Abstract
Vacuole-type ATPases (VoV1) and FoF1 ATP synthases couple ATP hydrolysis/synthesis in the soluble V1 or F1 portion with proton (or Na+) flow in the membrane-embedded Vo or Fo portion through rotation of one common shaft. Here we show at submillisecond resolutions the ATP-driven rotation of isolated V1 and the whole VoV1 from Thermus thermophilus, by attaching a 40-nm gold bead for which viscous drag is almost negligible. V1 made 120° steps, commensurate with the presence of three catalytic sites. Dwells between the steps involved at least two events other than ATP binding, one likely to be ATP hydrolysis. VoV1 exhibited 12 dwell positions per revolution, consistent with the 12-fold symmetry of the Vo rotor in T. thermophilus. Unlike F1 that undergoes 80°–40° substepping, chemo-mechanical checkpoints in isolated V1 are all at the ATP-waiting position, and Vo adds further bumps through stator–rotor interactions outside and remote from V1.
 Rotary ATPases FoF1 and VoV1 couple ATP hydrolysis with proton flow. Furuike et al. observe ATP-driven rotation in V1 and VoV1, at submillisecond resolution, and find that rate-limiting reactions in V1 all occur at the same angle, and stator–rotor interactions in Vo introduce additional checkpoints.
Rotary ATPases FoF1 and VoV1 couple ATP hydrolysis with proton flow. Furuike et al. observe ATP-driven rotation in V1 and VoV1, at submillisecond resolution, and find that rate-limiting reactions in V1 all occur at the same angle, and stator–rotor interactions in Vo introduce additional checkpoints.
The FoF1- and V-type ATPase/ATP synthase superfamily utilizes a rotary mechanism to perform their specific functions1,2,3. The basic structures of these ATPases/synthases are conserved among species. The soluble, cytoplasmic portion of FoF1- and V-type ATPases (called F1 and V1, respectively), responsible for ATP hydrolysis/synthesis, is connected via the central rotor stalk and the peripheral stator stalk to the transmembrane portion (Fo and Vo) that houses the ion-transporting pathway. In the bacterial V-type ATPase of Thermus thermophilus (VoV1), the V1 portion is composed of a hexameric A3B3 cylinder and a central shaft composed of D and F subunits4 (see Fig. 1a). The Vo portion of T. thermophilus is composed of two distinct domains: a hydrophobic rotor ring made of Vo-c subunits supplemented with a funnel shape Vo-d subunit and a stator apparatus composed of a transmembrane Vo-a subunit and EG subunits forming the peripheral stalk5,6 (see Fig. 1b). Cryoelectron micrographs of two-dimensional crystals of the Vo ring at 7.0 Å resolution showed the presence of 12 Vo-c subunits, each composed of two transmembrane helices7. The bacterial V-ATPase that we describe here works as an ATP synthase1, whereas its eukaryotic counterpart is vacuolar proton pump and thus some mechanistic differences may exist1,2,8. A number of researchers refer to the bacterial V-ATPase as archaeal-ATPase or AoA1-ATP synthase, but here we adopt the broader terminology.
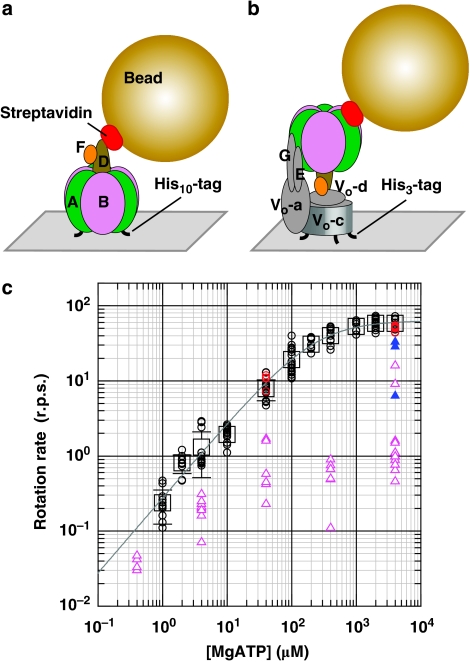
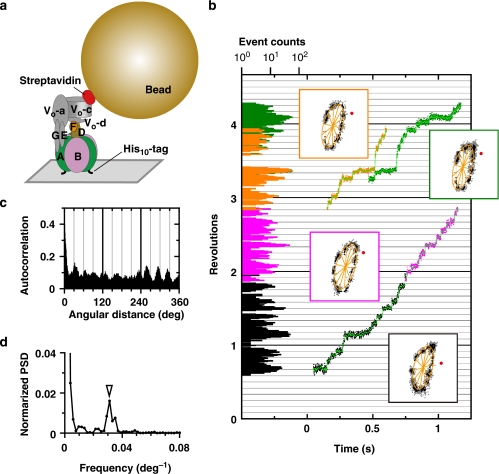
Figure 1. Rotation of V1 and VoV1 carrying a 40-nm bead.
Schematic observation systems for rotation of V1 (a) and VoV1 (b). (a) V1 was fixed to the Ni2+-NTA-coated glass surface with his10 tags at A subunits. A 40-nm bead (or duplex) was attached to the biotinylated cysteine residues (E48C/Q55C) of the D subunit via streptavidin. In this system, the central shaft composed of D and F subunits rotates relative to A3B3 subcomplex containing catalytic sites. (b) VoV1 was fixed to the Ni2+-NTA-coated glass surface with His3 tags at Vo-c subunits. In this system, the stator apparatus composed of A3B3, E, G and Vo-a subunit rotates relative to the fixed central rotor shaft composed of Vo-c ring, Vo-d, D and F subunits. A 40-nm bead (or duplex) was attached to the AviTag at A subunit(s) by biotin–streptavidin linkage. Bead rotation was observed under an optical microscope with dark-field illumination, and recorded with a high-speed camera at 250–8000 frames per s (fps). (c) Rotation rates of beads attached onto V1 (circles) and VoV1 (triangles) at the indicated ATP concentrations. Red and black circles indicate in the presence and absence of 0.05% (w/v) DDM, respectively. Squares indcate the averages of V1 rotation rates (n≥8; s.d. greater than the symbol size shown with bars). Line indicates the fit with Michaelis–Menten kinetics: V=Vmax·[ATP]/(Km + [ATP]), where Vmax and Km are 64 r.p.s. and 229 μM, respectively, giving the apparent ATP-binding rate kon of 0.84×106 M−1 s−1 (3×Vmax/Km). For VoV1, the rotation buffer contained 0.05% DDM. Time-averaged rotation rates of V1 or VoV1 were estimated over tens of consecutive revolutions as listed in Supplementary Table S1. The molecules of VoV1 which showed relatively clean 120° steps are shown as closed blue triangles.
It is believed that Vo (and Fo) is a rotary motor driven by the transmembrane flow of protons (or Na+) and V1 (and F1) is another rotary motor driven by ATP hydrolysis, and that the two motors have a common rotary shaft yet their genuine rotary directions are opposite to each other. Thus, when Vo (Fo) takes control, V1 (F1) is rotated in reverse direction, ending in the synthesis of ATP. Powering V1 (F1), on the other hand, results in proton pumping9. According to a model for Vo and Fo, a proton enters an access channel and binds to a glutamate on one of the c subunits in the rotor ring and after one revolution of the ring, the proton is released to the other side of the membrane via an exit channel10. In this model, the copy number of the c subunit of Vo or Fo in the rotor ring is equal to the number of transported protons per revolution. For the T. thermophilus V-ATPase, 12 protons are expected per revolution.
The ATP-driven rotation of the DF shaft in V1 has been observed directly11: a bead (nominal diameter 0.56 μm) attached to the D subunit rotated unidirectionally anticlockwise when viewed from the membrane side. At low ATP concentrations where ATP binding is rate limiting, the rotation proceeded in steps of 120°, commensurate with the presence of three catalytic sites at A–B interfaces12. Rotation of the Vo-c ring in VoV1 has also been observed13, with 120° steps at low ATP concentrations14.
For F1, which also undergoes anticlockwise 120° stepping at low ATP, high-speed imaging with 40-nm gold particles, with little drag, has revealed that a 120° step consists of 80–90° and 40–30° substeps15. F1 cycles through an ATP-waiting dwell, ∼80° substep rotation driven by ATP binding and subsequent ADP release, a catalytic dwell where ATP is hydrolyzed and the phosphate is released, and ∼40° substep rotation driven by the phosphate release16. ATP-driven rotation of FoF1 has also been demonstrated for Escherichia coli and thermophilic Bacillus PS3 enzymes, with features basically similar to those of F117,18,19. So far, ATP-driven rotation either in VoV1 or in FoF1 has failed to reveal a sign of specific interactions between a rotor and a stator subunit in the Vo/Fo portion, even in the high-resolution study17.
Here, we have analysed ATP-driven rotation of both V1 and VoV1 (holo V-ATPase) derived from T. thermophilus, using a 40-nm bead and a submillisecond fast camera. V1 molecules rotated with 120° steps without adopting the 80°–40° substep scheme of F1. VoV1, in contrast, showed ∼30° steps that likely reflect stator–rotor interactions in the Vo domain. All rate-limiting reactions in the V1 chemo-mechanical cycle occur in one angle, whereas stator–rotor interactions in Vo pose additional bumps that might check rotation depending on protonation/deprotonation.
Results
Stepwise rotation of V1
V1 was immobilized on a nickel-nitrilotriacetic acid (Ni2+-NTA )-coated glass surface through His (histidine)10-tags introduced at the amino terminus of the A subunits, and a 40-nm streptavidin-coated gold colloid (40-nm bead) was attached to the biotin-labelled D subunit (Fig. 1a). Bead rotation was imaged by laser dark-field microscope and recorded on a fast-framing CMOS camera at speeds up to 8,000 frames per s.
ATP dependence of the time-averaged rotation rate of V1 is shown in Figure 1c. Below 100 μM, ATP binding was rate limiting, the rotation speed being practically proportional to the ATP concentration ([ATP]). The rate constant for apparent, or effective, ATP binding was 0.8×106 M−1 s−1, assuming three ATP molecules consumed per revolution. Above 1 mM ATP, the rotary speed saturated, reaching Vmax of 64 revolutions per s (r.p.s.). This is the full speed of V1 rotation at 23 °C, not limited by the viscous drag on the bead (see below). The Michaelis–Menten constant, Km, of 229 μM (Fig. 1c) agrees with that for the bulk ATP hydrolysis assay without beads of 205 μM14, supporting the contention that Vmax above represents the speed of unloaded rotation (the reported maximal hydrolysis activity of 39.9 s−1 is lower than 180 s−1 expected for rotation at ∼60 r.p.s., because of MgADP inhibition14,15).
Even at saturating [ATP], all 40-nm beads rotated stepwise, pausing every 120° (Fig. 2a), reminiscent of the unloaded rotation of F1 at saturation. The 120° steps were completed within 0.25 ms (two frames), indicating that V1 can drive the 40-nm bead at >480° ms−1, and thus mechanical stepping does not limit the overall rotation rate. The average rotation speed of 64 r.p.s. at saturating [ATP] is limited by the ∼5 ms dwells where a reaction(s) that does not accompany rotation takes place. The 120° steps at saturating [ATP] were not resolved in the previous study with a 340-nm bead duplex12, where the time-averaged rotation speed at saturation was also low, limited by viscous drag on the large beads.
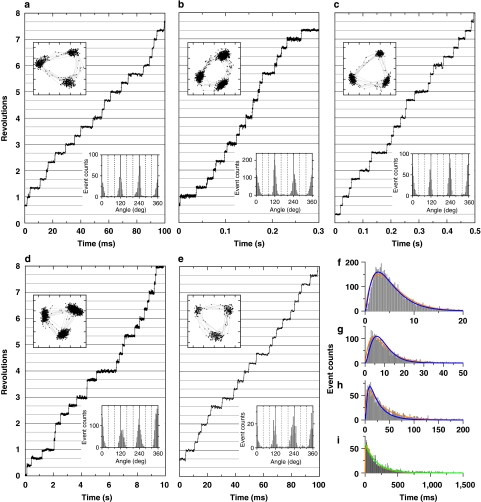
Figure 2. Rotation of V1.
(a–e) Typical time courses of rotation with a 40-nm bead (or duplex). (a) Rotation at 4 mM ATP captured at 8,000 fps; (b) 200 μM ATP at 2,000 fps; (c) 40 μM ATP at 250 fps; (d) 4 μM ATP at 4,000 fps and (e) 2 mM ATP at 4,000 fps, obtained from the same molecule as in d after medium exchange. Trajectories of the bead centroid (axis divisions: 11.1 nm) and histograms of angular positions, both for the indicated portion of the records, are shown in the upper and lower insets, respectively. (f–i) Histograms of dwell times between 120° steps. (f) Dwell times at 4 mM ATP with 125 μs bin size obtained from 6 molecules observed at 8,000 fps; (g) 200 μM ATP, 250 μs bin size, 6 molecules at 8,000 fps; (h) 40 μM ATP, 1 ms bin size, 6 molecules at 4,000 fps; (i) 4 μM ATP, 4 ms bin, 15 molecules at 2,000 fps. Orange curves show fit with the sequential two-reaction scheme with rates ka and kb: constant·(exp(−kat) − exp(−kbt)). At 4 mM ATP, the two rates turned out to be indistinguishable and thus the fit was made with two identical rates k: constant·t·exp(−kt). The estimated rates and associated s.e. are: k4 mM=0.36±0.01 ms−1, ka200 μM=0.17±0.02 ms−1, kb200 μM=0.28±0.03 ms−1, ka40 μM=31±1 s−1, kb40 μM=0.40±0.03 ms−1, and ka4 μM=6.1±0.1 s−1, kb4 μM=0.26±0.02 ms−1. If we assume that ka represents the rate of ATP binding (ka=kon[ATP]), kon is given as 0.85×106 M−1 s−1 at 200 μM ATP, 0.78×106 M−1 s−1 at 40 μM and 1.5×106 M−1 s−1 at 4 μM. At 4 μM, kon should dominate the histogram, and the green fit with constant·exp(−kon[ATP]t) gave kon of 1.5×106 M−1 s−1. Blue curves show a global fit to f–h (equal weight for each count), with sequential reactions starting with ATP binding at the rate kon[ATP] and two ATP-independent reactions with rates k1 and k2: constant·{(k2−k1)·exp(−kon[ATP]t) + (kon[ATP]−k2)·exp(−k1t) + (k1−kon[ATP])·exp(−k2t)} with kon=(1.2±0.1)×106 M−1 s−1, k1=0.49±0.05 ms−1, k2=0.34±0.04 ms−1.
At lower [ATP], we still observed 120° steps (Fig. 2b–d) without a clear sign of substeps as with F1 (refs 15, 16). Even at 200 μM ATP, around Km where F1 would repeat ∼80° and ∼40° substeps with equal dwells in between, V1 underwent 120° stepping (Fig.2b and insets therein). The V1 dwells at low [ATP] must be at ATP-waiting angles, implicating that the ∼5 ms dwells at saturating [ATP] were also at, or close to, ATP-waiting angles. This was also confirmed by solution exchange: Figure 2d,e show rotation of the same V1 molecule, showing that dwell positions at both high and low [ATP] do not differ significantly.
Events that underlie the V1 dwell
V1 dwells basically (see below) at every 120°, or once per catalytic cycle, irrespective of [ATP]. We now enquire what causes these dwells. At least four events occur in a catalytic cycle of V1: ATP binding, ATP hydrolysis, phosphate release and ADP release. Of these, ATP binding must trigger, and likely drives at least partially, the 120° step. Our previous study12 with a slowly hydrolyzed ATP analogue ATP-γ-S indicated that ATP hydrolysis occurs at an ATP-waiting angle, and thus the time required for hydrolysis is a determinant of the dwell.
To see whether hydrolysis alone is responsible for the dwell, we have analysed the distribution of dwell times, measured as the time between the midpoints of two successive 120° steps (Fig. 2 f–i). At all four [ATP] examined, the dwell-time histogram was not exponential and rose from the origin (not well resolved at 4 μM), indicating the involvement of two or more rate-limiting reactions. Sequential two-reaction scheme could reasonably fit the histograms (orange lines in Fig. 2f–i). At 4 mM ATP, the two rates seemed indistinguishable and were 0.36 ms−1. One rate should correspond to that of ATP hydrolysis, unless a third reaction is also involved. The nature of the other reaction is unknown, but it cannot be ATP binding, which must be rapid at 4 mM ATP (binding rate for ATP is calculated as 3.2 ms−1 by multiplying 4 mM by 0.8×106 M−1 s−1). Likely candidates are phosphate or ADP release (or both combined).
At and below 200 μM ATP, the dwells must also involve the time for ATP binding in addition to the two (or more) reactions at 4 mM. We therefore attempted a global fit to the three histograms (Fig. 2f–h, blue lines) around Km where the rise from the origin was well resolved, with a sequential scheme for three reactions, of which one is ATP binding with the apparent rate constant kon. Although the fit was not perfect, the recovered kon of 1.2×106 M−1 s−1 is consistent with that for 4 μM ATP, and with the estimate from Figure 1c above and a previous value of ∼1.3×106 M−1 s−1 obtained with 220-nm duplex beads14. The other two rates were 0.49 ms−1 and 0.34 ms−1, roughly consistent with the two-rate fit of the 4 mM dwells above.
In addition to the relatively clean 120° steps as in Figure 2, some beads (52 out of 169; see Supplementary Table S1) exhibited peculiar fluctuations such as jumping to and fro between two angles separated by ∼40° (see Supplementary Fig. S1). Because the basic 120° stepping feature was preserved, we ignore these minor fluctuating beads in the analyses above.
Rotation of VoV1
To examine the effect(s) of the Vo domain on the ATP-driven rotation of V1 in intact VoV1, we constructed the experimental system in Figure 1b. VoV1 was fixed, in the presence of 0.05% (w/v) N-dodecyl β-D-maltoside (DDM) upside down on a Ni2+-NTA-coated glass surface via His3 tags on the Vo-c subunits. A 40-nm gold bead was attached to V1-A subunit(s) through the Avitag–biotin–streptavidin linkage. Immediately after infusion of millimolar ATP, we found a few rotating beads per field of view (7.1×7.1 μm2). The number decreased with time, particularly at high [ATP] where finding the rotation became difficult after 1 h. Both VoV1 and V1 are highly susceptible to ADP inhibition even in the presence of an ATP-regeneration system14,20. Part of the dormant molecules was somehow reactivated by re-infusion of the observation buffer, allowing further observations.
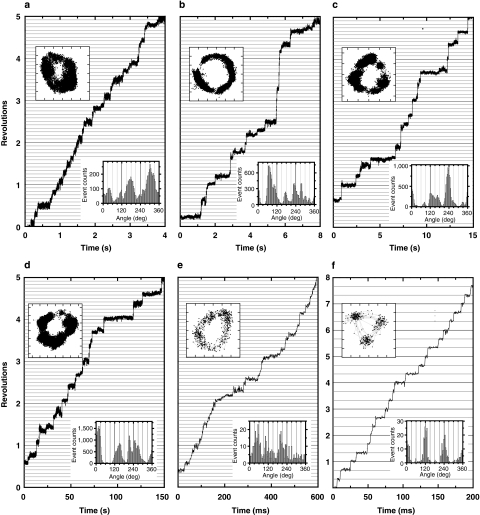
All molecules that rotated for many revolutions (as listed in Supplementary Table S1) without an obvious sign of obstruction at a particular angle were subjected to analysis. Rotation speed of VoV1 was variable and was distributed around 1–10 r.p.s. at 4 mM ATP (Fig. 1c). Typical rotation time courses are shown in Figure 3a–e. Unlike V1, which basically paused every 120°, VoV1 made short pauses at many angles at all [ATP] examined. A relatively fast rotation (∼10 r.p.s.) at 4 mM ATP is shown in Figure 3e, which still contains many pauses. At this [ATP], most V1 molecules rotated much faster, at ∼60 r.p.s. (Fig. 1c). The Vo domain seems to introduce bumps that lead to the small steps and the reduced average speed of VoV1 rotation. In this observation system, the whole stator apparatus (A3B3EGVo-a) rotates against the central rotor spanning the VoV1 (DFVo-d Vo-c ring). The bumps likely represent the interaction between Vo-c ring and Vo-a in the Vo domain. In 15 analysed molecules, we found three beads that showed clean 120° steps (Fig. 3f), and these beads (Fig. 1c, blue triangles) rotated fast (>∼30 r.p.s.). Detailed analyses of the short pauses in the presence of Triton below suggest that these 120° stepping beads are attached to defective VoV1 in which the Vo interaction is somehow impaired, although the opposite possibility of short pauses being an artefact cannot be ruled out.
Figure 3. Rotation of VoV1.
Typical time courses of the rotation of a 40-nm gold bead attached on VoV1 in the presence of 0.05% DDM. Horizontal lines are 30° apart, except in f. (a) Rotation at 4 mM ATP captured at 2,000 fps; (b) 40 μM ATP at 2,000 fps; (c) 4 μM ATP at 1,000 fps and (d) 400 nM ATP at 250 fps. (e) A relatively fast rotation (∼10 r.p.s.) with small substeps at 4 mM ATP captured at 1,000 fps. (f) A minor case of rotation with 120° steps at 4 mM ATP captured at 2,000 fps. Trajectories of the bead centroid (axis divisions: 11.1 nm) and histograms of angular positions for the indicated portion of the records are shown in the upper and lower insets, respectively.
Approximately 30° stepping
The detergent Triton X-100 (Triton) has been reported to be deleterious to the integrity of FoF1, presumably affecting stator–rotor interaction in Fo21. Unexpectedly, however, the substep behaviour of VoV1 above, indicative of rotor–stator interaction in the Vo domain, was enhanced when DDM was replaced with Triton. The small substeps could be more clearly discerned in the presence of Triton. When Triton-solubilized VoV1 was reconstituted into liposomes, it actively pumped protons, indicating that Triton treatment leaves VoV1 intact22. The same lot of VoV1 has also been shown to be inactivated by N,N′-dicyclohexylcarbodiimide14, another sign of integrity particularly in the Vo portion. Below, we analyze the clearer substeps observed in the presence of Triton.
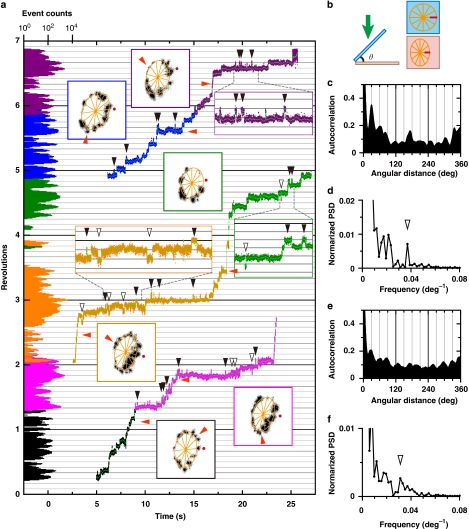
Somehow, rotation trajectory of VoV1 was unstable in the presence of a detergent, whether Triton or DDM, and gradually drifted both rotationally and translationally up to a few nanometres. Nevertheless, we could identify pauses clearly in trajectories of successive segments for one to two revolutions (Fig. 4a, square insets, with frames coloured as in the segmented time course). We could also estimate pausing angles by fitting an ellipse to each segmented trajectory and assuming that the ellipse represents the projection of a circular orbit oblique to the glass surface (Fig. 4b). The angular histogram of the time course is shown on the left axis of Figure 4a. In most parts, the histogram as well as the trajectories show dwells that occur every ∼30°, missing positions ascribed to rapid passage. An autocorrelation of the histogram, equivalent with the pairwise angular distribution function23,24, is shown in Figure 4c together with its power spectrum (Fig. 4d). The latter shows a peak at (27°)−1, indicated by the arrowhead at the resolution of ∼4°. In Figure 4e,f, the average of all autocorrelations of individual angular histograms and its power spectrum, including other examples of ∼30° step rotation shown in Figure 5 and Supplementary Figures S2a,b is shown. The power spectrum in Figure 4f shows a peak at (32°)−1.
Figure 4. Well-resolved substeps in VoV1.
(a) An expanded time course of the rotation of a 40-nm gold bead attached on a VoV1 at 40 μM ATP, in the presence of 0.1% (w/v) Triton captured at 2,000 fps. Horizontal lines are 30° apart. The time course is split into three and horizontally shifted (magenta and orange curves partially overlap). To minimize the effect of small, gradual drift on the angle analysis, the record was divided into six coloured portions (black, magenta, orange, green, blue and purple) covering ∼1 revolution and analysed as follows. First, the bead trajectory in each portion (coloured square insets; grey points show raw data and black after 21-point median filtering of x and y time courses) was fitted with an ellipsoid (orange). Rotary angle was calculated by assuming the ellipsoid to be a projection of a circular orbit (b). The angle 0, a start of a revolution on the vertical axis of the figure, was assigned to the red dot in each inset, chosen from the 12 orange spokes that fitted the dwells. The green line on the time courses shows 41-point (20 ms) median. The histograms on the left axis represent logarithm of the number of data points per 2°. Red arrowheads, dwells that are clearly out of the 30° periodicity. Black arrowheads, excursions to a neighbouring (closed, forward; open, backward) dwell position for >20° and >20 ms. Boxes enclosing trajectories show a fixed 89×89 nm2 area, such that drifts manifest as differences between insets. (b) Circular orbit (cyan) of a bead projected on the image plane (pink). Direction of observation is indicated by a green arrow. For the data in a, the angle θ ranged between 43° and 55°. (c) The autocorrelation of the angular histogram derived from a; the continuous time course over 2,500° was 21-point median filtered and then binned at 0.25° intervals. For this analysis, we calculated the angular histogram without adjusting the angular origins of the six portions, that is, without correction for the rotational drift, to eliminate possible subjectivity. (d) The power spectrum of c, the arrowhead showing a peak at (27°)−1. (e) The average of autocorrelations of individual angular histograms for Figures 4c, 5c and Supplementary Figures S2a,b. (f) The power spectrum of e, the arrowhead showing a peak at (32°)−1.
Figure 5. Substep rotation in upside up VoV1.
(a) Schematic observation system. VoV1 was fixed on a Ni2+-NTA-coated glass surface via His10 tags in the A subunits and a bead was attached to a biotinylated Vo-c subunit. This VoV1 had the TSSA mutation to suppress the ADPMg inhibition11. (b) Rotation observed at 2,000 fps at 40 μM ATP in the presence of 0.1% (w/v) Triton X-100. Horizontal lines are 30° apart. The time course is split into three and horizontally shifted (orange and green curves partially overlap). The record was divided into four coloured portions (black, magenta, orange and green) covering ∼1 revolution and analysed as described in Figure 4. The angle 0, a start of a revolution on the vertical axis of the figure, was assigned to the red dot in each inset, chosen from the 12 orange spokes that fitted the dwells. The green line on the time courses shows 21-point (10 ms) median. Boxes enclosing trajectories measure 66×66 nm2. Histogram bin size is 3°. (c) Autocorrelation of the angular histogram: the continuous time courses over 1,500° were 21-point median filtered and then binned at 0.25° intervals without the correction for rotational drift. (d) The power spectrum of c, the arrowhead showing a peak at (32°)−1.
In Figure 5, in particular, VoV1 was fixed upside up on a Ni2+-NTA-coated glass surface via His10 tags in the A subunits and beads were attached with biotinylated Vo-c subunit (see Fig. 5a). The ∼30° steps are not the consequences of the upside down configuration (Fig. 5b–d).
Taking into account the variations in the peak position in the individual power spectra, we conclude that substeps in VoV1 rotation are characterized by an amplitude between 27°–32°.
We noticed that some dwells were observed between two ∼30° dwell positions (Fig. 4, orange arrow heads). These may represent ATP-waiting dwells, because they were roughly 120° apart, taking the drift into account. If so, the ∼30° steps are not synchronous with ATP binding. This is not entirely unexpected, if the ∼30° steps arise from the stator–rotor interaction in the Vo domain, whereas ATP binding takes place in V1. As mentioned above, ATP-waiting dwells in VoV1 do not stand out even at low [ATP]. This suggests that the driving torque produced in the V1 portion, the torque that can drive the DF rotor of V1 over 120° in a matter of 0.25 ms or less, is sustained for many seconds while the Vo rotor slowly proceeds over the bumps presented by the Vo stator every ∼30°. An alternative, less likely scenario is that every ∼30° step is driven by ATP binding: because of friction in Vo, VoV1 works in a half-engaged clutch mode where 120° rotation in V1 results in ∼30° rotation in Vo.
We also noticed that, during a long dwell, momentary excursions to a neighbouring dwell position took place in either direction, mostly forward. In Figure 4a and Supplementary Figure S2, we indicate conspicuous excursions (amplitude >20° and duration >20 ms) with black arrowheads, counting 49 forward (closed arrowheads) and ten backward (open) ones in the total of 17 revolutions. The basically rectangular time courses seen in the expanded insets indicate metastable nature of the neighbouring dwell positions, consistent with bumps of structural origin as with the Vo-c and Vo-a interaction.
Discussion
We have characterized the ATP-driven rotation of both V1 and VoV1 under the conditions where the viscous drag between the probe and medium is negligible. For V1, the major results are that it pauses every 120° at all [ATP] (Fig. 2), implying that the pauses occur at ATP-waiting angles, and that at least two reactions other than ATP-binding limit each dwell. No dwells at other positions are resolved, at the resolution of ∼0.1 ms, in contrast to F1 that shows millisecond dwells at ∼80° past ATP-waiting angles.
The previous study using a mutated V1 and a slowly hydrolyzed ATP analogue suggested that hydrolysis in V1 occurs at 0° (ATP-waiting angle), as opposed to the 80° hydrolysis in F115, but absence of an 80° reaction(s) could not be demonstrated. In F1, another reaction, Pi release, takes place at ∼80°, contributing to the millisecond ∼80° dwells that are resolved even at saturating [ATP] if the temporal resolution is sufficiently high15,16. By contrast, the present results show that catalytic events in V1, at least those that take longer than a submillisecond, all occur at the ATP-binding position. At least two events other than ATP binding occur at this position, one likely to be ATP hydrolysis and the other phosphate or ADP release (or both combined). Together, it is safe to conclude that the canonical '80° and 40° scheme' for F1 does not apply to V1.
VoV1 shows significantly different rotation behaviours from that of V1. VoV1 rotated an order of magnitude slower. VoV1 did not show clear 120° steps as observed in V1, and instead exhibited short pauses separated by ∼30°. We could not judge whether the [ATP] dependence of the rotation speed of VoV1 follows simple Michaelis–Menten kinetics because of the large scatters in the data (Fig. 1c). At all [ATP], the rotary speed of VoV1 was significantly lower than that of V1. The bumps introduced by the Vo addition are high, such that passage has to wait for a rare thermal activation. The bumps also obscured ATP-waiting angles, although the angular histograms (Fig. 3, insets) indicate three broad peaks separated by ∼120°. Note that the ATP-waiting angles clearly observed in F1 or V1 represent the most stable orientation of the rotor in the ATP-waiting state. The rotor thermally fluctuates around this angle and actual ATP binding can take place at any point around the most stable angle25,26,27. In the presence of the Vo bumps, the motor would wait for ATP on either side of a bump16,28, resulting in more than three ATP-waiting angles.
The slow substep rotation of VoV1 observed here is at odds with our previous observation with a duplex of 220-nm beads on the same upside down system (the A subunits were mutated to render the enzyme less prone to MgADP inhibition)14: the average rate of rotation was ∼10 r.p.s. at saturating [ATP], and the molecules basically showed 120° stepwise rotation at low [ATP]. Defective interaction in the Vo domain could explain the discrepancy, although we are not sure if this was really the case.
The ∼30° steps that we resolve relatively clearly in the presence of Triton are commensurate with the periodicity of the Vo rotor ring in T. thermophilus V-ATPase7. It is highly likely that dwells result from specific interaction between a Vo-c subunit in the ring and the Vo-a subunit in the stator. When VoV1 works as an ATP-driven proton pump in a membrane, proton translocation occurs at the interface between Vo-c and Vo-a. It is possible that protons were also translocated in our experiment with detergent-solubilized VoV1 on a glass surface at one proton per ∼30° step.
The momentary excursions to a neighbouring ∼30° position reinforce that the ∼30° bumps are of structural origin. Presumably, ATP hydrolysis reaction in V1 domain sets up an energy slope that biases the thermal ride over bumps in the anticlockwise direction, and the elastic nature of the rotor29 helps go over the bumps. Note that this view alone does not account for the strong tendency to rotate back to the original dwelling position after an excursion: the original position is somehow more stable than that of its neighbours. An obvious explanation would be the stable positions being next to an ATP-waiting angle, which must pose an energy valley until the next ATP binds. Indeed, starting angles of the excursions are grossly clustered at ∼120° intervals, supporting this interpretation. The 120° intervals, however, were not strictly observed and there were excursions from other angles. These are likely statistical exceptions, but might point to a remote possibility that the rotor–stator interaction is not static and each time it is reconfigured, possibly accompanying protonation/deprotonation, to make the new position stable; until that happens, the previous position remains more stable.
Recently, Düser et al.30 have reported stepwise c-ring rotation relative to the stator a subunits, equivalent to Vo-a subunit of our VoV1, in E. coli FoF1 during ATP synthesis using single-molecule fluorescence resonance energy transfer. They estimate the step size as ∼36°, which is consistent with the proposed c subunit stoichiometry of 10 in E. coli FoF1. In their experiment, protons, presumably each one of them, directly drive the rotation of the Fo motor, whereas in our experiment the Vo motor is passively driven by the V1 motor and proton translocation would be the result and not the cause. The ∼30° steps we have observed indicate that passive interactions in the Vo domain, possibly coupled to proton translocation, check and set the pace of ATP-driven rotation.
Methods
Proteins
The His-tagged V1 (A(His−10/C28S/C508S)3B(C264S)3D(E48C/Q55C)F) was expressed in E. coli. After disruption of the cells by sonication, the his-tagged V1 was purified by Ni2+-affinity column (Qiagen) and RESOURCE Q column (GE healthcare)11. The purified his-tagged V1 was biotinylated at two cysteines using 6-[N-[2-(N-maleimide)ethyl]-N-piperazinylamide]hexyl-D-biotinamide (Dojindo). The VoV1 for rotation assay was obtained by reconstitution of the Vo containing a His3 tag in each Vo-c subunit and the Avi-Tagged V114. The bound ADP in V1 or VoV1 was partially removed by successive EDTA–heat treatments14.
Observation of rotation of 40-nm gold beads
Streptavidin-coated 40-nm gold beads and Ni2+-NTA-coated cover glass were prepared28,31. A flow cell (5–10 μl) was made of two coverslips: a Ni2+-NTA-coated bottom one (24×36 mm2) and an untreated top one (24×24 mm2) separated by two spacers of 50 μm thickness. The biotinylated V1 or Avitagged VoV1 (1–5 nM) in buffer A (50 mM Hepes-KOH, pH8.0, 100 mM KCl, with 0.05% (w/v) DDM only for VoV1) was applied to the flow cell and incubated for a few minutes. Unbound V1 or VoV1 was washed out with 20 μl of buffer A more than three times. Then, 20 μl of buffer A with 10 mg ml−1 BSA was infused to the flow cell and incubated for ∼30 s to prevent nonspecific binding. The BSA solution in the chamber was washed out with 20 μl of buffer A more than five times. Then, buffer A containing streptavidin-coated 40-nm beads (1010∼1011 particles per ml) were infused into the flow cell and incubated for a few min. Unbound gold beads were washed out with 20 μl of buffer A more than five times. After infusion of 80 μl of buffer A containing Mg-ATP at the indicated concentration, 2 mM MgCl2, 2.5 mM phosphoenol pyruvate and 0.5 mg ml−1 pyruvate kinase, bead rotation was observed at 23 °C by laser dark-field microscope15 on an inverted microscope (Olympus IX70) with a stable microscope stage (KS-O, Chuukoshaseisakujo), with some modifications28 (S. Furuike, unpublished): in place of the oblique laser-illumination15, the specimen was illuminated along the optical axis with parallel beam (diameter ∼10 μm, power <10 mW), by collimating a laser beam (Millennia IIs, Spectra Physics) with an objective placed just before the specimen. After the specimen was illuminated, the transmitted beam was let out through a pinhole at the centre of a mirror while the mirror deflected the scattered light to form a dark-field image of the beads. Images were captured with a high-speed CMOS camera (FASTCAM-DJV, Photron) at 250 to 8,000 frames per s as an 8-bit AVI file. Centroid of bead images was calculated15,16.
Author contributions
S.F. and K.Y. performed the experiments. M.N. and H.N. performed the sample preparation. S.F., K.K. and K.A. analysed the data. K.Y. designed the study. K.K., K.Y. and S.F. wrote the paper.
Additional information
How to cite this article: Furuike, S. et al. Resolving stepping rotation in Thermus thermophilus H+-ATPase/synthase with an essentially drag free probe. Nat. Commun. 2:233 doi: 10.1038/ncomms1215 (2011).
Supplementary Material
Supplementary Figures S1-S2 and Supplementary Table S1.
Acknowledgments
We thank M. Shio for the microscope technique; R. Chiwata and T. Ogawa for technical assistance; Y. Onoue, Y. Maki, H. Yoshida, H. Imamura and E. Saita for critical discussion; S. Takahashi for lab management; and members of Yoshida and Kinosita labs, ICORP in Odaiba for help and advice. This work was partly supported by Grant-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan to K.Y. (No. 21023009 and 21370042), Targeted Proteins Research Program (TPRP; B-37 to K.Y.), Young Scientists (B) to S.F., and Specially Promoted Research to K.K.
References
- Yokoyama K. & Imamura H. Rotation, structure, and classification of prokaryotic V-ATPase. J. Bioenerg. Biomembr. 37, 405–410 (2005). [DOI] [PubMed] [Google Scholar]
- Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 (2007). [DOI] [PubMed] [Google Scholar]
- Yoshida M., Muneyuki E. & Hisabori T. ATP synthase–a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2, 669–677 (2001). [DOI] [PubMed] [Google Scholar]
- Yokoyama K., Oshima T. & Yoshida M. Thermus thermophilus membrane-associated ATPase. Indication of a eubacterial V-type ATPase. J. Biol. Chem. 265, 21946–21950 (1990). [PubMed] [Google Scholar]
- Iwata M. et al. Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc. Natl Acad. Sci. USA 101, 59–64 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K. et al. Subunit arrangement in V-ATPase from Thermus thermophilus. J. Biol. Chem. 278, 42686–42691 (2003). [DOI] [PubMed] [Google Scholar]
- Toei M. et al. Dodecamer rotor ring defines H+/ATP ratio for ATP synthesis of prokaryotic V-ATPase from Thermus thermophilus. Proc. Natl Acad. Sci. USA 104, 20256–20261 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüber G., Wieczorek H., Harvey W. R. & Müller V. Structure-function relationships of A-, F- and V-ATPases. J. Exp. Biol. 204, 2597–2605 (2001). [DOI] [PubMed] [Google Scholar]
- Boyer P. D. The binding change mechanism for ATP synthase–some probabilities and possibilities. Biochim. Biophys. Acta 1140, 215–250 (1993). [DOI] [PubMed] [Google Scholar]
- Junge W., Lill H. & Engelbrecht S. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem. Sci. 22, 420–423 (1997). [DOI] [PubMed] [Google Scholar]
- Imamura H. et al. Evidence for rotation of V1-ATPase. Proc. Natl Acad. Sci. USA 100, 2312–2315 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura H. et al. Rotation scheme of V1-motor is different from that of F1-motor. Proc. Natl Acad. Sci. USA 102, 17929–17933 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K., Nakano M., Imamura H., Yoshida M. & Tamakoshi M. Rotation of the proteolipid ring in the V-ATPase. J. Biol. Chem. 278, 24255–24258 (2003). [DOI] [PubMed] [Google Scholar]
- Nakano M. et al. ATP hydrolysis and synthesis of a rotary motor V-ATPase from Thermus thermophilus. J. Biol. Chem. 283, 20789–20796 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda R., Noji H., Yoshida M., Kinosita K. Jr & Itoh H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 (2001). [DOI] [PubMed] [Google Scholar]
- Adachi K. et al. Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 130, 309–321 (2007). [DOI] [PubMed] [Google Scholar]
- Ueno H., Suzuki T., Kinosita K. Jr & Yoshida M. ATP-driven stepwise rotation of FoF1-ATP synthase. Proc. Natl Acad. Sci. USA 102, 1333–1338 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez M. et al. Proton-powered subunit rotation in single membrane-bound FoF1-ATP synthase. Nat. Struct. Mol. Biol. 11, 135–141 (2004). [DOI] [PubMed] [Google Scholar]
- Sambongi Y. et al. Mechanical rotation of the c subunit oligomer in ATP synthase (FoF1): direct observation. Science 286, 1722–1724 (1999). [DOI] [PubMed] [Google Scholar]
- Yokoyama K. et al. V-ATPase of Thermus thermophilus is inactivated during ATP hydrolysis but can synthesize ATP. J. Biol. Chem. 273, 20504–20510 (1998). [DOI] [PubMed] [Google Scholar]
- Tsunoda S. P., Aggeler R., Yoshida M. & Capaldi R. A. Rotation of the c subunit oligomer in fully functional FoF1 ATP synthase. Proc. Natl Acad. Sci. USA 98, 898–902 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K. et al. V-type H+-ATPase/synthase from a thermophilic eubacterium, Thermus thermophilus. Subunit structure and operon. J. Biol. Chem. 275, 13955–13961 (2000). [DOI] [PubMed] [Google Scholar]
- Svoboda K., Schmidt C.F., Schnapp B.J. & Block S.M. Direct observation of kinesin stepping by optical trapping interferometry. Nature 365, 721–727 (1993). [DOI] [PubMed] [Google Scholar]
- Abbondanzieri E. A., Greenleaf W. J., Shaevitz J. W., Landick R. & Block S. M. Direct observation of base-pair stepping by RNA polymerase. Nature 438, 460–465 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe-Nakayama T. et al. Effect of external torque on the ATP-driven rotation of F1-ATPase. Biochem. Biophys. Res. Commun. 366, 951–957 (2008). [DOI] [PubMed] [Google Scholar]
- Hirono-Hara Y., Ishizuka K., Kinosita K. Jr, Yoshida M. & Noji H. Activation of pausing F1 motor by external force. Proc. Natl Acad. Sci. USA 102, 4288–4293 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iko Y. et al. Acceleration of the ATP-binding rate of F1-ATPase by forcible forward rotation. FEBS Lett. 583, 3187–3191 (2009). [DOI] [PubMed] [Google Scholar]
- Hossain M. D. et al. Stimulation of F1-ATPase activity by sodium dodecyl sulfate. Biochim. Biophys. Acta 1797, 435–442 (2010). [DOI] [PubMed] [Google Scholar]
- Junge W., Sielaff H. & Engelbrecht S. Torque generation and elastic power transmission in the rotary FoF1-ATPase. Nature 459, 364–370 (2009). [DOI] [PubMed] [Google Scholar]
- Düser M. G. et al. 36 degrees step size of proton-driven c-ring rotation in FoF1-ATP synthase. EMBO J. 28, 2689–2696 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H. et al. Mechanically driven ATP synthesis by F1-ATPase. Nature 427, 465–468 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-S2 and Supplementary Table S1.