944 with the CGG spacer appeared to be 2:1, as no further qualitative change was observed in the 1D NMR spectra of XR5944-ERE complexes at the drug equivalence higher than 2 (Figs. 2A–D). While some imino proton peaks from the drug-DNA complex are located in the same region as those of the free DNA (12–13.5 ppm), some of the new emerging imino peaks from the drug-DNA complex were upfield-shifted, e.g., those observed at 10–11 ppm (Fig 2A). The upfield-shifting of the DNA imino protons is characteristic of an intercalating drug binding mode [6, 12]. At the drug equivalence of 2, the imino proton peaks from the free DNA almost completely vanished, leaving the new set of well-resolved imino proton peaks from the drug-DNA complex (Fig 2A). The total number of the imino proton peaks of the drug-DNA complex was 15 (second from the top spectrum, Fig. 2A and Fig S1A-bottom), indicating that the imino protons from the two terminal base pairs were also observed in the XR5944-DNA complex. This observation suggests that the binding of XR5944 rescued the end-fraying effect by stabilizing the ERE DNA duplex.
The ERE DNA duplex with the AGG spacer also showed a similarly specific binding with XR5944 (Fig. 2B). It’s spectrum at a 2:1 (drug:DNA) complex with XR5944 also showed 15 imino proton peaks (Fig. S1B-bottom). However, the ERE DNA duplexes with spacers CTG and TTT were found to be less specific as to their binding with XR5944 (Figs. 2C, D). At the drug equivalence of 2 for these two spacers, the imino proton peaks from the free DNA did not completely vanish, and a larger number of imino proton peaks with broader line-widths were observed, indicating more promiscuous drug binding to multiple binding sites for these EREs. For the CTG-spaced ERE, as for those with CGG and AGG spacers, a major set of imino proton peaks from the drug-ERE complex was seen at a higher drug equivalence (3N) when the drug was in excess. These data indicate that there still exits preferred drug binding sites for this sequence although the drug binding specificity appears to be lower than those with the CGG or AGG spacers. By contrast, a discernable set of complex imino protons was not evident for the TTT-spaced ERE even at the drug equivalence of 3. Thus, the binding of XR5944 with the TTT-spaced ERE appeared to be least specific among those sequences that were tested. While high specificity of binding requires high affinity of XR5944 to discrete sites, the apparent binding affinity of the compound to a DNA sequence may be influenced by multiple non-specific interactions. As such, it is difficult to compare the binding affinity of XR5944 to different ERE sequences based on the NMR titration data. As XR5944 has been shown to interact in the bis-intercalating mode that spans four nucleotides [6], our results indicate that one of the two functional sites of XR5944 is likely to intercalate in the tri-nucleotide spacer region of ERE sequences. It is clear from the present findings that variations in this spacer region can affect the specificity of the XR5944-ERE interaction.
3.2. Binding of XR5944 with natural promoters
We used 1H NMR titration experiments to investigate the interactions of XR5944 with various naturally occurring and bona fide ERE sequences, including those for the estrogen-responsive target genes trefoil factor 1 (TFF1, previously designated PS2), growth regulation by estrogen in breast cancer 1 (GREB1), cathepsin D (CTSD), lactoferrin (LFN), and transforming growth factor-α (TGF-α) (Fig. 1B). These sequences were chosen because they are ERα-bound in vivo and the target genes are estrogen-responsive [13, 14]. The 1H NMR spectra of the free DNA duplexes containing those natural ERE sequences are shown at the bottom of titration profiles, while the 1H NMR spectra of drug-DNA complexes with drug equivalences of 0.5 to 3 are shown at the top (Fig. 3A–E). As observed for the consensus ERE duplexes with variable tri-nucleotide spacer sequences, XR5944 binding resulted in the loss of imino peaks measured for the free DNA and the generation of a new set of imino peaks produced by the drug-DNA complex (top four spectra of Fig 3A–E). XR5944 bound these naturally occurring EREs in a slow-exchange binding mode, and the binding stoichiometry of XR5944 with natural EREs was 2:1 (drug:DNA) (Fig. 3A–E).
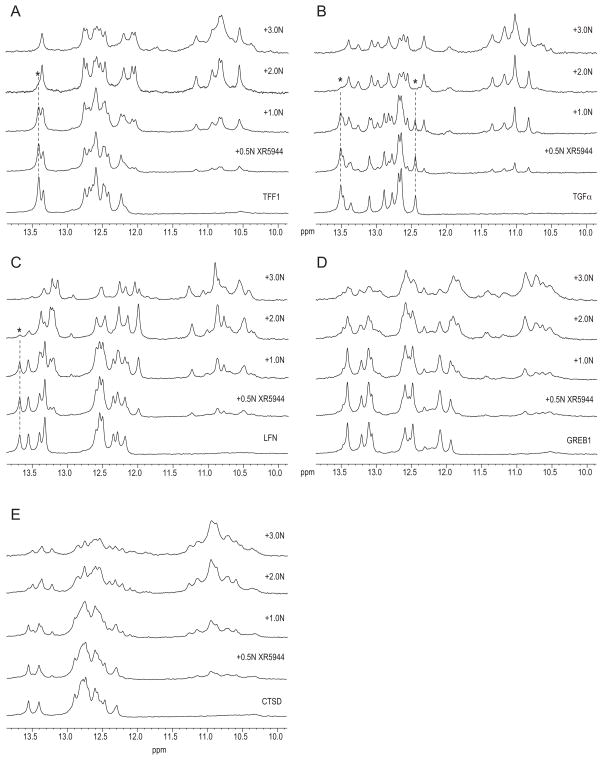
Figure 3.
Binding of XR5944 with some natural EREs by NMR. The EREs tested were those located in the promoters of TFF1 (A), TGF-α(B), LFN (C), GREB1 (D), and CTSD (E) titrated with XR5944 at drug equivalence from 0 (bottom) to 3 (top). Upon addition of increasing concentrations of XR5944, the imino proton peaks of the free EREs started to vanish while a new set of imino proton peaks reflecting the drug-DNA complexes emerged. Several isolated imino proton peaks observed with the free DNAs are indicated by asterisks (*) and dashed lines (---).
Our NMR results indicated that natural ERE sequences bind to XR5944 with different specificities as evident by differences in the 1H NMR spectra of their respective drug-DNA complexes, such as their unique spectral resolution and linewidths (Fig. 3A–E). Of the sequences tested, NMR results showed that XR5944 binds with the highest specificity to the ERE sequence of TFF1 (Fig 3A) and TGF-α(Fig 3B), followed by that of the LFN gene ERE (Fig 3C). The 1H NMR spectrum with integration of the free DNA duplex of the TFF1 ERE sequence is shown in Fig S1C-top. As expected, three thymine imino protons were observed for the three non-terminal T:A/A:T base pairs (13.2–13.6 ppm) and ten guanine imino protons were observed for ten G:C/C:G base pairs (12–12.8 ppm). After titration of the TIFF1 ERE with XR5944, a new set of imino proton peaks with sharp linewidths emerged, whereas the imino proton peaks of the free DNA started to vanish as indicated by the peak followed with a dotted line and asterisk (*) in Fig 3A. At the drug equivalence of 2, the imino proton peaks from the free DNA were almost completely lost, leaving a new set of well-resolved imino proton peaks from the drug-DNA complex (Fig 3A). As shown in Fig S1C-bottom, the total number of the imino proton peaks of the 2:1 drug-DNA complex for the TFF1 ERE sequence was 15, as expected.
Interestingly, the EREs for TGF-αand TFF1, which demonstrated the best binding specificity for XR5944, both contain a CGG spacer, while the ERE for LFN, which also demonstrated high drug binding specificity, contains an AGG-spaced ERE. Both of these spacer sequences were shown to favor interaction with XR5944 in the consensus ERE duplexes described above. The ERE showing the least specific interaction with XR5944 was that for CTSD (Fig 3E). The shifted imino peaks from the drug complex of the CTSD ERE sequence showed broad and less-resolved linewidths, indicating less specific drug binding. It should be noted that although both the TGF-αand TFF1 EREs, and the consensus DNA ERE with CGG spacer all contain the identical tri-nucleotide spacer sequence, there are clear differences in their respective NMR spectra when complexed with drug (Figs 2A, 3A, and 3B). This observation indicates that each molecule of XR5944 intercalates at both half-site and spacer residues and that differences in either the half-site or spacer sequences of an ERE can modify ERE interaction with XR5944.
3.3. Inhibition by XR5944 of ERE reporter gene activity
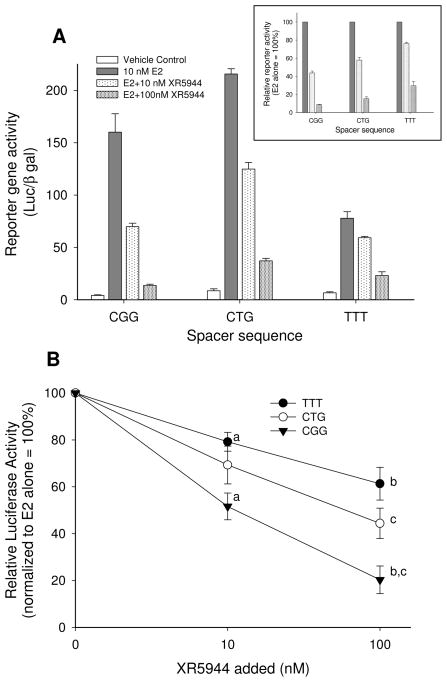
Having shown that the tri-nucleotide spacer sequence of the consensus ERE modulates the characteristics of its binding with XR5944, we sought to determine whether the spacer also affects the ability of XR5944 to inhibit transactivation of the consensus ERE. To test this, we cloned single-copy consensus EREs with different spacers into a luciferase reporter vector (pGL2-promoter). ERα-expressing MCF-7 cells were transfected with each variant ERE-reporter construct and data were normalized for transfection efficiency using β-galactosidase-expressing plasmid. For these studies, we utilized reporter constructs containing ERE’s with tri-nucleotide spacers CGG, CTG, and TTT (ERE-CGG, ERE-CTG and ERE-TTT, respectively) as representative of strong, intermediate, and weak ERE-XR5944 binders in our 1D 1H NMR titration studies (above). Results of luciferase reporter assays in MCF-7 cells indicated that XR5944 was significantly more potent in inhibiting E2-stimulated activity of reporters containing ERE-CGG than those containing ERE-TTT. Thus, 10 nM XR5944 inhibited reporter activity of ERE-CGG and ERE-TTT by 50% and 20%, respectively, while 100 nM XR5944 inhibited their activity by 80% and 40%, respectively (Fig. 4B). The tri-nucleotide spacer-dependent order of potency of inhibition by XR5944 was CGG>CTG>TTT. These functional consequences of the spacer sequence are consistent with our NMR results indicating substantially greater specificity of binding of XR5944 with EREs spaced by CGG versus TTT, with CTG-spaced EREs showing intermediate binding characteristics to XR5944.
Figure 4.
The tri-nucleotide spacer sequence modulates ERE sensitivity to repression by XR5944. Luciferase reporter assays of single copy consensus EREs with variable tri-nucleotide spacer sequences were performed in MCF-7 cells. Basal and E2-stimulated luciferase values were normalized to co-transfected β-galactosidase-expressing plasmid and expressed as mean ± SEM of the indicated biological replicates. (A) Representative experiment performed in triplicate showing that E2-stimulated luciferase activities were highest for the ERE with the spacer sequence CTG, followed by spacer CGG and then spacer TTT (CTG>CGG>TTT). The inset in (A) shows relative reporter activity of this experiment where each reporter is normalized to its corresponding E2 treatment alone in order to emphasize differential sensitivity of the reporters to inhibition by XR5944. (B) Results of three independent experiments, each performed in triplicate showing comparative sensitivity of each ERE sequence to XR5944 (shown relative to E2-stimulated = 100%). At the two doses of XR5944, all EREs were significantly inhibited by the compound. At the lower XR5944 concentration (10 nM), CGG-spaced EREs were significantly more repressed than TTT-spaced EREs. At the higher XR5944 concentration (100 nM), the order of sensitivity to suppression was CGG>CTG>TTT. Values represented by symbols in (B) that are labeled with identical letters showed statistically significant differences.
Our previous work showed that the tri-nucleotide spacer sequence modulates the transcriptional response to estrogen when evaluated in reporter assays of single copy consensus EREs transfected into MCF-7 cells [9]. Furthermore, our data indicated that the tri-nucleotide spacer can influence reporter sensitivity to the selective estrogen receptor modulator hydroxytamoxifen (OHT), which antagonizes many receptor functions in MCF-7 cells [9]. In those studies, there was not a strict concordance between the spacer-dependent order of potency to stimulate E2 transcriptional responses (CTG>GCC>TTT) with that showing inhibition of E2 responses by OHT (CTG>TTT>GCC). We observed a similar lack of concordance between E2-mediated transactivation potential and inhibition by XR5944 using variably-spaced ERE sequences (Fig. 4A). Specifically, E2-stimulated ERE-driven reporter activity was highest for the sequence containing CTG as the tri-nucleotide spacer, intermediate for that containing CGG, and lowest for that containing TTT (CTG>CGG>TTT; fig. 4A). This order of potency did not strictly correspond to the spacer-dependent sensitivity of reporters to inhibition by XR5944 (CGG>CTG>TTT; Fig. 4A inset and 4B). These data confirm that the tri-nucleotide spacer sequence modulates ERE responses to E2 as previously reported [9] and demonstrate that the spacer sequence also modulates transcriptional repression mediated by XR5944. Further, the sensitivity of consensus EREs with variable tri-nucleotide spacer sequences to these distinct activities (transactivation in response to E2, transrepression in response to OHT or XR5944) is not always congruous. These findings predict additional specificity to the anti-estrogenic effects of XR5944 that will depend not only on the ERE half-site composition [15] but also upon the tri-nucleotide spacer sequences of EREs in the promoters/enhancers of diverse endogenous target genes.
We were also interested to test whether the interaction of XR5944 with natural EREs, as assessed by 1D 1H NMR, could predict the ability of XR5944 to inhibit E2-stimulated ERE transactivation in a reporter assay. To address this question, we developed reporter constructs containing single copies of the five natural ERE sequences utilized in the NMR study. Unfortunately, with the exception of the construct containing the GREB1 ERE, which is a consensus ERE spaced by TCA (Fig. 1), these single-copy but imperfect (i.e. non-consensus) ERE reporters did not show robust E2-driven transactivation in reporter assays. In this regard, EREs with reduced transactivation potential (e.g. non-consensus EREs) are often assayed as tandem repeats (i.e. multiple copies of the sequence) in order to increase the magnitude of their transactivation responses in reporter gene assays [16]. Although reporters containing multiple copies of our natural ERE sequences were dependably stimulated in our reporter assays (data not shown), we concluded that results of XR5944 inhibition experiments using multiple copies of ERE sequences would not provide meaningful comparisons to the 1D 1H NMR results since the latter assessed the interaction of XR5944 with single (15-mer) ERE motifs.
3.4. Conclusions
A major mechanism of de novo or acquired resistance to antiestrogen therapy involves estrogen-independent receptor activation that still requires ER-ERE binding to control the expression of estrogen-regulated target genes [17, 18]. An ERE intercalator that binds and occupies the ER binding site and inhibits receptor-DNA interactions might be a useful therapeutic agent and may overcome resistance to existing endocrine therapies which is observed in a considerable percentage of ER-positive patients [19]. A major challenge in our work to develop ERα inhibitors that utilize a DNA-binding mechanism of action has been to understand and delineate factors that influence the specificity of drug-ERE interaction. In the present work, we have determined that the binding stoichiometry of XR5944 with ERE duplex DNAs is 2:1, and that the specificity and affinity of the XR5944-ERE complex is influenced by the nucleotide sequence of the half-sites and the sequence of the tri-nucleotide spacer. These data suggest that the estrogen response of endogenous genes may be differentially subject to regulation by XR5944 depending not only on the ERE half-site composition but also upon the tri-nucleotide spacer sequences of their respective estrogen responsive elements. This finding may be of clinical significance for future anti-estrogenic applications of XR5944 and similar compounds since the tri-nucleotide spacer has been shown to be non-random at ERα-bound genomic loci while only a minority of predicted ERE sequences is operative in any given cell type [8, 9, 13]. In addition, although the preferred intercalation sites of XR5944 [(5′-TpG):(CpA)] can be found in the regulatory elements of a number of transcription factors (e.g. AP-1)[6], the spacing, stoichiometry, and promoter contexts of potential target sites are additional variables that may dictate sensitivity to XR5944. As such, the involvement of the tri-nucleotide spacer sequence in the functional effects of XR5944 reveals additional variables predictive of its specificity of action and may prove useful in identifying and developing new derivatives that demonstrate still greater specificity of action for EREs compared with other cis-regulatory motifs.
Supplementary Material
Spectral integration of 1D NMR spectra of some ERE sequences. Shown are the spectra of EREs containing the consensus sequence with the CGG spacer (A-top) and the AGG spacer (B-top), the natural ERE sequence in the promoter of the TFF1 gene (C-top), and their complexes with XR5944 at the drug equivalence of 2 [(A-bottom) (C-bottom)], respectively. The number of protons for each peak is shown above the spectra based on the integration values shown underneath the NMR spectra.
Acknowledgments
The authors are indebted to Kelly Shen for her outstanding technical support. This work was supported by the NCI/NIH through Grant RO1-CA129424 (to NS) and the Research Scientist Development Program/NIH K12-HD000849 (to CBK).
Abbreviations
- E2
17-β-estradiol/estrogen
- ERα
estrogen receptor-α
- ERE
estrogen response element
- 1D 1H NMR
one dimensional nuclear magnetic resonance
- TFF1
trefoil factor 1
- GREB1
growth regulation by estrogen in breast cancer 1
- CTSD
cathepsin D
- LFN
lactoferrin
- TGF-α
transforming growth factor-α
- OHT
hydroxytamoxifen
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 2.Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19:833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- 3.Kurebayashi J. Resistance to endocrine therapy in breast cancer. Cancer Chemother Pharmacol. 2005;56(Suppl 1):39–46. doi: 10.1007/s00280-005-0099-z. [DOI] [PubMed] [Google Scholar]
- 4.Schiff R, Massarweh S, Shou J, Osborne CK. Breast cancer endocrine resistance: how growth factor signaling and estrogen receptor coregulators modulate response. Clin Cancer Res. 2003;9:447S–454S. [PubMed] [Google Scholar]
- 5.Punchihewa C, De Alba A, Sidell N, Yang D. XR5944: A potent inhibitor of estrogen receptors. Mol Cancer Ther. 2007;6:213–219. doi: 10.1158/1535-7163.MCT-06-0392. [DOI] [PubMed] [Google Scholar]
- 6.Dai J, Punchihewa C, Mistry P, Ooi AT, Yang D. Novel DNA bis-intercalation by MLN944, a potent clinical bisphenazine anticancer drug. J Biol Chem. 2004;279:46096–46103. doi: 10.1074/jbc.M404053200. [DOI] [PubMed] [Google Scholar]
- 7.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason CE, Shu FJ, Wang C, Session RM, Kallen RG, Sidell N, Yu T, Liu MH, Cheung E, Kallen CB. Location analysis for the estrogen receptor-alpha reveals binding to diverse ERE sequences and widespread binding within repetitive DNA elements. Nucleic Acids Res. 2010;38:2355–2368. doi: 10.1093/nar/gkp1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shu FJ, Sidell N, Yang D, Kallen CB. The tri-nucleotide spacer sequence between estrogen response element half-sites is conserved and modulates ERalpha-mediated transcriptional responses. J Steroid Biochem Mol Biol. 2010;120:172–179. doi: 10.1016/j.jsbmb.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C, Yu J, Kallen CB. Two estrogen response element sequences near the PCNA gene are not responsible for its estrogen-enhanced expression in MCF7 cells. PLoS One. 2008;3:e3523. doi: 10.1371/journal.pone.0003523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang D, van Boom SS, Reedijk J, van Boom JH, Wang AH. Structure and isomerization of an intrastrand cisplatin-cross-linked octamer DNA duplex by NMR analysis. Biochemistry. 1995;34:12912–12920. doi: 10.1021/bi00039a054. [DOI] [PubMed] [Google Scholar]
- 12.Yang D, Wang AH. Structure by NMR of antitumor drugs aclacinomycin A and B complexed to d(CGTACG) Biochemistry. 1994;33:6595–6604. doi: 10.1021/bi00187a029. [DOI] [PubMed] [Google Scholar]
- 13.Hua S, Kallen CB, Dhar R, Baquero MT, Mason CE, Russell BA, Shah PK, Liu J, Khramtsov A, Tretiakova MS, Krausz TN, Olopade OI, Rimm DL, White KP. Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol Syst Biol. 2008;4:188. doi: 10.1038/msb.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 15.Krieg AJ, Krieg SA, Ahn BS, Shapiro DJ. Interplay between estrogen response element sequence and ligands controls in vivo binding of estrogen receptor to regulated genes. J Biol Chem. 2004;279:5025–5034. doi: 10.1074/jbc.M307076200. [DOI] [PubMed] [Google Scholar]
- 16.Tyulmenkov VV, Jernigan SC, Klinge CM. Comparison of transcriptional synergy of estrogen receptors alpha and beta from multiple tandem estrogen response elements. Mol Cell Endocrinol. 2000;165:151–161. doi: 10.1016/s0303-7207(00)00250-1. [DOI] [PubMed] [Google Scholar]
- 17.Auchus RJ, Fuqua SA. Clinical syndromes of hormone receptor mutations: hormone resistance and independence. Semin Cell Biol. 1994;5:127–136. doi: 10.1006/scel.1994.1016. [DOI] [PubMed] [Google Scholar]
- 18.Michalides R, Griekspoor A, Balkenende A, Verwoerd D, Janssen L, Jalink K, Floore A, Velds A, van’t Veer L, Neefjes J. Tamoxifen resistance by a conformational arrest of the estrogen receptor alpha after PKA activation in breast cancer. Cancer Cell. 2004;5:597–605. doi: 10.1016/j.ccr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Jordan VC. How is tamoxifen’s action subverted? J Natl Cancer Inst. 2000;92:92–94. doi: 10.1093/jnci/92.2.92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spectral integration of 1D NMR spectra of some ERE sequences. Shown are the spectra of EREs containing the consensus sequence with the CGG spacer (A-top) and the AGG spacer (B-top), the natural ERE sequence in the promoter of the TFF1 gene (C-top), and their complexes with XR5944 at the drug equivalence of 2 [(A-bottom) (C-bottom)], respectively. The number of protons for each peak is shown above the spectra based on the integration values shown underneath the NMR spectra.