Abstract
The molecular complexity of biological tissue and the spatial and temporal variation in the biological processes involved in human disease requires new technologies and new approaches in order to provide insight into disease processes. Imaging mass spectrometry is an effective tool that provides molecular images of tissues in the molecular discovery process. The analysis of human tissue presents special challenges and limitations because the heterogeneity among human tissues and diseases is much greater than that observed in animal models, and discoveries made in animal tissues might not translate well to the human counterpart. In this article, we briefly review the challenges of imaging human tissue using mass spectrometry and suggest approaches to address these issues.
Introduction
Matrix-assisted laser desportion/ionization (MALDI) imaging mass spectrometry (IMS) is a powerful tool for the analysis of a variety of different endogenous and exogenous molecules directly in tissue sections. Applications of IMS include analysis of proteins [1–3], peptides [4–7], lipids [8–10], drugs and metabolites [11–13], and oligonucleotides [14]. Although image analysis of samples by MS dates back several decades and employs different ionization methods, it is beyond the scope of this article to review this broad field. Instead, this article will focus on analyzing peptides and proteins from human tissues using MALDI IMS. More detailed reviews of IMS technologies have recently been published [15,16].
Advantages of IMS
IMS provides many advantages over other classical protein analysis approaches, and can provide high-throughput capabilities. Often 15–20 tissue samples can be collected on the same target plate and imaged in a batch mode. Direct tissue analysis is quite complementary to 2D gel approaches in that it is it most sensitive for visualization of low molecular weight (MW <30 kDa) proteins. Most classical proteomic technologies require extraction and homogenization for sample preparation, which results in a loss of spatial relationships within tissue sections. Immunohistochemical approaches allow for preservation of spatial information, but have limitations inasmuch as the target protein must be known in advance in order to use the correct antibody for staining, and generally only one or two antibodies may be applied simultaneously. Conversely, IMS can be used in a discovery mode to determine proteins that are characteristic of a disease state; no target-specific reagents are needed, and hundreds of analytes can be detected simultaneously from a single tissue section.
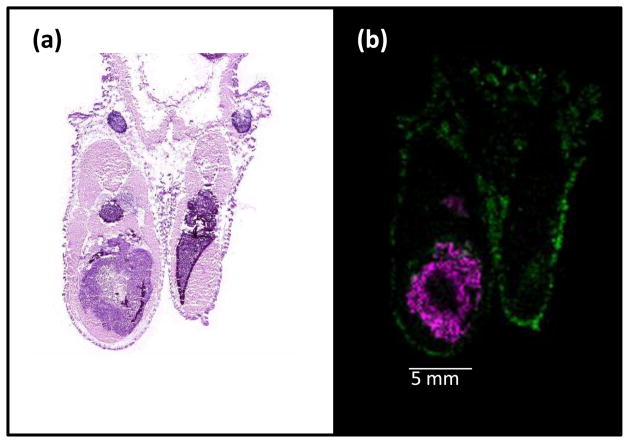
Antibodies can suffer from poor specificity, leading to a high level of background staining or cross-reaction with multiple analytes. Moreover, antibodies are seldom able to distinguish between multiple isoforms of the same protein; yet multiple isoforms of the same protein may play different roles in the disease process. For example, the neutrophil defensins are a set of three small proteins that differ in MW, but not in antibody recognition. In a recent study of human breast tissue by IMS, all three defensins were found to be differentially expressed, but at different significance levels in the tissue of patients who responded to treatment (Box 1) [17]. A second example involves a xenograph model of cancer where a human metastatic breast cancer cell line was implanted into the tibia of a mouse. Using MALDI IMS, expression of both human and mouse calcyclin, which can be distinguished by MW, are observed within the tissue section (R. Caprioli, unpublished) (Figure 1), whereas antibody detection methods are unable to differentiate between the two forms of the protein.
Box 1. IMS in the clinic.
Imaging mass spectrometry is currently used in clinical diagnostic and prognostic applications. We have recently shown the application of IMS to predict responsiveness to neoadjuvant taxane therapy and radiation in breast cancer [17]; diagnosis of gastrointestinal cancer using pinch biopsies [35]; and determination of molecular tumor margins in clear cell renal cell carcinoma [40].
Histology-directed proteomic analysis of 19 human breast cancer biopsies (13 non-responders and 6 pathological complete responders) has resulted in the discovery of a set of 3 proteins (the neutrophil defensins) that correlate with therapeutic responsiveness, and 4 other proteins that correlate with non-responsiveness [17]. These data in combination with gene array data could be used to predict treatment outcome, and furthermore could avoid the use of agents that would not be effective–information that is difficult to obtain from histological evaluation alone. Histology-directed analysis has also been used to identify proteomic markers in tumor and normal endothelial tissue from endoscopic biopsy specimens that could potentially be used to aid in the diagnosis of gastric cancer [35]. Proteomic profiles were able to distinguish between control and disease and between early and late stage cancers. The profiles were also able to predict treatment outcome.
Local recurrence after tumor resection is a major problem in cancer treatment. In an attempt to determine the underlying cause of recurrence, we examined 34 renal cell carcinoma samples with a large amount of adjacent normal tissue attached [40]. IMS of the tumor margin region indicated that several of the tumor-specific proteins exhibit an expression gradient outside of the histologically defined margins. In some cases, molecular distributions similar to the tumor were observed up to 1.5 cm outside of the tumor. These data indicate that tissue distant from the tumor margin may, in some cases, already be compromised. Molecular margins determined by IMS might be an effective way to determine successful tumor excision.
Figure 1.
Mass spectrometry can distinguish the protein calcyclin from different species. (a) Hematoxylin and eosin (H&E)-stained section of a metastatic human breast cancer cell line growing in the hind limb of a mouse. (b) In this false-colored MS image, pink is human calcyclin (m/z 10090), produced by the human tumor cells, and green is mouse calcyclin (m/z 9960), produced by mouse tissue.
Instrumentation
MALDI IMS is typically carried out on an instrument with a TOF mass analyzer that allows detection of high-MW analytes (>200 kDa) once ionized [18], although these types of measurements are not routine. For protein analyses (typically 2–80 kDa), the instrument is operated in linear mode, optimal for higher mass resolution and increased sensitivity for these species. Peptides are typically analyzed in reflection mode on a tandem (MS/MS) instrument; this allows for greater resolution and mass accuracy as well as fragmentation of an ion-of-interest, ultimately leading to identification of the parent protein. Other low-MW species, such as lipids, drugs and metabolites, are also amenable to analysis and identification through imaging MS/MS platforms.
Sample preparation
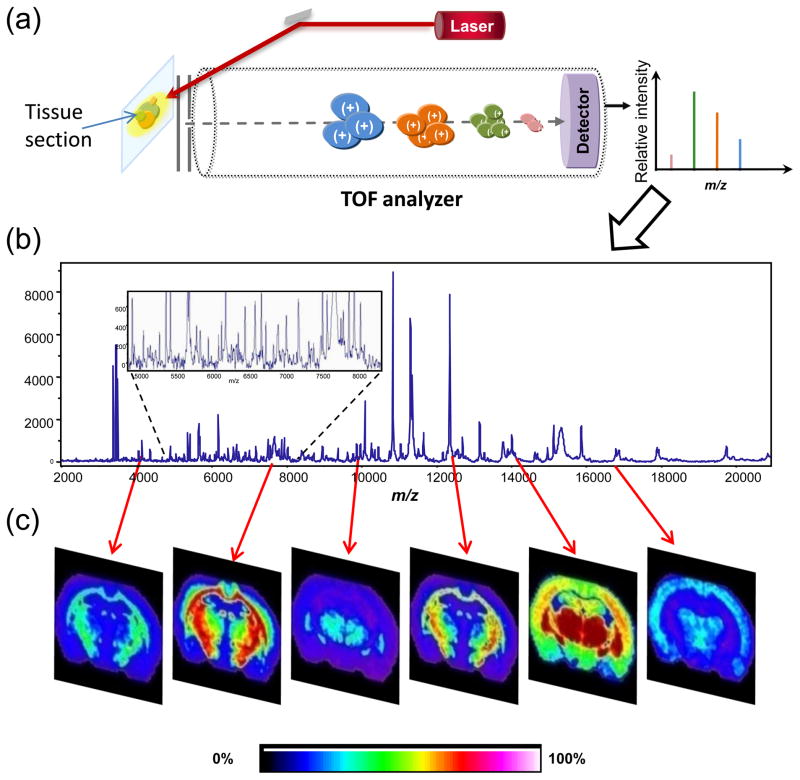
For optimal spectral quality for peptides and proteins, the tissue section is often fixed in graded alcohol to help remove species that unnecessarily complicate the spectrum (e.g. salts) or that cause ion suppression (e.g. lipids) [19]. To generate an image, matrix is applied over the entire tissue section, by robotic spotting [20], sublimation [21] or nebulization, and mass spectra are acquired across the entire tissue section in an ordered array. Using this method, spatial resolution typically ranges from 10–100 μm. With currently available lasers that have repetition rates of >1 kHz, images can be generated in minutes to hours, depending on the sample size and spatial resolution. Each individual spot has an associated mass spectrum (Figure 2a) analogous to a pixel in a digital image. One can generate an average spectrum from the entire tissue section to aid in the generation of MS images (Figure 2b). Each peak from the average spectrum can be displayed as a function of location and relative intensity across the tissue section. In this way, hundreds of analytes can be visualized from a single raster of a tissue section (Figure 2c). When full images are not required, but rather data are needed from discrete targeted areas within the tissue, a histology-directed profiling approach can be employed [22]. This approach is conducive to high-throughput imaging analysis of many samples as well as statistical analysis for biomarker discovery from areas of the tissue determined to be of interest upon microscopic (i.e. histological) examination. Under carefully controlled experimental conditions, the reproducibility of mass spectra between samples can be quite high, with relative standard errors ranging from 4.7–9.6% [13]. For a detailed explanation of the imaging process, the reader is referred to several review articles [16,23,24].
Figure 2.
Schematic of the MALDI IMS process. (a) A tissue section on a conductive slide is placed in the source of the instrument and a laser is fired at the surface to desorb and ionize molecules. Analytes from each spot are separated in the time of flight (TOF) analyzer, their flight times converted to a mass-to-charge ratio (m/z), and a spectrum recorded. (b) A representative mass spectrum collected from a tissue section. Inset shows detail and complexity of the spectrum collected. A tissue imaging experiment might result in thousands of such spectra. (c) Using an average spectrum from the entire section, ion images can be generated from each peak. Each m/z value of interest can be displayed as a function of position in the tissue section and relative intensity. Hundreds of such images can be created from a single tissue section.
Challenges with human tissues
The study of human tissue brings a unique set of challenges. Compared to laboratory mice, humans are genetically diverse. We come from different backgrounds and environments and have different levels of personal care. Factors such as genetic diversity and disease etiology [25] lead to high variability among samples from different individuals with the same nominal disease. Additionally, a person’s lifestyle, diet and therapeutic regimen can have a marked effect on protein expression. For example, the breast proliferation index is a measure of the number of Ki-67-positive nuclei and can be affected by a woman’s use of oral contraceptives that alter estrogen and progesterone levels [26]. Human samples must also be controlled for a variety of other factors, such as gender, race, age and menopausal status, to achieve a uniform patient population. Human samples are subject to strict Institutional Review Board (IRB) approval protocols, and permission must be obtained from the patient for use of their tissue for research purposes. It should be kept in mind that permission may be withdrawn at any time, resulting in the loss of the samples and data from the study.
Fresh frozen tissues
Often, human samples are not harvested specifically for IMS or other research applications. As such, tissue specimens may be handled in a manner that can lead to poor compatibility with mass spectrometric analysis. For example, and perhaps most importantly, the time between harvesting of the tissue and freezing can be critical [27,28]. The tissue specimen may sit on ice (or worse, at room temperature) for minutes or hours until it is frozen. This provides time for enzymatic degradation to take place, leading to inconsistencies in proteins observed between different samples, especially if this warm ischemia time is not constant from one sample to the next. For optimal analysis, tissue should be immediately snap frozen in liquid nitrogen to minimize proteolytic degradation.
Care should be taken when freezing samples to ensure that they maintain their original shape. If samples are placed in a rigid storage device prior to freezing, they are likely to take on the shape of the container, potentially skewing or destroying any spatial relationships within the sample. Tissue should be set on a piece of aluminum foil and then floated on the surface of liquid nitrogen to freeze the tissue. Plunge freezing of tissue chunks or blocks should be avoided because it tends to produce cracks and tears. Once frozen, the sample can be transferred to another vessel (e.g. Eppendorf or Falcon tube) for storage. Tissue samples collected in this manner must be stored in a freezer at −80°C or in liquid nitrogen to prevent proteolytic activity.
Follow-up clinical information on patient samples is often limited or difficult to match to frozen research samples. Human tissue specimens should be de-identified from the patient and robustly labeled for accurate data tracking. Fresh frozen samples have a limited lifetime, especially if they are not stored in an embedding medium. This can be attributed to several factors, including freezer failure, limited space for sample storage, or the fact that tissues tend to dry out and get ‘freezer burn’ with prolonged storage, rendering them unusable for sectioning and analysis.
There have been numerous successful studies reported using IMS with fresh frozen biopsies (Box 1). We have previously shown feasibility of the histology directed protein profiling for breast [29,30], lung [31], colon [32], brain [33,34], gastric [35] cancers, as well as other diseases, such as inflammatory bowel diseases [36]. Several reports have involved studies to predict response to neoadjuvant chemotherapy and radiation [17] as well as HER2 status in breast cancer [37], molecular diagnosis of prostate cancer [3,38], classification [6] and survival prediction [39] in lung cancer, and determination of molecular tumor margins [40,41]. These approaches provide us with valuable information about the molecular aspects of disease in a histology-related manner that would not be obtainable by other technologies.
Profiling of frozen human tissue has been reported for the analysis of human non-small cell lung cancer (NSCLC) tumors in order to determine a signature predictive of patient survival [39]. These investigators evaluated a total of 174 NSCLC cases and 27 normal lung specimens. This sample number allowed robust calculation of a classification model and subsequent independent validation of the model. Intact protein profile data were collected using a histology-directed approach with manual matrix application and spectral acquisition. Only homogenous areas of tumor or normal lung were targeted to avoid ambiguity in the observed mass spectral peaks. A signature of 25 proteins within the tumors was found to be predictive of both disease-free survival and overall survival. The protein signature was successfully used in the survival classification of stage I or stage II-III cancers, but did not discriminate between stages of the disease. Nevertheless, these results could be used to guide treatment of newly diagnosed cancers to avoid over-treatment of tumors unlikely to recur or under-treatment of tumors with aggressive characteristics. Studies such as these provide insight that is not possible with standard histological approaches.
Formalin-fixed, paraffin-embedded tissues
Most clinical tissue specimens are stored as formalin-fixed paraffin-embedded (FFPE) samples, which allow an extraordinarily long lifetime and storage at room temperature [42]. Formalin fixation involves the dehydration of the tissue sample through graded ethanol and xylene followed by partial protein crosslinking in 10% neutral buffered formalin. The sample is embedded in paraffin wax attached to a cassette for sectioning. Until recently, it was believed that the proteins in these samples were inaccessible for direct mass spectral analysis of tissue sections. Our group [6] and others [4] have shown that it is possible to gain access to the proteins through a process of antigen retrieval [43] (heating the sample section in buffer) and in situ enzymatic digestion prior to matrix application. This process denatures some of the proteins, thus allowing controlled enzymatic breakdown of proteins to peptides within accessible areas. Tissue sections can then subjected to in situ tryptic digestion, resulting in hundreds of non-crosslinked peptides that are analyzed by mass spectrometry. Peptides from distinct cellular regions are analyzed by intact mass. Peptides of interest (i.e. those shown to be statistically significant within the area of interest) are then isolated for sequencing by MS/MS. Using database programs, such as Mascot or Sequest, the amino acid sequence can be determined from the fragmentation patterns and matched to the parent protein, thus correlating protein expression to the histology. With this approach, peptides can be identified directly from the tissue sections without the need for additional separation steps.
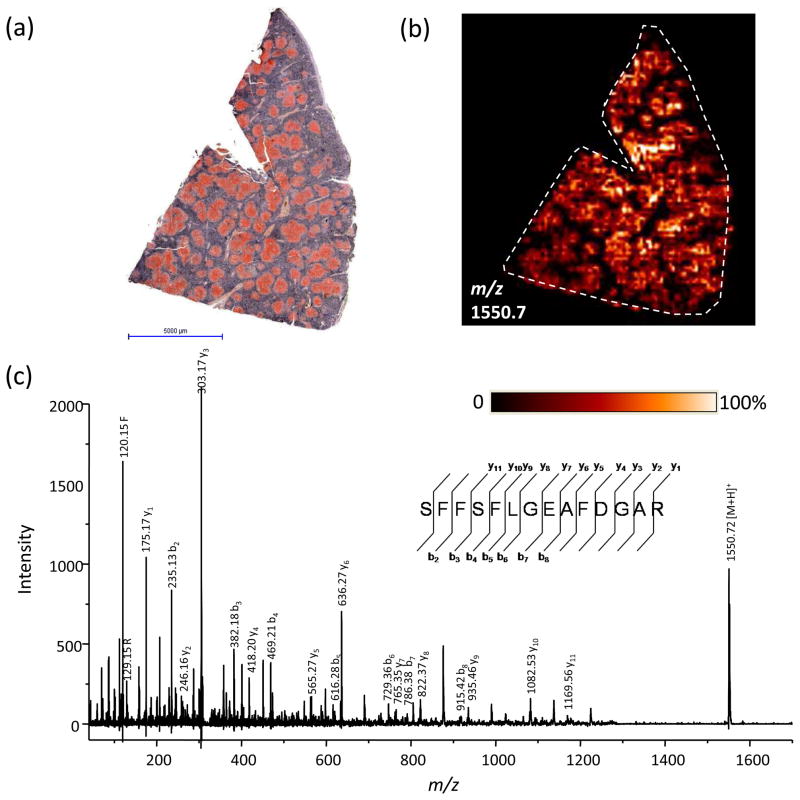
The use of FFPE tissue helps to alleviate many of the problems associated with frozen tissues. They are much easier to store and transport because they are stable at room temperature. Collection and storage of FFPE tissue is therefore easier and less expensive, even in facilities that do not have ready access to large freezers or liquid nitrogen. The first step, the preservation process, is dehydration by ethanol; this can be performed immediately in the operating room, eliminating the differences in warm ischemia time that can lead to proteomic degradation. The long lifetime of the FFPE sample also makes it is possible to collect long-term follow up information from the patients. Prospective studies can therefore be performed to look for proteomic markers of outcome, such as disease progression or long-term survival. Recently, a biopsy from the spleen of a patient who died of tuberculosis and secondary amyloidosis in 1899 was analyzed. Antigen retrieval and on tissue digestion allowed for analysis and identification of several peptides of serum amyloid A (Figure 3), the primary component of the amyloid deposits, as previously determined [44].
Figure 3.
IMS analysis of a 109-year-old FFPE sample. (a) Contrast-enhanced Congo Red staining of a human spleen biopsy shows extensive amyloid deposits (in red) throughout the section. Scale bar, 5 mm. (b) MS image of a peptide at m/z 1550.7 from serum amyloid A localized to the areas of amyloid deposition. (c) MS/MS of this peptide directly from the tissue section resulted in nearly complete sequence coverage and identification of the peptide SFFSLGEAFDGAR.
There is a considerably larger library of available FFPE samples. This is important for acquiring a large enough number of samples for statistical confidence from a highly variable human population pool. Our experience has been that a minimum of 50 to 60 samples is needed for robust statistical classification of disease models for eventual diagnostics. These numbers are often difficult to achieve with frozen samples.
Although FFPE tissue has been shown to be amenable to the analysis of peptides and allows mapping of peptides to their parent proteins, this type of tissue has not yet been shown to work for the analysis of lipids or exogenous pharmaceuticals. These molecules are likely to be washed away, at least in part, during the fixation process and during the deparaffinization and antigen-retrieval processes. To date, there is no literature demonstrating the analysis of these molecules from fixed tissues and analyses of these classes of molecules must be performed from frozen tissue specimens. However, the analyses of lipids in tissue are of great interest since they play important roles in many biological and disease processes [45,46]. Recently, sulfatides were evaluated by MALDI IMS for their role and localization in ovarian cancer [45]. Three species (d18:1/C16:0, d18:1/C24:1, and d18:1/C24:0) were found to be elevated in tumor tissue as compared to the surrounding normal tissue. Thus far, imaging of pharmaceutical compounds has only been carried out in animal tissues [11,13] or xenographs [47]. These studies have provided insight into delivery of drugs to specific locations in tissues as well their concomitant metabolism.
Tissue microarrays
One of the additional benefits of FFPE tissue is their suitability for assembling tissue microarrays (TMAs). While some fresh frozen TMAs have been reported [48], FFPE tissue microarrays are most common. Hundreds of tissue cores can be placed into a single paraffin block. These can be sectioned and the cores analyzed simultaneously, thus allowing for increased throughput and consistency relative to individual analysis of single biopsies. Recently, a TMA comprising 112 needle cores was analyzed for peptides that could distinguish between adenocarcinomas and squamous cell lung carcinomas [6]. Sample sections were prepared by deparaffinization and antigen retrieval prior to application of trypsin and matrix in an array over the entire TMA using an acoustic robotic microspotter. Spectra were collected from the matrix spots using a MALDI TOF/TOF mass spectrometer. Areas from the tissue cores were classified by a pathologist as cancer (adenocarcinoma or squamous cell carcionoma), non-cancer (i.e. stroma, connective tissue, etc.), or normal endothelium. Spectra from these areas were subjected to a support vector machine algorithm using ClinProTools software and resulted in a signature of 73 peaks, with a minimum 3-fold change in intensity that could successfully classify squamous cell carcinoma and adenocarcinoma with a respective 98.6% and 97.9% spectral classification accuracy. Approximately 50 of the 73 peaks in the signature were identified directly from the TMA surface through the use of MS/MS. In many cases, multiple tryptic peptides from the same parent protein were detected and their identities were further validated by visualization of their similar distribution throughout the TMA. Additionally, a peptide at m/z 1410.7 belonging to cytokeratin 5 was found to be specific to a subset, but not all of the squamous cell carcinoma cores. Reproducibility of the technology was verified by statistical comparisons between multiple needle cores from the same patient that were part of the TMA block.
Histological staining and IMS
A final requirement when working with any tissue samples is the use of histological stains (either organic or antibody) to determine a diagnosis and to guide analysis. Previous work has shown MS analysis of frozen tissue to be incompatible with the most commonly used histological stain, hematoxylin and eosin (H&E) [49]. A light cresyl violet or methylene blue stain can be applied to the tissue section being analyzed, but these generally do not provide enough cellular detail for clinical diagnosis. For this reason, when analyzing fresh frozen tissue, a serial section stained with H&E is used to guide matrix application to an unstained section.
The same issues surrounding histological stain compatibility with MS analysis do not apply to FFPE tissue (R. Caprioli, unpublished). Many of the more commonly used stains, including H&E, have been shown to be compatible with FFPE tissue analysis. Figure 4 shows a comparison of spectra from a stained and unstained section of a human kidney tumor. A high degree of similarity is observed between the two spectra. The ability to stain the same section of tissue used for MS analysis is important for two reasons. First, using two serial sections, the cells seen in one section are not necessarily the same as those seen in the next section; the types of cells being analyzed are inferred from what is seen in an adjacent section. Second, there is inherent error associated with the process of matching two serial sections, and the wrong area may be targeted. This matching process (between serial sections) also increases the total analysis time and therefore lowers the throughput.
Figure 4.
H&E staining coupled with MS of an FFPE human kidney tumor. (a) A mass spectrum obtained from an H&E-stained section of a human kidney tumor. The inset shows the tissue that was analyzed, with 5× magnification of the area from which the spectrum was acquired. (b) The mass spectrum from an unstained serial section.
Conclusions
There are enormous benefits to performing IMS on human tissue samples, despite the many challenges outlined above. Molecular analyses provide new insight into disease processes that are not attainable by standard histological analyses. The ability to work with FFPE tissue as well as fresh frozen tissue greatly increases the number and quality of samples available for study. Additionally, samples with long-term follow up information allow for prospective studies on disease progression and outcome. Such molecular analyses are vital for achieving personalized medicine, wherein each individual’s tissue is characterized at the molecular level. IMS allows for high-throughput analysis while preserving spatial relationships within the tissue in healthy and diseased states. IMS can also be combined with other imaging modalities, such as MRI, to achieve additional information about a tissue at the molecular level (Box 2). The IMS platform provides a significant level of diversity in terms of molecular analysis and promises to have animpact on the characterization of disease and the effectiveness of treatment in the coming years.
Box 2. Multi-modality imaging: MRI meets IMS.
One of the exciting new advances in the field of clinical mass spectrometry is the ability to co-register multiple imaging modalities in the same three-dimensional space [50]. In a recent study, a mouse with a human glioma cell line implanted in its brain was subjected to high-resolution MRI. Subsequently, the mouse was sacrificed, embedded in an ice block, and the whole head sectioned on a cryomacrotome. Blockface digital images were acquired throughout the head, and selected sections were subjected to IMS for protein analysis of the tumor and surrounding normal brain. The digital blockface images serve as an intermediary upon which MS and MR imaging data can be matched. The imaging modalities were then co-registered to each other for visualization in a 3D model that can be viewed in any spatial orientation. Combining these data sets provides insight into the molecular changes that are occurring that allow for the visualization of a tumor by non-invasive MRI. Thus far, such studies of co-registration have only been performed in the head due to the rigid structure of the skull that allows for more facile alignment. Studies are currently underway in our lab to perform multi-modality imaging experiments in the hind limb and abdomen of a mouse: areas that contain much less rigid structure. These analyses require more detailed algorithms for co-registration, but will provide important information about disease processes through the correlation of molecular changes via IMS and proton relaxation measurements using MRI.
Acknowledgments
We would like to thank Kristina Schwamborn and Jamie Allen for help with figures and Peggi Angel for critical reading and editing of the manuscript. We would also like to acknowledge funding from NIH/NIGMS 5R01 GM58008, DOD W81XWH-05-1-0179, Vanderbilt Ingram Cancer Center Core Support Grant P30 CA68485, and the National Foundation for Cancer Research: Vanderbilt Center for Proteomics and Drug Action.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Andersson M, et al. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat Methods. 2008;5:101–108. doi: 10.1038/nmeth1145. [DOI] [PubMed] [Google Scholar]
- 2.Burnum KE, et al. Imaging mass spectrometry reveals unique protein profiles during embryo implantation. Endocrinology. 2008;149:3274–3278. doi: 10.1210/en.2008-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cazares LH, et al. Imaging mass spectrometry of a specific fragment of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 2 discriminates cancer from uninvolved prostate tissue. Clin Cancer Res. 2009;15:5541–5551. doi: 10.1158/1078-0432.CCR-08-2892. [DOI] [PubMed] [Google Scholar]
- 4.Lemaire R, et al. Direct Analysis and MALDI Imaging of Formalin-Fixed, Paraffin-Embedded Tissue Sections. Journal of Proteome Research. 2007;6:1295–1305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- 5.Groseclose MR, et al. Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J Mass Spectrom. 2007;42:254–262. doi: 10.1002/jms.1177. [DOI] [PubMed] [Google Scholar]
- 6.Groseclose MR, et al. High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics. 2008;8:3715–3724. doi: 10.1002/pmic.200800495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoeckli M, et al. MALDI MS imaging of amyloid. Methods in Enzymology. 2006;412:94–106. doi: 10.1016/S0076-6879(06)12007-8. [DOI] [PubMed] [Google Scholar]
- 8.Burnum KE, et al. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J Lipid Res. 2009;50:2290–2298. doi: 10.1194/jlr.M900100-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puolitaival SM, et al. Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J Am Soc Mass Spectrom. 2008;19:882–886. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meriaux C, et al. Liquid ionic matrixes for MALDI mass spectrometry imaging of lipids. J Proteomics. 73:1204–1218. doi: 10.1016/j.jprot.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 11.Stoeckli M, et al. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. International Journal of Mass Spectrometry. 2007;260:195–202. [Google Scholar]
- 12.Reyzer ML, et al. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 2003;38:1081–1092. doi: 10.1002/jms.525. [DOI] [PubMed] [Google Scholar]
- 13.Khatib-Shahidi S, et al. Direct Molecular Analysis of Whole-Body Animal Tissue Sections by Imaging MALDI Mass Spectrometry. Analytical Chemistry. 2006;78:6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y, et al. Improving spot homogeneity by using polymer substrates in matrix-assisted laser desorption/ionization mass spectrometry of oligonucleotides. Anal Chem. 2001;73:2617–2624. doi: 10.1021/ac001392v. [DOI] [PubMed] [Google Scholar]
- 15.McDonnell LA, Heeren RM. Imaging mass spectrometry. Mass Spectrom Rev. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 16.Rubakhin SS, Sweedler JV, editors. Mass Spectrometry Imaging. Springer; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bauer JA, et al. Identification of markers of taxane sensitivity using proteomic and genomic analyses of breast tumors from patients receiving neoadjuvant paclitaxel and radiation. Clin Cancer Res. 2010;16:681–690. doi: 10.1158/1078-0432.CCR-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaurand P, et al. Exploring the potential of cryodetectors for the detection of matrix-assisted laser desorption/ionization produced ions: application to profiling and imaging mass spectrometry. Zhipu Xuebao. 2004;25:205–206. 216. [Google Scholar]
- 19.Seeley EH, et al. Enhancement of Protein Sensitivity for MALDI Imaging Mass Spectrometry after Chemical Treatment of Tissue Sections. Journal of the American Society for Mass Spectrometry. 2008;19:1069–1077. doi: 10.1016/j.jasms.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aerni HR, et al. Automated acoustic matrix deposition for MALDI sample preparation. Anal Chem. 2006;78:827–834. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- 21.Hankin JA, et al. Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom. 2007;18:1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornett DS, et al. A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Molecular and Cellular Proteomics. 2006;5:1975–1983. doi: 10.1074/mcp.M600119-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Seeley EH, Caprioli RM. Molecular imaging of proteins in tissues by mass spectrometry. Proc Natl Acad Sci U S A. 2008;105:18126–18131. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonnell LA, Heeren RMA. Imaging mass spectrometry. Mass Spectrometry Reviews. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 25.Lerescu L, et al. Primary cell culture of human adenocarcinomas--practical considerations. Roum Arch Microbiol Immunol. 2008;67:55–66. [PubMed] [Google Scholar]
- 26.Garcia y Narvaiza D, et al. Effect of combined oral contraceptives on breast epithelial proliferation in young women. Breast J. 2008;14:450–455. doi: 10.1111/j.1524-4741.2008.00621.x. [DOI] [PubMed] [Google Scholar]
- 27.Spruessel A, et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. BioTechniques. 2004;36:1030–1032. 1034–1037. doi: 10.2144/04366RR04. [DOI] [PubMed] [Google Scholar]
- 28.Grundmann R, et al. Relationship between the prolongation of warm ischemia and the maximum available preservation period. Surgery. 1977;81:542–550. [PubMed] [Google Scholar]
- 29.Reyzer ML, et al. Early Changes in Protein Expression Detected by Mass Spectrometry Predict Tumor Response to Molecular Therapeutics. Cancer Research. 2004;64:9093–9100. doi: 10.1158/0008-5472.CAN-04-2231. [DOI] [PubMed] [Google Scholar]
- 30.Sanders ME, et al. Differentiating proteomic biomarkers in breast cancer by laser capture microdissection and MALDI MS. J Proteome Res. 2008;7:1500–1507. doi: 10.1021/pr7008109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahman SM, et al. Proteomic patterns of preinvasive bronchial lesions. Am J Respir Crit Care Med. 2005;172:1556–1562. doi: 10.1164/rccm.200502-274OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu BJ, et al. Identification of early intestinal neoplasia protein biomarkers using laser capture microdissection and MALDI MS. Mol Cell Proteomics. 2009;8:936–945. doi: 10.1074/mcp.M800345-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz SA, et al. Protein profiling in brain tumors using mass spectrometry: feasibility of a new technique for the analysis of protein expression. Clin Cancer Res. 2004;10:981–987. doi: 10.1158/1078-0432.ccr-0927-3. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz SA, et al. Proteomic-based prognosis of brain tumor patients using direct-tissue matrix-assisted laser desorption ionization mass spectrometry. Cancer Res. 2005;65:7674–7681. doi: 10.1158/0008-5472.CAN-04-3016. [DOI] [PubMed] [Google Scholar]
- 35.Kim HK, et al. Gastric Cancer-Specific Protein Profile Identified Using Endoscopic Biopsy Samples via MALDI Mass Spectrometry. J Proteome Res. 2010;9:4123–4130. doi: 10.1021/pr100302b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.M'Koma AE, et al. Proteomic profiling of mucosal and submucosal colonic tissues yields protein signatures that differentiate the inflammatory colitides. Inflamm Bowel Dis. 2010 doi: 10.1002/ibd.21442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauser S, et al. Classification of HER2 receptor status in breast cancer tissues by MALDI imaging mass spectrometry. J Proteome Res. 2010;9:1854–1863. doi: 10.1021/pr901008d. [DOI] [PubMed] [Google Scholar]
- 38.Schwamborn K, et al. Identifying prostate carcinoma by MALDI-Imaging. International Journal of Molecular Medicine. 2007;20:155–159. [PubMed] [Google Scholar]
- 39.Yanagisawa K, et al. A 25-signal proteomic signature and outcome for patients with resected non-small-cell lung cancer. J Natl Cancer Inst. 2007;99:858–867. doi: 10.1093/jnci/djk197. [DOI] [PubMed] [Google Scholar]
- 40.Oppenheimer SR, et al. Molecular analysis of tumor margins by MALDI mass spectrometry in renal carcinoma. J Proteome Res. 2010;9:2182–2190. doi: 10.1021/pr900936z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caldwell RL, et al. Molecular assessment of the tumor protein microenvironment using imaging mass spectrometry. Cancer Genomics and Proteomics. 2006;3:279–288. [Google Scholar]
- 42.Fox CH, et al. Formaldehyde fixation. J Histochem Cytochem. 1985;33:845–853. doi: 10.1177/33.8.3894502. [DOI] [PubMed] [Google Scholar]
- 43.Yamashita S. Heat-induced antigen retrieval: mechanisms and application to histochemistry. Prog Histochem Cytochem. 2007;41:141–200. doi: 10.1016/j.proghi.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Westermark P, Nilsson GT. Demonstration of Amyloid Protein AA in Old Museum Specimens. Arch Pathol Lab Med. 1984;108:217–219. [PubMed] [Google Scholar]
- 45.Liu Y, et al. Elevation of sulfatides in ovarian cancer: an integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol Cancer. 2010;9:186–199. doi: 10.1186/1476-4598-9-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eberlin LS, et al. Discrimination of human astrocytoma subtypes by lipid analysis using desorption electrospray ionization imaging mass spectrometry. Angew Chem Int Ed Engl. 49:5953–5956. doi: 10.1002/anie.201001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atkinson SJ, et al. Examination of the distribution of the bioreductive drug AQ4N and its active metabolite AQ4 in solid tumours by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Communications in Mass Spectrometry. 2007;21:1271–1276. doi: 10.1002/rcm.2952. [DOI] [PubMed] [Google Scholar]
- 48.Fejzo MS, Slamon DJ. Tissue microarrays from frozen tissues-OCT technique. Methods Mol Biol. 2010;664:73–80. doi: 10.1007/978-1-60761-806-5_8. [DOI] [PubMed] [Google Scholar]
- 49.Chaurand P, et al. Integrating histology and imaging mass spectrometry. Analytical Chemistry. 2004;76:1145–1155. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- 50.Sinha TK, et al. Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nat Methods. 2008;5:57–59. doi: 10.1038/nmeth1147. [DOI] [PMC free article] [PubMed] [Google Scholar]