Abstract
Isosphaera pallida (ex Woronichin 1927) Giovannoni et al. 1995 is the type species of the genus Isosphaera. The species is of interest because it was the first heterotrophic bacterium known to be phototactic, and it occupies an isolated phylogenetic position within the Planctomycetaceae. Here we describe the features of this organism, together with the complete genome sequence and annotation. This is the first complete genome sequence of a member of the genus Isosphaera and the third of a member of the family Planctomycetaceae. The 5,472,964 bp long chromosome and the 56,340 bp long plasmid with a total of 3,763 protein-coding and 60 RNA genes are part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: thermophilic, aerobic, filamentous, budding, gliding motility, Gram-negative, phototactic comets, gas vesicles, chemoheterotrophic, hot spring, Planctomycetaceae, GEBA
Introduction
Strain IS1BT (= ATCC 43644) is the type strain of Isosphaera pallida which in turn is the type and sole species of the genus Isosphaera [1,2]. The genus Isosphaera is one out of nine genera in the family Planctomycetaceae [3]. The genus name is derived from the Greek adjective isos, equal and sphaera, a ball, globe, yielding Isosphaera, sphere of equal size [4]. The species epithet pallida is derived from the Latin adjective pallida, pale [1]. Strain IS1BT was isolated from a hot spring in Kah-nee-tah, Oregon, USA [1]. Other closely related strains belonging to the species were isolated from several warm springs in North America [1]. The cells resemble Isocystis pallida Worochin 1927 [5] which was previously described as a cyanobacterium and later as a yeast. Here we present a summary classification and a set of features for I. pallida strain IS1BT, together with the description of the complete genomic sequencing and annotation.
Classification and features
A representative genomic 16S rRNA sequence of strain IS1BT was compared using NCBI BLAST under default values (e.g., considering only the best 250 hits) with the most recent release of the Greengenes database [6] and the relative frequencies, weighted by BLAST scores, of taxa and keywords (reduced to their stem [7]) were determined. The five most frequent genera were Isosphaera (35.4%), Nostocoida (26.4%; a genus with Candidatus status [8]), Singulisphaera (20.4%), 'Isophaera' (15.9%; a misspelling of Isosphaera) and Planctomyces (1.9%). The species yielding the highest score was Candidatus Nostocoida limicola [8]. The five most frequent keywords within the labels of environmental samples which yielded hits were 'skin' (3.9%), 'soil' (3.0%), 'fossa' (2.2%), 'adult/zebrafish' (2.2%) and 'microbi' (1.9%). The two most frequent keywords within the labels of environmental samples which yielded hits of a higher score than the highest scoring species were 'adult, zebrafish' (10.0%) and 'conventionally-rais, digest, gender, germ-fre, gut, habitat, host, mice, micro-biota, mix, pool, recipi, reciproc, select, tract, transplant' (5.0%), i.e. many ties occurred, rendering it difficult to ecologically interpret this outcome.
Figure 1 shows the phylogenetic neighborhood of I. pallida IS1BT in a 16S rRNA based tree. The sequences of the three copies in the genome do not differ from each other, and differ by two nucleotides from the previously published 16S rRNA sequence (AJ231195).
Figure 1.
Phylogenetic tree highlighting the position of I. pallida relative to the other type strains within the class family Planctomycetacia. The tree was inferred from 1,362 aligned characters [9,10] of the 16S rRNA gene sequence under the maximum likelihood criterion [11] and rooted in with members of the class Phycisphaerae. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 450 bootstrap replicates [12] if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [13] are shown in blue, published genomes in bold [14,15].
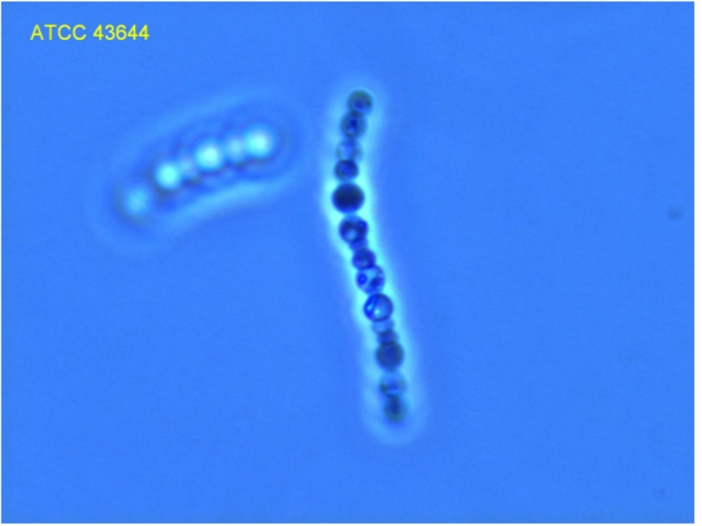
Cells of strain IS1BT are spherical with 2.5 to 3 µm in diameter (Figure 2 and Table 1), with cell growth and division occurring by intercalary budding, resulting in filaments [1]. The cells are salmon-colored (caused by carotenoids), contain gas vesicles and resemble Isocystis pallida Worochin 1927 [5]. Ultra-thin sections observed by TEM revealed pit-like ultrastructural features in the cell wall [1,24]. The cells contain numerous pili (not visible in Figure 2) but no flagella, and form motile phototactic “comets” in liquid cultures or on media containing Gelrite® as the solidifying agent [1].
Figure 2.
Photomicrograph (1000 x) of I. pallida IS1BT (provided by ATCC)
Table 1. Classification and general features of I. pallida IS1BT according to the MIGS recommendations [16].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [17] | |
| Phylum Planctomycetes | TAS [18] | ||
| Class Planctomycetacia | TAS [19] | ||
| Order Planctomycetales | TAS [3,20-22] | ||
| Family Planctomycetaceae | TAS [3,20] | ||
| Genus Isosphaera | TAS [1,2] | ||
| Species Isosphaera pallida | TAS [1,2] | ||
| Type strain IS1B | TAS [1] | ||
| Gram stain | variable to negative | TAS [1] | |
| Cell shape | coccoid, chain-forming | TAS [1] | |
| Motility | motile by gliding | TAS [1] | |
| Sporulation | not reported | ||
| Temperature range | 40–55°C, thermophile | TAS [1] | |
| Optimum temperature | 41°C | TAS [1] | |
| Salinity | about 0.1% NaCl | TAS [1] | |
| MIGS-22 | Oxygen requirement | obligately aerobic | TAS [1] |
| Carbon source | glucose, lactate | TAS [1] | |
| Energy source | chemoheterotrophic | TAS [1] | |
| MIGS-6 | Habitat | algal mat, fresh water | TAS [1] |
| MIGS-15 | Biotic relationship | not reported | |
| MIGS-14 | Pathogenicity | none | NAS |
| Biosafety level | 1 | NAS | |
| Isolation | hot spring | TAS [1] | |
| MIGS-4 | Geographic location | Kah-nee-tah Hot Spring, Oregon,USA | TAS [1] |
| MIGS-5 | Sample collection time | 1987 or before | TAS [1] |
| MIGS-4.1 MIGS-4.2 |
Latitude Longitude |
44.86 -121.20 |
TAS [1] |
| MIGS-4.3 | Depth | 0 m, probably surface waters | NAS |
| MIGS-4.4 | Altitude | not reported |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [23]. If the evidence code is IDA, the property was observed by one of the authors or an expert mentioned in the acknowledgements.
Chemotaxonomy
Muramic acid and diaminopimelic acid are absent from the cell wall [1,24], like in other members of the Planctomycetes. Cells stain Gram-negative but lack an outer membrane [1]. Cells possess a proteinaceous cell wall structure without cysteine, methionine, proline and tryptophan [24]. Ester-linked lipids with predominantly unbranched C14 and C18 fatty acids, traces of C18:1 acids, no hydroxyl-fatty acids [24].
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position [25], and is part of the Genomic Encyclopedia of Bacteria and Archaea project [26]. The genome project is deposited in the Genomes OnLine Database [13] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Three genomic libraries: one 454 pyrosequence standard library, one 454 PE library (11 kb insert size), one Illumina library |
| MIGS-29 | Sequencing platforms | Illumina GAii, 454 GS FLX Titanium |
| MIGS-31.2 | Sequencing coverage | 109.5 × Illumina; 31.2 × pyrosequence |
| MIGS-30 | Assemblers | Newbler version 2.0.00.20-PostRelease-11-05-2008-gcc-3.4.6, Velvet, phrap |
| MIGS-32 | Gene calling method | Prodigal 1.4, GenePRIMP |
| INSDC ID | CP002353 (chromosome) CP002354 (plasmid) |
|
| Genbank Date of Release | January 26, 2011 | |
| GOLD ID | Gc01591 | |
| NCBI project ID | 32825 | |
| Database: IMG-GEBA | 2503538023 | |
| MIGS-13 | Source material identifier | ATCC 43644 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
I. pallida IS1BT, ATCC 43644, has been in the American Type Culture Collection since July 1987. The culture used at ATCC to prepare genomic DNA (gDNA) for sequencing was only two transfers away from the original deposit. The purity of the culture was determined by growth in ATCC medium 1962 Broth [27] at 45oC under aerobic conditions. Cells were harvested by centrifugation after 72 hours of incubation. The cell pellet exhibited a salmon color. Genomic DNA was extracted from lysozyme-treated cells using a standard CTAB and phenol-chloroform protocol. The purity, quality and size of the bulk gDNA preparation were assessed according to DOE-JGI guidelines. Amplification and partial sequencing of the 16S rRNA gene confirmed the isolate as I. pallida. The quantity of the DNA was determined on a 1% agarose using gel mass markers of known concentration supplied by JGI. The average fragment size of the purified gDNA determined to be ~43 kb by pulsed-field gel electrophoresis.
Genome sequencing and assembly
The genome was sequenced using a combination of Illumina and 454 sequencing platforms. All general aspects of library construction and sequencing can be found at the JGI website [28]. Pyrosequencing reads were assembled using the Newbler assembler version 2.0.00.20-PostRelease-11-05-2008-gcc-3.4.6 (Roche). The initial Newbler assembly, consisting of 36 contigs in 1 scaffold, was converted into a phrap assembly by making fake reads from the consensus [29], to collect the read pairs in the 454 paired end library. Illumina GAii sequencing data (461 Mb) was assembled with Velvet [30] and the consensus sequences were shredded into 1.5 kb overlapped fake reads and assembled together with the 454 data. The 454 draft assembly was based on 172.7 Mb of 454 draft data and all of the 454 paired end data. Newbler parameters are -consed -a 50 -l 350 -g -m -ml 20. The Phred/Phrap/Consed software package [29] was used for sequence assembly and quality assessment in the subsequent finishing process. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with gapResolution [28], Dupfinisher, or sequencing cloned bridging PCR fragments with subcloning or transposon bombing (Epicentre Biotechnologies, Madison, WI) [31]. Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR primer walks (J.-F.Chang, unpublished). A total of 411 additional reactions and 14 shatter libraries were necessary to close gaps and to raise the quality of the finished sequence. Illumina reads were also used to correct potential base errors and increase consensus quality using a software Polisher developed at JGI [32]. The error rate of the completed genome sequence is less than 1 in 100,000. Together, the combination of the Illumina and 454 sequencing platforms provided 140.7 × coverage of the genome. The final assembly contained 764,175 pyrosequence and 16,816,247 Illumina reads.
Genome annotation
Genes were identified using Prodigal [33] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [34]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review (IMG-ER) platform [35].
Genome properties
The genome consists of a 5,472,964 bp long chromosome with a 62% GC content and a 56,340 bp plasmid with 67% GC content (Table 3 and Figures 3a and 3b). Of the 3,823 genes predicted, 3,763 were protein-coding genes, and 60 RNAs; 41 pseudogenes were identified. The majority of the protein-coding genes (59.7%) were assigned with a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 5,529,304 | 100.00% |
| DNA coding region (bp) | 4,671,376 | 84.48% |
| DNA G+C content (bp) | 3,455,288 | 62.49% |
| Number of replicons | 2 | |
| Extrachromosomal elements | 1 | |
| Total genes | 3,823 | 100.00% |
| RNA genes | 60 | 1.57% |
| rRNA operons | 3 | |
| Protein-coding genes | 3,763 | 98.43% |
| Pseudo genes | 41 | 1.07% |
| Genes with function prediction | 2,284 | 59.74% |
| Genes in paralog clusters | 227 | 5.94% |
| Genes assigned to COGs | 2,408 | 62.99% |
| Genes assigned Pfam domains | 2,563 | 67.04% |
| Genes with signal peptides | 792 | 20.72% |
| Genes with transmembrane helices | 967 | 25.29% |
| CRISPR repeats | 3 |
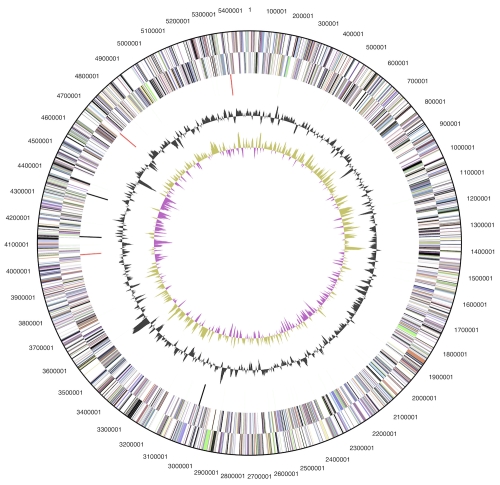
Figure 3a.
Graphical circular map of the chromosome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Figure 3b.
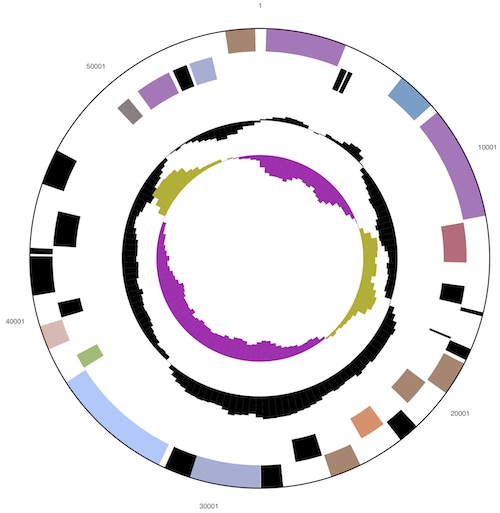
Graphical circular map of the plasmid (not drown to scale with chromosome). From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 138 | 4.7 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.0 | RNA processing and modification |
| K | 166 | 5.7 | Transcription |
| L | 165 | 5.7 | Replication, recombination and repair |
| B | 1 | 0.0 | Chromatin structure and dynamics |
| D | 24 | 0.8 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.0 | Nuclear structure |
| V | 54 | 1.9 | Defense mechanisms |
| T | 187 | 6.5 | Signal transduction mechanisms |
| M | 196 | 6.8 | Cell wall/membrane/envelope biogenesis |
| N | 77 | 2.7 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 128 | 4.4 | Intracellular trafficking and secretion, and vesicular transport |
| O | 132 | 4.5 | Posttranslational modification, protein turnover, chaperones |
| C | 157 | 5.4 | Energy production and conversion |
| G | 176 | 6.1 | Carbohydrate transport and metabolism |
| E | 197 | 6.8 | Amino acid transport and metabolism |
| F | 57 | 2.0 | Nucleotide transport and metabolism |
| H | 152 | 5.3 | Coenzyme transport and metabolism |
| I | 83 | 2.9 | Lipid transport and metabolism |
| P | 110 | 3.8 | Inorganic ion transport and metabolism |
| Q | 70 | 2.4 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 435 | 15.0 | General function prediction only |
| S | 190 | 6.6 | Function unknown |
| - | 1,415 | 37.0 | Not in COGs |
Acknowledgements
This work was performed under the auspices of the US Department of Energy Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, UT-Battelle, and Oak Ridge National Laboratory under contract DE-AC05-00OR22725.
References
- 1.Giovannoni SJ, Schabtach E, Castenholtz RW. Isosphaera pallida, gen. and comb. nov., a gliding, budding eubacterium from hot springs. Arch Microbiol 1987; 147:276-284 10.1007/BF00463488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Validation List No 54. Int J Syst Bacteriol 1995; 45:619-620 10.1099/00207713-45-3-619 [DOI] [Google Scholar]
- 3.Schlesner H, Stackebrandt E. Assignment of the genera Planctomyces and Pirella to a new family Planctomycetaceae fam. nov. and description of the order Planctomycetales ord. nov. Syst Appl Microbiol 1986; 8:174-176 [Google Scholar]
- 4.Euzéby JP. List of Bacterial Names with Standing in Nomenclature: a folder available on the internet. Int J Syst Bacteriol 1997; 47:590-592 10.1099/00207713-47-2-590 [DOI] [PubMed] [Google Scholar]
- 5.Woronichin NN. Materiali kagologitscheskoj flore i rastitjelnosti mineralnich istotschnikov gruppi Kaukaskich mineralnich wod. Trav Inst Balneol aux Eaux Miner du Caucase 1927; 5:90-121 [Google Scholar]
- 6.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie E, Keller K, Huber T, Dalevi D, Hu P, Andersen G. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-5072 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porter MF. An algorithm for suffix stripping. Program: electronic library and information systems 1980; 14:130-137 [Google Scholar]
- 8.Backall LL, Seviour EM, Bradford D, Rossetti S, Tandoi V, Sviour RJ. 'Candidatus Nostocoida limicola', a filamentous bacterium from activated sludge. Int J Syst Evol Microbiol 2000; 50:703-709 [DOI] [PubMed] [Google Scholar]
- 9.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 10.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 11.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 12.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [PubMed] [Google Scholar]
- 13.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346-D354; . 10.1093/nar/gkp848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clum A, Tindall BJ, Sikorski J, Ivanova N, Mavromatis K, Lucas S, Glavina Del Rio T, Nolan M, Chen F, Tice H, et al. Complete genome sequence of Pirellula staleyi type strain (ATCC 27377T). Stand Genomic Sci 2009; 1:308-316 10.4056/sigs.51657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaButti K, Sikorski J, Schneider S, Nolan M, Lucas S, Glavina Del Rio T, Tice H, Cheng JF, Goodwin L, Pitluck S, et al. Complete genome sequence of Planctomyces limnophilus type strain (290T). Stand Genomic Sci 2010; 3:47-56 10.4056/sigs.1052813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garrity GM, Holt JG. The Road Map to the Ma-nual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriolo-gy, Second Edition, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 19.Garrity GM, Holt JG. Taxonomic Outline of the Archaea and Bacteria In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 155-166. [Google Scholar]
- 20.List Editor Validation List no. 23. Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1987; 37:179 10.1099/00207713-37-2-179 [DOI] [Google Scholar]
- 21.Euzéby JP, Tindall BJ. Nomenclatural type of orders: corrections necessary according to Rules 15 and 21a of the Bacteriological Code (1990 Revision), and designation of appropriate nomenclatural types of classes and subclasses. Request for an opinion. Int J Syst Evol Microbiol 2001; 51:725-727 [DOI] [PubMed] [Google Scholar]
- 22.Judicial Commission of the International Committee on Systematics of Prokaryotes The nomenclatural types of the orders Acholeplasmatales, Halanaerobiales, Halobacteriales, Methanobacteriales, Methanococcales, Methanomicrobiales, Planctomycetales, Prochlorales, Sulfolobales, Thermococcales, Thermoproteales and Verrucomicrobiales are the genera Acholeplasma, Halanaerobium, Halobacterium, Methanobacterium, Methanococcus, Methanomicrobium, Planctomyces, Prochloron, Sulfolobus, Thermococcus, Thermoproteus and Verrucomicrobium, respectively. Opinion 79. Int J Syst Evol Microbiol 2005; 55:517-518 10.1099/ijs.0.63548-0 [DOI] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giovannoni SJ, Godchaux W, III, Schabtach E, Castenholtz RW. Cell wall and lipid composition of Isosphaera pallida, a budding eubacterium from hot springs. J Bacteriol 1987; 169:2702-2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klenk HP, Göker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 2010; 33:175-182 10.1016/j.syapm.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 26.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.http://www.atcc.org/Attachments/3128.pdf
- 28.The DOE Joint Genome Institute http://www.jgi.doe.gov/
- 29.Phrap and Phred for Windows. MacOS, Linux, and Unix. www.phrap.com
- 30.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821-829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han C, Chain P. 2006. Finishing repeat regions automatically with Dupfinisher. in Proceeding of the 2006 international conference on bioinformatics & computational biology. Edited by Hamid R. Arabnia & Homayoun Valafar, CSREA Press. June 26-29, 2006: 141-146. [Google Scholar]
- 32.Lapidus A, LaButti K, Foster B, Lowry S, Trong S, Goltsman E. POLISHER: An effective tool for using ultra short reads in microbial genome assembly and finishing. AGBT, Marco Island, FL, 2008 [Google Scholar]
- 33.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A gene prediction improvement pipeline for microbial genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 35.Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]