Abstract
Ubiquitin chains of different topologies trigger distinct functional consequences, including protein degradation and reorganization of complexes. The assembly of most ubiquitin chains is promoted by E2s, yet how these enzymes achieve linkage specificity is poorly understood. We have discovered that the K11-specific Ube2S orients the donor ubiquitin through an essential non-covalent interaction that occurs in addition to the thioester bond at the E2 active site. The E2-donor ubiquitin complex transiently recognizes the acceptor ubiquitin, primarily through electrostatic interactions. The recognition of the acceptor ubiquitin surface around Lys11, but not around other lysines, generates a catalytically competent active site, which is composed of residues of both Ube2S and ubiquitin. Our studies suggest that monomeric E2s promote linkage-specific ubiquitin chain formation through substrate-assisted catalysis.
Introduction
By regulating protein stability, activity, or localization, ubiquitination exerts control over almost every cellular process. As this includes pathways responsible for the duplication and separation of genetic material, aberrant ubiquitination often results in tumorigenesis. Despite the importance for cellular regulation, the mechanisms determining the specificity and efficiency of ubiquitination reactions are still incompletely understood.
Ubiquitination requires at least three enzymatic activities. An E1 enzyme forms a thioester between a cysteine at its active site and the C-terminus of ubiquitin (Schulman and Harper, 2009). The activated ubiquitin is transferred to a cysteine of an E2 (Ye and Rape, 2009). In the third step, the charged E2s cooperate with E3s to catalyze formation of an isopeptide bond between the C-terminus of ubiquitin and the ε-amino group of a substrate lysine (Joazeiro and Deshaies, 2009). The ~600 human RING-E3s interact with E2s and substrates at the same time, allowing them to promote the transfer of ubiquitin directly from the E2 to the substrate.
The modification of a substrate with a single ubiquitin usually leads to changes in protein interactions (Dikic et al., 2009). In many cases, additional ubiquitin molecules are attached to a substrate-linked ubiquitin, giving rise to polymeric ubiquitin chains. Such chains can be connected through the N-terminus of ubiquitin or through one of its seven Lys residues, and all linkages have been detected in cells (Ye and Rape, 2009; Xu et al., 2009). Ubiquitin chains of different topologies can have distinct structures and functions (Dikic et al., 2009; Matsumoto et al., 2010). K48-linked chains, for example, drive protein degradation, while K63-linked chains regulate the assembly of protein complexes (Ye and Rape, 2009). Thus, the efficiency and specificity of chain formation have profound consequences for the modified protein.
We recently identified K11-linked ubiquitin chains as critical cell cycle regulators in human cells (Jin et al., 2008). Most K11-linked chains are synthesized during mitosis by the E3 APC/C and its E2s Ube2C/UbcH10 and Ube2S (Williamson et al., 2009; Matsumoto et al., 2010; Wu et al., 2010). Together, these enzymes modify mitotic regulators, such as cyclin B, securin, or HURP, to trigger their degradation (Jin et al., 2008; Song and Rape, 2010). As a result, inhibiting the formation of K11-linked chain blocks mitotic progression in Xenopus, Drosophila, and humans (Jin et al., 2008; Williamson et al., 2009; Garnett et al., 2009), while their untimely assembly causes inaccurate cell division and tumorigenesis (Wagner et al., 2004; Jung et al., 2006). How the APC/C and its E2s assemble K11-linked chains, however, is poorly understood.
Linkage between two ubiquitin moieties involves the covalent connection of one ubiquitin, the ‘donor’, to the active site cysteine of the E2, followed by nucleophilic attack by a lysine of an acceptor ubiquitin (Figure 1A). Much of our knowledge about the basis of linkage specificity is limited to the E2 Ube2N-Uev1A (Ubc13-Mms2; VanDemark et al., 2001). In this system, the catalytically inactive Uev1A orients the acceptor ubiquitin, such that Lys63 of the acceptor is at the active site of Ube2N charged with the donor (Eddins et al., 2006). In contrast, K11- and K48-linkage-specific E2s promote chain elongation in reconstituted systems lacking UEVs (Li et al., 2009; Pierce et al., 2009; Williamson et al., 2009). Although kinetic analyses suggest that these E2s also engage acceptor ubiquitin residues (Petroski and Deshaies, 2005; Rodrigo-Brenni et al., 2010), the molecular details of acceptor recognition by monomeric E2s and its importance for linkage-specific chain formation have not been established.
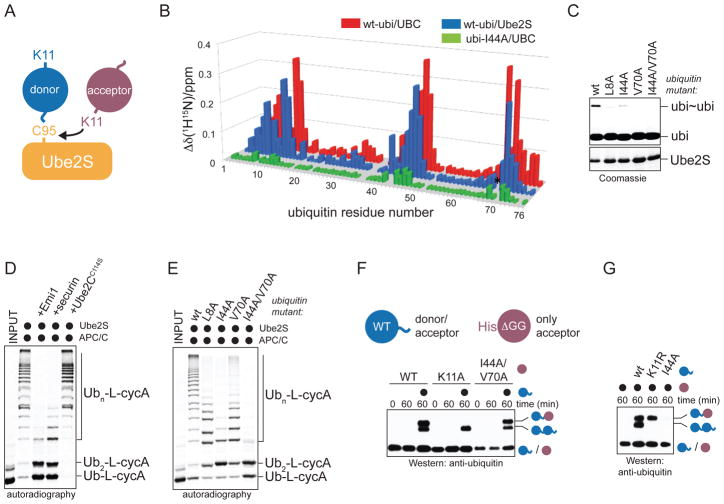
Figure 1. Ube2S recognizes the hydrophobic patch of donor ubiquitin.
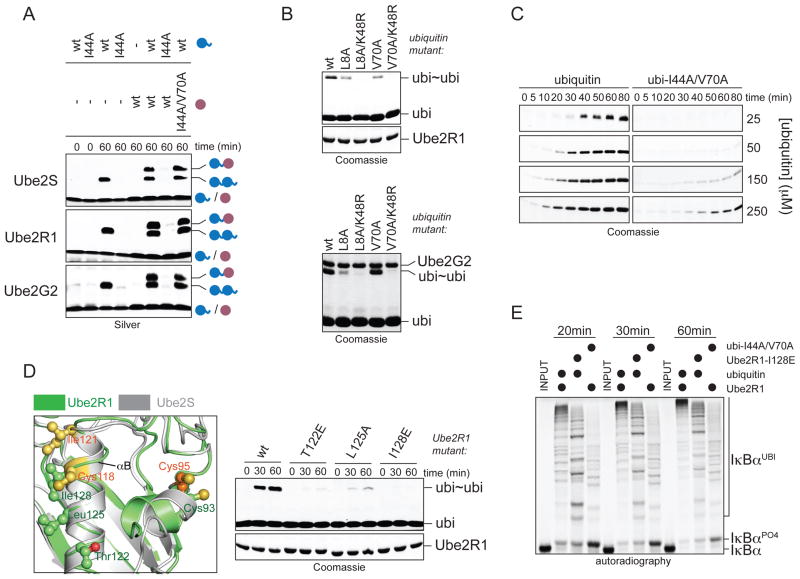
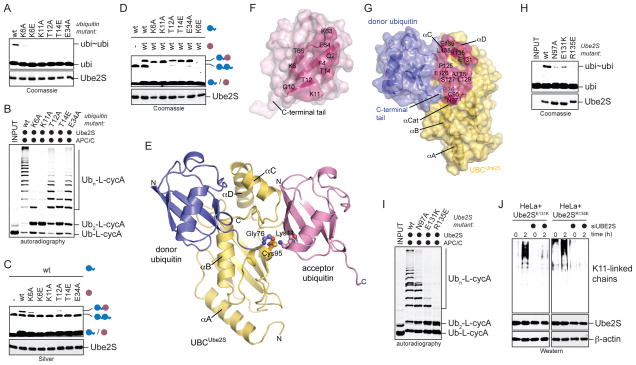
A. Overview of K11-specific linkage formation. Lys11 of acceptor ubiquitin attacks the thioester bond between Cys95 of Ube2S and the C-terminus of the donor ubiquitin. B. Ube2S interacts with ubiquitin non-covalently. Weighted combined chemical shift perturbations, ⊗™(1H15N), are plotted over residue number. The asterisk indicates the disappearance of the resonance for His68 of ubiquitin in the presence of Ube2S due to intermediate exchange. C. Mutation of the hydrophobic patch in ubiquitin interferes with formation of K11-linked ubiquitin dimers (ubi~ubi) by Ube2S, as monitored by Coomassie-staining. D. Ube2S and APC/C extend ubiquitin chains on a fusion between ubiquitin and cyclin A (Ub-L-cycA), as analyzed by autoradiography. E. The hydrophobic patch of ubiquitin is required for chain elongation by APC/CCdh1 and Ube2S, as analyzed by autoradiography. F. The hydrophobic patch is not required on acceptor ubiquitin. Ube2S was mixed with acceptor His6ubiquitinΔGG mutants (ubiΔGG; purple) and wt-ubiquitin (blue), and analyzed by αubiquitin-Western. G. The hydrophobic patch is required on the donor ubiquitin. Ube2S was mixed with wt-ubiΔGG and ubiquitin mutants, and analyzed by αubiquitin-Western.
Here, we have combined functional studies with NMR and computational docking to dissect the mechanism of linkage-specific chain formation by single-subunit E2s. We show that the K11-specific Ube2S orients the donor ubiquitin by a non-covalent interaction that is in addition to the flexible covalent linkage between these molecules at the E2 active site. We find that a similar tethering mechanism is used by other E2s independently of linkage specificity. The Ube2S-donor ubiquitin complex transiently engages the acceptor ubiquitin through electrostatic interactions. As indicated by our analysis, only binding of the acceptor surface around Lys11, but not around other lysines, leads to formation of a catalytically competent active site composed of residues of both Ube2S and ubiquitin. Hence, linkage-specific ubiquitin chain formation by Ube2S is the result of substrate-assisted catalysis.
Results
A non-covalent interaction with ubiquitin is required for Ube2S-activity
In the absence of the APC/C, Ube2S generates K11-linked ubiquitin dimers (ubi2) and chains attached to Lys residues of its UBC-domain and its C-terminal tail (Figure S1A). The UBC-domain of Ube2S (UBCUbe2S) promotes ubi2-formation with similar kinetics and specificity as Ube2S (Figure S1A). As Ube2S, UBCUbe2S is monomeric in ubiquitination buffers, as suggested by gel filtration, SAXS and other biophysical techniques (Figure S1B, C; data not shown). Thus, UBCUbe2S contains all elements required for the synthesis of K11-linkages, making it an appropriate system for analyzing the mechanism of linkage-specific chain formation.
The prevailing model of linkage-specific chain formation posits that an elongating E2, charged with the donor ubiquitin, binds the acceptor in such a way that a preferred acceptor lysine is at the E2 active site (Eddins et al., 2006). To test for such a non-covalent interaction between Ube2S and ubiquitin, we performed titrations of 15N-enriched ubiquitin with Ube2S and UBCUbe2S, respectively, and monitored 1H-15N HSQC spectra. The presence of either E2 caused significant resonance-specific chemical shift perturbations in the ubiquitin spectrum, indicating a specific interaction (Figure 1B).
Chemical shift mapping on the surface of ubiquitin revealed that the hydrophobic patch surrounding Ile44 is involved in the non-covalent interaction with Ube2S (Figure S2B). Mutation of the isoleucine to alanine (ubiI44A) disrupted the interaction with Ube2S (Figure 1B and 2A). Furthermore, mutating residues in the hydrophobic patch (L8A, I44A, V70A) interfered strongly with the formation of K11-linked ubi2 by Ube2S (Figure 1C).
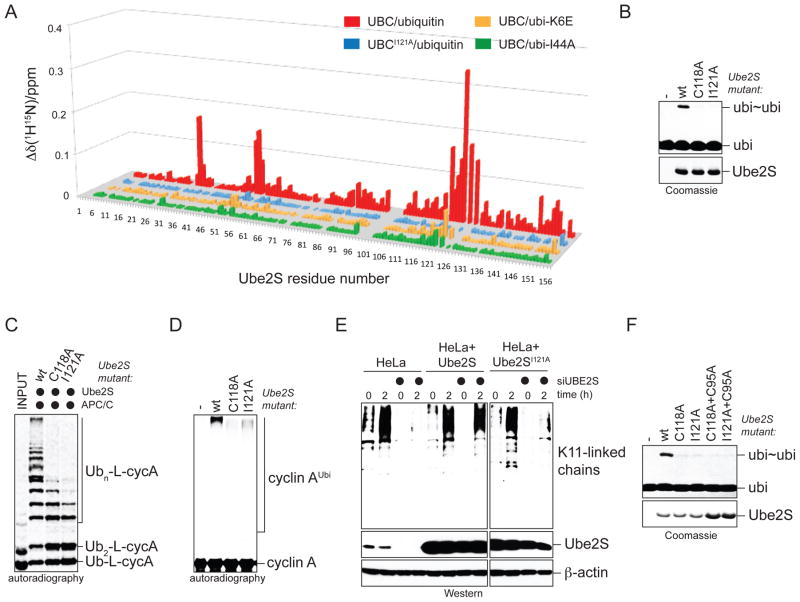
Figure 2. Non-covalent donor ubiquitin binding is required for Ube2S activity.
A. Identification of Ube2S-residues involved in non-covalent binding of ubiquitin. Weighted combined chemical shift perturbations, ⊗™(1H15N), are plotted over the residue number. B. Donor-binding is required for formation of ubi2-dimers (ubi~ubi) by Ube2S mutants, as analyzed by Coomassie-staining. C. Donor-binding by Ube2S is required for chain elongation on Ub-L-cycA with APC/C, as analyzed by autoradiography. D. Donor-binding by Ube2S is required for chain formation in a full APC/C-assay. Ubiquitination of cyclin A by APC/C, Ube2C, and Ube2S mutants was analyzed by autoradiography. E. Donor- binding is required for Ube2S-activity in vivo. HeLa cell lines expressing Ube2S or Ube2SI121A were treated with siRNAs against the 3′-UTR of Ube2S, which specifically depletes endogenous Ube2S. Cells were synchronized in prometaphase (t=0h) or late mitosis (t=2h), and K11-linked ubiquitin chains were detected by αK11-Western. F. Donor-binding occurs in cis. Ube2SC118A or Ube2SI121A (lack the non-covalent ubiquitin-binding site) and Ube2SC95A (no active site) were mixed, and ubi2-formation was monitored by Coomassie staining.
Ube2S extends K11-linked chains on APC/C-substrates after initiation by Ube2C (Williamson et al., 2009; Wu et al., 2010). To test whether mutations in ubiquitin interfere with chain elongation by Ube2S and APC/C, we bypassed the need for Ube2C by generating a fusion between ubiquitin and the APC/C-substrate cyclin A (Ub-L-cycA). Ube2S and APC/C rapidly elongated ubiquitin chains on Ub-L-cycA, which did not require Ube2C and was not inhibited by an excess of inactive Ube2CC114S (Figure 1D). Mutation of the hydrophobic patch of ubiquitin interfered strongly with this activity of Ube2S (Figure 1E; Figure S1D), without affecting charging by E1 (Figure S1F). The same mutations in ubiquitin blocked ubiquitination of APC/C-substrates in an assay containing both Ube2C and Ube2S (Figure S1E). Thus, a non-covalent interaction with ubiquitin is required for the ability of Ube2S to assemble K11-linked ubiquitin chains.
The hydrophobic patch is required on the donor ubiquitin
To determine whether Ube2S interacts with donor or acceptor ubiquitin, we made a ubiquitin mutant lacking its two C-terminal Gly residues, ubiΔGG. UbiΔGG is not activated by E1 and can only act as acceptor. Ube2S produced dimers between ubiΔGG and ubiquitin (ubiΔGG-ubi; Figure 1F), and mutation of Lys11 on the acceptor ubiΔGG, but not the donor ubiquitin, blocked this reaction (Figure 1F, G). The mutation of Leu8, Ile44, or Val70 on the acceptor ubiΔGG had no effect on the formation of ubiΔGG-ubi dimers (Figure 1F; Figure S1G). Instead, when the hydrophobic patch was mutated on the donor ubiquitin, dimer formation was prevented (Figure 1G).
We conclude that Ube2S recognizes the donor ubiquitin. Several aspects of our analysis indicate that this is the only thermodynamically stable interaction between ubiquitin and Ube2S in solution. The NMR-derived binding isotherms are well described by a single-site binding model (Figure S2D); significant chemical shift perturbations map to one contiguous binding region (Figure S2B); and disruption of the donor interface did not result in the population of an alternate binding site (Figure 1B). Variation of the experimental conditions, such as ionic strength and pH, also did not provide evidence for a second binding site (data not shown). Thus, Ube2S forms a non-covalent interface with the hydrophobic patch of the donor ubiquitin, which is required for its activity to promote the formation of K11-linked ubiquitin chains.
The donor ubiquitin interacts with helix αB of Ube2S
We identified the donor-binding site on Ube2S by titrating 15N-enriched UBCUbe2S with ubiquitin and measuring 1H-15N HSQC spectra. Significant ubiquitin-induced chemical shift perturbations mapped to a surface region around the C-terminal part of helix αB of Ube2S (Figure 2A, S2B). The same region was found to interact with covalently bound donor ubiquitin (Figure S2A). Mutation of two Ube2S-residues in this region (C118A, I121A) impaired ubiquitin binding (Figure 2A, S2C), without affecting the structural integrity of Ube2S (data not shown). The KD for this interaction (1.11 +/− 0.08 mM or 1.7 +/− 0.08 mM for ubiquitin binding to UBCUbe2S or Ube2S; Figure S2D) was comparable to the estimated concentration of donor ubiquitin linked to the Ube2S active site (~3mM; Petroski and Deshaies, 2005). Our results, therefore, suggest that covalently linked donor ubiquitin occupies the non-covalent binding site on Ube2S around helix αB.
Based on our previous results, we expected the donor-interface of Ube2S to be required for activity. Indeed, Ube2SC118A and Ube2SI121A were strongly impaired in ubi2-formation (Figure 2B); K11-linked chain assembly on Ub-L-cycA with APC/C (Figure 2C); or modification of cyclin A in an APC/C-assay containing Ube2C and Ube2S (Figure 2D). Disrupting this Ube2S surface did not affect charging by E1 (Figure S2E) or binding to the APC/C (Figure S2F).
To test for the importance of donor-binding at physiological ubiquitin levels, we generated HeLa cell lines that stably express Ube2S or Ube2SI121A. Endogenous Ube2S was specifically depleted from cells by siRNAs against the 3′-UTR of the Ube2S mRNA, and formation of K11-linked chains was monitored upon exit from mitosis by a K11-linkage specific antibody. As expected, long K11-linked chains were absent from cells lacking Ube2S, which was rescued by expression of siRNA-resistant wt-Ube2S (Figure 2E). By contrast, the donor-binding deficient Ube2SI121A failed to promote K11-linked chain formation. Thus, recognition of the donor ubiquitin by Ube2S is essential for K11-linked ubiquitin chain formation in vitro and in vivo.
NMR-based docking of the donor ubiquitin on Ube2S
The binding site for the donor ubiquitin on Ube2S, as defined by chemical shift mapping, makes it plausible that this interaction occurs in cis, i.e. involves the same E2 that the donor is covalently attached to. To test this idea, we determined whether a catalytically inactive Ube2S-mutant with an intact donor-binding site (Ube2SC95A) could complement the loss-of-function phenotype of a Ube2S-mutant with a defective non-covalent interface (Ube2SC118A, Ube2SI121A). As this was not case (Figure 2F), the observed non-covalent interaction most likely occurs in cis.
To obtain a structural model of the interaction between UBCUbe2S and donor ubiquitin, we used the docking program HADDOCK (de Vries et al., 2007). The NMR chemical shift data were used to specify residues at the interface, and we defined only one explicit distant restraint that required the C-terminal carbon atom of Gly76 of ubiquitin to be close to the Sγ atom of Cys95 in Ube2S. HADDOCK produced an ensemble of 200 structures after automated refinement, which were clustered using a backbone RMSD cut-off of 7.5 Å. The resulting three clusters contain 71%, 25.5%, and 2.5% of all docked models, respectively (Table S1). As shown later, structures in cluster 1, but not those of clusters 2 and 3, could be validated by biochemical data.
As a representative structure of the Ube2S-donor ubiquitin complex, we selected a model from cluster 1 that among the top 3 according to HADDOCK scoring had the largest buried surface area, the most negative interaction energy and the smallest number of distant restraint violations (Figure 3A; Table S1). A similar model with a low backbone RMSD of 1.1Å was obtained by a different docking program, Cluspro, without restraints (Figure S3A; Comeau et al., 2007). Our model resembles the structure of the charged E2 Ubc9, when bound to its E3 (Reverter and Lima, 2005), and an NMR-based, docked model of the E2 Ubc1 and ubiquitin (Hamilton et al., 2001).
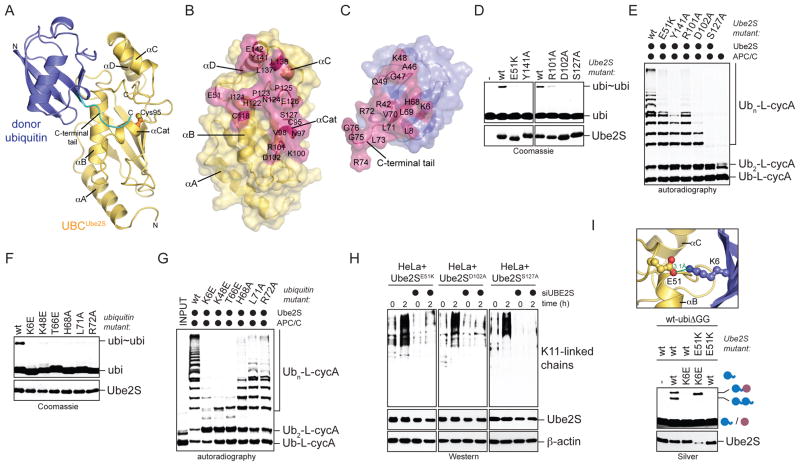
Figure 3. Structural model of the Ube2S-donor ubiquitin complex.
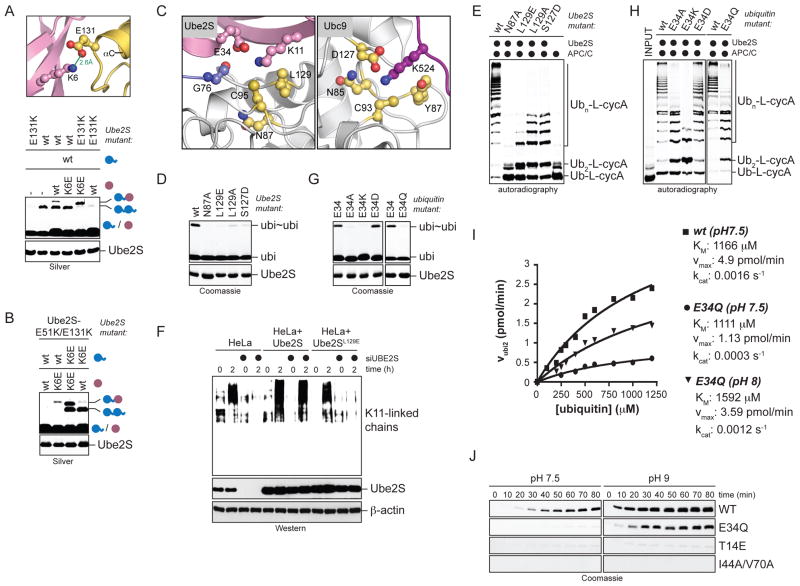
A. NMR-based HADDOCK model of the UBCUbe2S-donor ubiquitin complex (cluster 1, no. 3; see Table S1). The C-terminal tail of ubiquitin (cyan) was allowed full flexibility during docking. B, C. Surface representation of the binding interface on UBCUbe2S (B) and donor ubiquitin (C). Residues that make intermolecular contacts within a radius of 4 Å are shown in pink. D. Ube2S-residues at the donor-binding interface are required for formation of ubi2-dimers (ubi~ubi), as monitored by Coomassie-staining. E. Donor-binding deficient Ube2S-mutants do not promote chain elongation on Ub-L-cycA with APC/C, as analyzed by autoradiography. F. Ubiquitin-residues at the Ube2S-interface are required for linkage formation, as seen by Coomassie staining. G. Ubiquitin residues at the Ube2S-interface are required for chain elongation on Ub-L-cycA with APC/C, as analyzed by autoradiography. H. Donor-binding is required for Ube2S-activity in cells. HeLa cell lines expressing donor-binding deficient Ube2S (E51K; D102A; S127A) were depleted of endogenous Ube2S, synchronized in prometaphase (t=0) or allowed to exit mitosis (t=2h), and tested for K11-linked ubiquitin chains by αK11-Western. I. Charge-swap analysis of the ionic interaction between Lys6 of donor ubiquitin and Glu51 on Ube2S. ubiΔGG was mixed with ubiquitin or ubiK6E in the presence of Ube2S or Ube2SE51K. Reactions were monitored by Silver staining.
Within our model, the donor ubiquitin docks onto a hydrophobic area on Ube2S, comprising the N-terminal half of helix αB, the C-terminal part of helix αC and the N-terminal part of helix αD (Figure 3A, B). The corresponding interaction surface on donor ubiquitin contains the hydrophobic patch (Figure 3C), which is extended to form a contact area that buries a total of ~830 Å2 on ubiquitin. The model includes ionic contacts between Lys6, Arg42, and Lys48 of ubiquitin, and Glu51, Glu126, and Glu142 of Ube2S (Figure S3B). An ionic contact between Arg74 of ubiquitin and Asp102 of Ube2S serves as a linchpin to guide the C-terminus of ubiquitin towards the active site of Ube2S (Figure S3C). The ubiquitin tail is also anchored by hydrogen bonds between the peptide backbone and Ube2S residues close to the active site. The distance between the Sγ atom of Cys95 of Ube2S and the C-terminal carbon atom of ubiquitin, 3.9 Å, is too long for a covalent bond, but small adjustments around the active site of Ube2S could readily close this gap.
Validation of the Ube2S-donor ubiquitin model
Based on the selected model for this complex, we designed additional mutations to test the structural details of the predicted interface. We found that altering residues at the binding interface (Ube2S: E51K, R101A, D102A, S127A, Y141A; ubiquitin: K6E, K48E, T66E, H68A, L71A, R72A) interfered with Ube2S-activity in vitro (Figure 3D–G), and as seen for ubiK6E, disrupted the Ube2S-donor ubiquitin interaction (Figure 2A). Residues that do not make intermolecular contacts (Ube2S: D29, G30, L114, E142; ubiquitin: A46) were not required for activity (data not shown). Introducing mutations into ubiΔGG showed that most ubiquitin residues were required in the donor, but not the acceptor (Figure S3D, S3E). The role of Lys6 in the acceptor ubiquitin is described below. With the exception of ubiR72A, no Ube2S or ubiquitin mutant was impaired in charging by E1 (Figure S3F, S3G). To confirm this analysis in vivo, we generated cell lines that express Ube2S-mutants with defective binding donor-interfaces (E51K; D102A; S127A). Importantly, all of these failed to promote the formation of K11-linked ubiquitin chains in HeLa cells that lacked endogenous Ube2S (Figure 3H).
To further test our model, we analyzed the role of the predicted ion pair between Glu51 of Ube2S and Lys6 of ubiquitin by charge-swap analysis. While the K6E-mutation in donor ubiquitin interfered with formation of ubiΔGG-ubi dimers, this was rescued by a complementary mutation in Ube2S, Ube2SE51K (Figure 3I). Ube2SE51K did not establish ubi2-formation for other ubiquitin mutants, such as ubiI44A, attesting to the specificity of this rescue (Figure S3H). Together, the mutational studies, charge-swap analysis, and in vivo experiments validate the selected NMR-based model for the Ube2S-donor ubiquitin interaction and show its importance for chain formation by this E2.
Non-covalent donor ubiquitin binding is required for processive chain formation
We next determined the role of donor-binding for catalysis by Ube2S. It was unlikely that recognizing the donor ubiquitin was important for specificity, and indeed, any ubi2 formed in the presence of ubiI44A was lost upon mutation of K11 (Figure 4A). The same was observed when Ube2S-mutants with a defective donor-binding interface (Ube2SI121A; Ube2SC118A; Ube2SE51K) were analyzed for ubi2-formation (Figure 4A). Thus, donor-binding does not determine the K11-specificity of Ube2S.
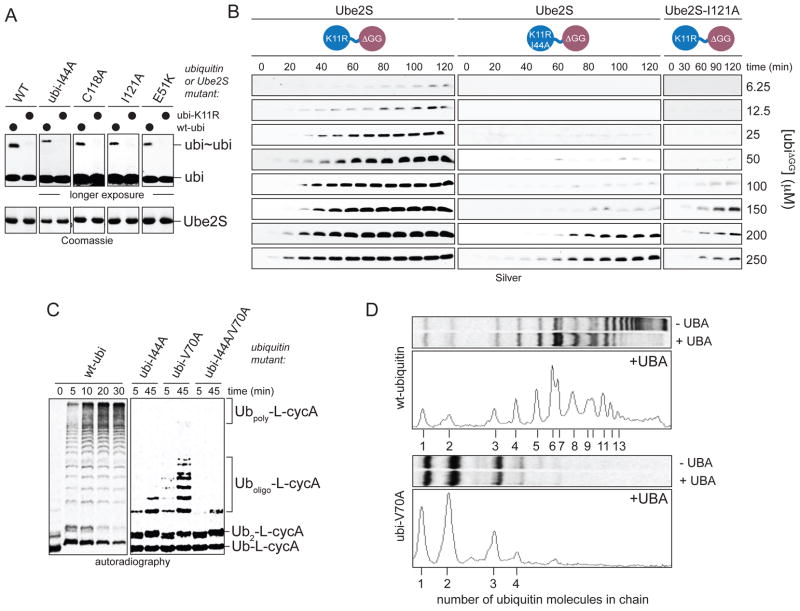
Figure 4. Non-covalent donor-binding increases the processivity of Ube2S.
A. Donor-binding is not required for the K11-linkage specificity of Ube2S. Ube2S or donor-binding mutants were incubated with ubiquitin or ubiK11R, or as indicated with ubiI44A and ubiI44A/K11R. Reactions were incubated longer and at higher ubiquitin concentrations to observe formation of ubi2 and analyzed by Coomassie staining. B. Donor-binding promotes catalysis at low acceptor concentrations. Dimer formation between increasing levels of ubiΔGG and ubiK11R or ubiK11R/I44A, respectively, by Ube2S was monitored by Silver staining. C. Time-course analysis of chain elongation on Ub-L-cycA by APC/CCdh1 and Ube2S in the presence of ubiquitin mutants, as analyzed by autoradiography. D. Donor binding is required for processive chain formation by Ube2S. Chain elongation on Ub-L-cycA by APC/C and Ube2S was monitored in the presence of the UBA-domains of Rad23A. Reactions were performed with ubiquitin or ubiV70A and analyzed by autoradiography (top) and line-scanning (bottom).
Alternatively, the non-covalent interaction between the donor and Ube2S might prevent a flexible donor molecule from interfering with acceptor recognition. If this were the case, higher concentrations of the acceptor ubiΔGG should rescue the defect in ubi2-formation when the Ube2S-donor ubiquitin interface is disturbed. Consistent with this hypothesis, high levels of ubiΔGG allowed linkage-formation with ubiI44A or Ube2SI121A (Figure 4B). The acceptor concentration required under these conditions was above the endogenous ubiquitin levels in HeLa cells (90 μM; Ryu et al, 2006), consistent with the lack of Ube2SI121A-activity in vivo. These findings suggest that non-covalent binding of the donor ubiquitin facilitates acceptor recognition by Ube2S.
Based on these observations, we expected that donor binding would increase the processivity of chain formation by Ube2S. Indeed, a time-resolved analysis of chain elongation on Ub-L-cycA suggested that Ube2S assembles chains with high processivity (Figure 4C), whereas chain formation occurred in a step-like, distributive fashion if donor-binding was impaired (Figure 4C). To directly measure the processivity of chain elongation, we supplied the reactions with a UBA-domain. As previously described (Rape et al., 2006), the UBA-domain captures any substrate dissociating from the APC/C, thereby preventing it from re-binding the E3 and revealing the number of ubiquitin molecules transferred in a single substrate binding event. Ube2S could transfer up to ~13 ubiquitin molecules to Ub-L-cycA per binding event (Figure 4D, top), while less than four molecules of the hydrophobic patch mutant ubiV70A were transferred (Figure 4D, bottom). As this UBA-domain only recognizes K11-chains with at least ~5 ubiquitin moieties (data not shown), the number of ubiV70A-molecules transferred in a single binding event is likely even smaller. Thus, the non-covalent interaction between Ube2S and donor-ubiquitin increases the processivity of chain formation, at least in part by facilitating acceptor ubiquitin recognition.
Non-covalent donor ubiquitin binding is a feature of chain elongation in other E2s
To test whether other E2s bind the donor ubiquitin non-covalently, we turned to Ube2R1 and Ube2G2, which extend K48-linked chains (Li et al., 2009; Pierce et al., 2009). Disruption of the hydrophobic patch on ubiquitin strongly impaired the formation of K48-linkages by these E2s (Figure S4A), while having no effect on their charging by E1 (Figure S4B). Analogous to our results for Ube2S, we found that the activity of Ube2R1 and Ube2G2 was dependent on recognition of the hydrophobic patch on the donor, but not the acceptor ubiquitin (Figure 5A). As revealed by ubiI44A/K48R- and ubiV70A/K48R-double mutants, non-covalent donor-binding did not determine linkage-specificity (Figure 5B), but was required for catalysis at low substrate concentrations (Figure 5C).
Figure 5. Non-covalent donor-binding is utilized by E2s independently of linkage specificity.
A. Ube2R1 and Ube2G2 require the hydrophobic patch in the donor, but not acceptor ubiquitin, for K48-linkage formation. Ube2S, Ube2R1, or Ube2G2 and its E3 gp78, were incubated with ubiΔGG or ubiΔGG/I44A/V70A (purple) and ubiquitin or ubiI44A (blue). Reactions were analyzed by Silver staining. B. The hydrophobic patch of donor ubiquitin is not required for K48-specificity of Ube2R1 or Ube2G2. ubi2-formation by Ube2R1 or Ube2G2/gp78 with ubiquitin mutants was analyzed by Coomassie staining. C. Donor-binding is required for rapid catalysis by Ube2R1. Time courses of ubi2-formation by Ube2R1 in the presence of increasing concentrations of ubiquitin or ubiI44A were analyzed by Coomassie staining. D. A similar surface as the donor-binding interface of Ube2S (grey) is required in Ube2R1 (green; PDB ID: 2OB4). Ube2R1 mutants were analyzed for K48-specific ubi2-formation by Coomassie staining. E. Donor-binding is required for processive chain formation by SCFβTrCP and Ube2R1. Phosphorylated IκBα was incubated with SCFβTrCP, Ube2R1 or Ube2R1I128E, and ubiquitin or ubiI44A/V70A and analyzed by autoradiography.
To test whether a common E2-surface recognizes the donor ubiquitin, we studied Ube2R1 mutations of sites that are structurally homologous to the Ube2S-donor interface (Figure 5D). These mutations (Ube2R1T122E; Ube2R1L125A; Ube2R1I128E) strongly inhibited the formation of K48-linkages (Figure 5D; Figure S4C), without affecting charging by E1 (Figure S4D). The same mutations also impaired the SCF- and UbeR1-dependent formation of ubiquitin chains on IκBα (Figure 5E). Thus, similar surfaces on the conserved E2 fold and ubiquitin are used for chain elongation by E2s of different linkage specificity. The tethering of the donor ubiquitin by an E2, therefore, provides a conserved mechanism to facilitate acceptor recognition.
Ube2S recognizes the TEK-box in acceptor ubiquitin
We next identified the binding site for the acceptor ubiquitin on Ube2S. Consistent with previous analyses (Petroski and Deshaies, 2005; Rodrigo-Brenni et al., 2010), acceptor binding was too transient to be detected by NMR, independently of whether the donor ubiquitin had been linked to the Ube2S active site or not (data not shown). We therefore used HADDOCK to dock a second ubiquitin molecule, the acceptor, onto the validated Ube2S-donor ubiquitin complex. The only restraint used for this docking defined the Nz-atom of the acceptor Lys11 to be close to the Sγ atom of Cys95 at the Ube2S active site. HADDOCK generated two clusters of models, which were similar in terms of energy and buried surface area (Figure S5; Table S2).
Intriguingly, models in cluster 1 orient the TEK-box of ubiquitin towards the active site of Ube2S. The TEK-box is a surface region of ubiquitin that was previously identified to mediate the preference of Ube2C/UbcH10 for assembling K11-linked chains (Jin et al., 2008). We found that mutation of the TEK-box strongly interfered with the ability of Ube2S to synthesize K11-linked ubi2 (Figure 6A), to elongate chains on Ub-L-cycA (Figure 6B), and to modify cyclin A in a full APC/C-assay (Figure S6A). Residues outside of the TEK-box (Thr9, Glu16, Lys33) were not required for activity (data not shown). Introducing mutations into ubiΔGG revealed that the TEK-box was essential on the acceptor (Figure 6C), but with exception of Lys6, not the donor ubiquitin (Figure 6D). The TEK-box was dispensable for Ube2S-charging by E1 (Figure S6B) or binding of donor ubiquitin to Ube2S (Figure S6C). Thus, the TEK-box of the acceptor ubiquitin is required for K11-linkage formation by Ube2S.
Figure 6. Acceptor ubiquitin recognition by the Ube2S-donor ubiquitin complex.
A. The TEK-box in ubiquitin is required for K11-linkage formation by Ube2S. Ube2S was incubated with ubiquitin mutants, and ubi2-formation (ubi~ubi) was monitored by Coomassie staining. B. TEK-box mutants in ubiquitin inhibit chain formation on Ub-L-cycA by Ube2S and APC/C, as analyzed by autoradiography. C. The TEK-box is required on acceptor ubiquitin. ubiΔGG mutants (purple) were incubated with ubiquitin (blue) and Ube2S, and analyzed by Silver staining. D. The TEK-box is not required in donor ubiquitin. TEK-box mutants of ubiquitin were mixed with wt-ubiΔGG and Ube2S, and reactions were analyzed by Silver staining. E. HADDOCK-based model of the ternary complex between the UBCUbe2S (yellow), donor ubiquitin (blue), and acceptor ubiquitin (pink; cluster 1, no. 1; see Table S2). F. Surface representation of the Ube2S-binding interface on acceptor ubiquitin. Contact residues within a radius of 4 Å are shown in pink. G. Surface representation of the acceptor-binding interface on the Ube2S-donor complex. H. Acceptor binding is required for Ube2S-activity. Ube2S mutants were incubated with ubiquitin and analyzed by Coomassie-staining. I. Ube2S-residues at the acceptor-interface are required for chain elongation by APC/C. The modification of Ub-L-cycA by APC/C and Ube2S mutants was analyzed by autoradiography. J. Ube2S-residues at the acceptor-binding interface are required in vivo. HeLa cell lines expressing Ube2SE131K and Ube2SR135E were treated with siRNAs to deplete endogenous Ube2S, arrested in prometaphase (t=0h) or allowed to exit mitosis (t=2h), and tested for K11-linked chains by αK11-Western.
On the basis of these results, we performed another docking run that defined four TEK-box residues to be at the interface (Table S2). All HADDOCK solutions grouped into a single cluster reproducing the binding topology of cluster 1 of the previous run (Figure 6E). With backbone RMSD values of ~1 Å, the best models of this ensemble were remarkably similar and we focused on the top-scoring model (cluster 1, no. 1; Table S2).
In this model of the ternary complex, the acceptor ubiquitin interacts mostly with Ube2S (Figure 6E), with a total buried surface area between the acceptor ubiquitin and the Ube2S-donor complex of ~980 Å2 (Figure 6F, G). The acceptor ubiquitin uses its βstrand region to bind a surface of Ube2S comprising the active site helix αCat, the loop region between helix αB and αC, and helix αC (Figure 6F). Although the interaction leads to the burial of few hydrophobic residues, these are not closely packed, and the interface is predominantly electrostatic. In particular, a network of ionic contacts involving Lys6 and Lys63 of ubiquitin and Glu131 and Glu139 of Ube2S, and a series of hydrogen bonds including Glu64 of ubiquitin and Arg135 of Ube2S are key features of the interface (Figure S6D). The electrostatic Ube2S-acceptor interface is consistent with the low affinity between these molecules (Sheinerman and Honig, 2002).
Mutating residues in the predicted binding site on Ube2S (N97A, E131K, R135E), but not outside of this interface (K76, N91, E93, K100, E126, E132), impaired production of ubi2 (Figure 6H; data not shown), chain elongation on Ub-L-cycA (Figure 6I), and substrate-modification in a full APC/C-assay (Figure S6E). As seen in cells expressing Ube2SE131K or Ube2SR135E, these mutations also inhibited formation of K11-linked chains in vivo (Figure 6J). Asn97, Glu131, and Arg135 were not required for Ube2S-charging by E1 (Figure S6F) or Ube2S-binding to the APC/C (Figure S2F).
We next probed the predicted ionic contacts between Ube2S and the acceptor ubiquitin by charge-swap analyses. Our model found the acceptor Lys6 to face Glu131 of Ube2S (Figure 7A). Consistent with this, the loss of ubiΔGG-ubi formation caused by a K6E-mutation in the acceptor ubiΔGG could be recued by Ube2SE131K or Ube2SE131A (Figure 7A; Figure S7A), but not by other Ube2S-mutants in this interface (N97A, R135E; Figure S7B). Thus, the acceptor Lys6 is recognized by Glu131 of Ube2S, while the donor Lys6 contacts Glu51 of Ube2S (Figure 3); indeed, Ube2SE51K/E131K completely rescued ubi2-formation by ubiK6E (Figure 7B; Figure S7C). Our model also showed Arg135 of Ube2S in proximity of Glu64 of ubiquitin. Accordingly, the diminished activity of Ube2SR135E was significantly rescued by ubiE64K (Figure S7D), but not other ubiquitin mutants in the proximity of this surface (Figure S7E). Finally, less ubi2 was formed in the presence of ubiK63E, which could be rescued by mutation of the opposing Glu139 of Ube2S (Figure S7F). The mutational and charge-swap analyses provide strong support for our model of acceptor recognition by Ube2S.
Figure 7. Substrate-assisted catalysis contributes to the K11-linkage specificity of Ube2S.
A. Charge swap analysis of the ionic contact between acceptor Lys6 and Glu131 of Ube2S. Ube2S or Ube2SE131K were mixed with ubiΔGG or ubiΔGG/K6E and reactions were analyzed by Silver staining. B. Ube2SE51K/E131K rescues mutation of Lys6 in both acceptor and donor ubiquitin. Lys6 was mutated in acceptor ubiΔGG (purple) or donor ubiquitin (blue), and ubiΔGG-ubi formation by Ube2S or Ube2SE51K/E131K was analyzed by Silver staining. C. Ube2S (left) and Ubc9 (PDB ID: 2GRN; right) show similar active site constellations. The highest scoring Ube2S-model of the HADDOCK run in the absence of ambiguous restraints is shown (Table S2 top; cluster 1, no 1). D. Candidate active site-residues are required for the activity of Ube2S to catalyze ubi2-formation (ubi~ubi), as analyzed by Coomassie staining. E. Active site-residues in Ube2S are required for chain elongation by APC/C. Ub-L-cycA was incubated with APC/CCdh1 and Ube2S mutants and analyzed by autoradiography. F. Leu129 is required for Ube2S-activity in vivo. HeLa cell lines expressing Ube2S or Ube2SL129 were tested for formation of K11-linked chains after endogenous Ube2S was depleted by siRNAs. K11-chain formation in cells arrested in prometaphase or exiting mitosis was monitored by αK11-Western. G. Glu34 of acceptor ubiquitin is required for K11-linkage formation. Ubiquitin mutants were incubated with Ube2S and analyzed by Coomassie staining. H. Glu34 of acceptor ubiquitin is required for chain elongation by APC/C and Ube2S. The modification of Ub-L-cycA by APC/C, Ube2S, and ubiquitin mutants was analyzed by autoradiography. I. ubiE34Q displays catalytic, but not binding defects. The rates of ubi2-formation at different concentrations of ubiquitin and ubiE34Q at the indicated pH were determined from two or three independent time-courses. Apparent kinetic constants were obtained by fitting the rate constants to a Michaelis-Menten equation. J. Rescue of ubiE34Q, but not other TEK-box or hydrophobic patch mutants, by increasing the reaction pH. Ubiquitin or indicated mutants were incubated with Ube2S at pH 7.5 (left) or pH 9 (right) and analyzed by Coomassie staining.
Linkage specificity is determined by substrate-assisted catalysis
The low affinity of Ube2S for acceptor ubiquitin raised the question of how this E2 achieves the stringent selection of K11 over other linkages. In one scenario, recognition of other Lys residues would be even less favored, with differences in binding energies accounting for the K11-specificity of Ube2S. To address this issue, we carried out docking calculations that placed each of the other Lys residues of ubiquitin in proximity to the active site of charged Ube2S. Among the clusters returned by HADDOCK, several had buried surface areas and energy characteristics comparable to our K11-centered model (Figure S8; Table S3). This suggests that other Lys residues can be exposed to the active site of Ube2S, yet these binding events do not result in linkage formation. Thus, selective acceptor binding is not sufficient to explain the K11-specificity of Ube2S.
Alternatively, the composition of the active site of Ube2S might force the reaction towards K11-linkages. Our models of the ternary complex found that the active site of Ube2S was similar to the E2 Ubc9 (Reverter and Lima, 2005; Figure 7C). For Ubc9, several residues in addition to the active site cysteine were attributed roles in catalysis: Tyr87 and Asn85 of Ubc9 contribute to pKa suppression of the substrate lysine through desolvation. Further, Asn85 serves to stabilize the oxyanion intermediate during ubiquitin transfer, and Tyr87 provides a hydrophobic platform to position the attacking lysine side chain (Yunus and Lima, 2006). While Asn85 of Ubc9 is conserved in Ube2S (Asn87; Figure 7C), the attacking lysine is likely positioned by Leu129 of Ube2S (Figure 7C). Asn87 and Leu129 are essential for Ube2S-activity in vitro (Figure 7D, E) and, as seen with HeLa cell lines expressing Ube2SL129E, in vivo (Figure 7F). The mutation of Asn87 or Leu129 did not impede charging of Ube2S by E1 (Figure S7G).
In addition to Asn85 and Tyr87, Asp127 of Ubc9 was assigned a catalytic role in reducing the pKa of the substrate lysine (Yunus and Lima, 2006). In Ube2S, this residue is replaced by serine (Ser127), which our models place into a position to interact with the donor ubiquitin rather than activate an acceptor lysine (Figure 3). Strikingly, instead of an E2-residue, our models show an amino acid of ubiquitin, Glu34, to be in an appropriate position to orient the acceptor Lys11 and to promote its desolvation (Figure 7C). The mutation of Glu34 inhibited K11-linkage formation (Figure 7G, H), and kinetic analyses found this to be due to a strong reduction in the apparent kcat, but not KM (Figure 7I). Thus, a residue in the substrate, Glu34 of ubiquitin, plays an important role in catalysis by Ube2S.
Due to its position in the ternary complex, Glu34 is expected to orient Lys11, but not other Lys residues, and to suppress its pKa. The E34Q mutant of ubiquitin might maintain the position of Lys11, but fail to promote its deprotonation. If this assumption were correct, the inability of ubiE34Q to produce ubi2 may be rescued by increasing the pH of the reaction, which facilitates lysine deprotonation. Indeed, Ube2S efficiently linked ubiE34Q molecules at higher pH (Figure 7J), which was due to a change in the apparent kcat, but not KM (Figure 7I). By contrast, an acceptor TEK-box mutant defective in Ube2S-binding (ubiT14E) or a donor ubiquitin mutant (ubiI44A/V70A) was inactive at pH9 (Figure 7J). These findings support the notion that Glu34 of ubiquitin participates in catalysis by suppressing the pKa of the acceptor Lys11.
As the catalytic role of Glu34 could be bypassed by increasing the pH, the same treatment might reduce the specificity of Ube2S. Consistent with this hypothesis, Ube2S modified Lys residues in a peptide derived from its C-terminal tail much more efficiently at pH9 than at pH7.5 (Figure S7H). Moreover, at pH9, but not at pH7.5, Lys63 and Lys48 of ubiquitin could act as acceptor for Ube2S (Figure S7I), although the bulk of linkage formation still occurred through K11 (Figure S7J). These findings further suggest that Ube2S requires a residue in ubiquitin, Glu34, for specific formation of K11-linkages. We conclude that Ube2S promotes linkage-specific ubiquitin chain formation by substrate-assisted catalysis.
Conclusions
K11- and K48-linked ubiquitin chains are often assembled by single-subunit E2s that cooperate with RING-E3s (Ye and Rape, 2009). Most of these E2s are specific and processive, but how these properties are achieved in the absence of co-factors was poorly understood. Here, we addressed this question by dissecting the mechanism of ubiquitin chain assembly by the K11-specific E2 Ube2S.
We found that Ube2S requires a non-covalent interaction with the donor ubiquitin for chain formation. Although the affinity of Ube2S for the donor ubiquitin is weak, this interaction occurs in addition to the covalent thioester-bond at the E2 active site. It tethers the donor ubiquitin to the E2, thereby restricting its flexibility and facilitating acceptor recognition. It also places the C-terminus of the donor ubiquitin in an optimal position for nucleophilic attack by the acceptor lysine.
The Ube2S-donor ubiquitin complex binds the acceptor ubiquitin very transiently through primarily electrostatic interactions. Ube2S recognizes the TEK-box on acceptor ubiquitin, a motif previously identified as being required for formation of K11-linkages by Ube2C (Jin et al., 2008). As seen in crystal structures of K11-linked ubiquitin dimers (Matsumoto et al., 2010; Bremm et al., 2010), all TEK-box residues in the distal ubiquitin are fully accessible for recognition by Ube2S.
The low affinity of Ube2S for the acceptor is in agreement with observations for other E2s (Petroski and Deshaies, 2005; Rodrigo-Brenni et al., 2010) and likely protects cells from spurious chain elongation in the absence of E3s. However, together with our comparative docking analysis, the transient nature of acceptor binding suggests that selective acceptor recognition does not explain the linkage specificity of Ube2S.
Indeed, our model of the ternary complex between Ube2S, donor, and acceptor ubiquitin revealed that Ube2S lacks a residue required for suppressing the pKa of the substrate lysine. This function is instead provided by Glu34 of ubiquitin, which is in direct proximity to Lys11. Mutation of Glu34 had strong effects on the apparent kcat, but not the KM, of linkage formation by Ube2S, supporting a role in catalysis. Other Lys residues of ubiquitin do not have a suitably positioned acidic residue when docked into the active site of Ube2S or display features incompatible with catalysis (Figure S8). The same likely applies to Lys residues of APC/C-substrates, which may explain why Ube2S is unable to promote chain initiation (Garnett et al., 2009; Williamson et al., 2009). Thus, our findings suggest that formation of a competent catalytic center requires residues of Ube2S and ubiquitin, which only occurs when K11 of the acceptor is exposed to the active site of Ube2S. We conclude that linkage-specific chain assembly by Ube2S occurs through substrate-assisted catalysis.
Do other E2 enzymes use similar mechanisms for ubiquitin transfer? We found that non-covalent donor binding is a property shared by E2s with different linkage specificity. Ube2R1 and Ube2G2 also tether the donor ubiquitin for efficient catalysis, but not for K48-specificity, and Ube2R1 uses a similar surface on its UBC-domain as Ube2S for donor recognition. In addition, the HECT-E3 Nedd4L binds E2-linked donor ubiquitin, a feature required for rapid ubiquitin transfer to the catalytic cysteine of the E3 (Kamadurai et al., 2009); the E3 RanBP2 binds SUMO to restrict its conformational freedom (Reverter and Lima, 2005); and in some cases, a ubiquitin-binding domain can promote E2-dependent ubiquitination reactions (Hoeller et al., 2007). We, therefore, propose that non-covalent donor-binding is a general property of ubiquitination enzymes to increase the processivity of substrate modification.
Other E2s may also use substrate-assisted catalysis for chain assembly. Mms2-Ubc13 positions Glu64 of ubiquitin close to the E2 active site, and mutation of this residue resulted in a decrease of K63-linkage formation (Eddins et al., 2006). Thus, although acceptor binding to Mms2 helps to orient Lys63 towards the active site of Ubc13, a catalytic ubiquitin residue might increase the specificity of chain formation. Moreover, mutation of a Tyr residue in ubiquitin reduced the catalytic rate of K48-linkage formation by yeast Ubc1 (Rodrigo-Brenni et al., 2010). Together, these findings allow us to propose that several E2 enzymes achieve linkage-specific ubiquitin chain formation through a mechanism of substrate-assisted catalysis.
Methods
A detailed methods description can be found in the supplementary information.
Reagents
Table S4 shows a complete list of all constructs.
Protein Purification
Most proteins were purified from BL21/DE3 (RIL) cells. E1 was purified from Sf9 cells. For uniform isotopic enrichment, Ube2S and ubiquitin were expressed in M9-medium using 15N-enriched (NH4)2SO4 and/or 13C-enriched glucose.
To generate ester-linked complex, 75μM 15N-enriched UBCUbe2S/C95S and 230μM unlabelled ubiquitin were incubated at 37°C for 3h. The diluted reaction was subjected to two rounds of anion exchange chromatography.
Formation of ubi2
60μM ubiquitin and/or ubiΔGG and 5μM E2s were incubated with E1 and energy mix at 30°C for 1h and analyzed by Coomassie or Silver staining. In assays comparing activity at different pH, Tris/HCl was replaced with 50mM Bis-tris propane, pH7.5, 8, or 9.
Ube2S kinetics assays
Time courses of ubi2-formation were performed with different concentrations of wt- or E34Q-ubiquitin. Levels of ubi2 were quantified by Quantity One and compared to a known amount of Ube2S on each gel. Initial velocity rates and kinetic constants were calculated with GraphPad Prism and Michaelis-Menten equations.
APC/C Ubiquitination Assays
35S-substrates were synthesized by IVT/T. Ub-L-CycA was synthesized in the presence of 175μM ubiK29R to inhibit the UFD-pathway, which is active in reticulocyte lysate. To purify 35S-Ub-L-cycA, HisCdk2 was bound to NiNTA. IVT/T was added to beads for 3h at 4°C. Beads were eluted with imidazole, and Ub-L-cycA/Cdk2-complexes were concentrated with 30 MWCO Microcon filters. APC/C was purified from G1-HeLa extracts and used for ubiquitination as described (Rape et al., 2006).
Analysis of Ube2S-activity in vivo
HeLa cells were transfected in 6-well plates with 4μg Ube2S-vectors and Lipofectamine 2000. 24h later, 10% of transfected cells were expanded to 10cm dishes and selected with hygromycin B. Individual hygromycin-resistant colonies were picked with cloning discs and tested for Ube2S-expression by Western.
Cells expressing Ube2S or mutants were transfected with 100nM siRNA with Oligofectamine and synchronized in prometaphase by thymidine/nocodazole. Samples were taken at 0h and 2h post release and processed for αK11-Western.
Computational docking
Donor docking was performed with HADDOCK 2.1, using crystal structures of UBCUbe2S (pdb ID: 1ZDN, chain A) and ubiquitin (pdb ID: 1UBQ). Active residues were based on chemical shift data and solvent accessibility. For thioester linkage between donor and Ube2S, we applied an unambiguous intermolecular distance restraint between C95 of Ube2S and G76 of ubiquitin. Residues 70–76 of ubiquitin were defined fully flexible.
To generate a model of the ternary complex, we docked a second ubiquitin onto the selected E2-donor complex (cluster1 no. 3; Table S1). We initially applied a single unambiguous restraint between Nz of acceptor-K11 and C95 of Ube2S (Table S2, top), followed by a refined run with ambiguous restraints based on functional data (Table S2).
Additional donor docking experiments used ClusPro 2.0 and default parameters.
NMR
Data were recorded at 25°C on Bruker DRX spectrometers (500, 600, 800, and 900 MHz) and processed with NMRPipe. Backbone chemical shift assignments for UBCUbe2S and ubiquitin were obtained by standard triple resonance experiments. Titration experiments were performed by mixing stock solutions in containing 240μM 15N-enriched UBCUbe2S or 200μM 15N-enriched ubiquitin, and a ~5.6-fold molar excess of unlabelled partner. Phase-sensitive gradient-enhanced 1H-15N HSQC spectra were recorded. To compare chemical shift perturbations a weighted combined chemical shift difference Δδ(1H15N) was calculated.
1H-15N HSQC experiments of ester-linked complex between 15N-enriched UBCUbe2S/C95S and ubiquitin were recorded with 22μM complex.
Supplementary Material
Figure S1: Characterization of Ube2S and its interaction with donor ubiquitin. A. The UBC-domain of Ube2S (UBCUbe2S) promotes formation of K11-linkages between ubiquitin molecules. UBCUbe2S or Ube2S were incubated with E1, ubiquitin or ubiK11R, and ATP for the indicated times. Reaction products were analyzed by SDS-PAGE and Coomassie staining. B. Radius of gyration, Rg, of Ube2S and UBCUbe2S at various protein concentrations, as derived from Guinier analysis in PRIMUS (Konarev et al., 2003). Analysis with the indirect transform package GNOM (Svergun et al., 1988) yielded similar Rg values (data not shown). For both proteins, Rg does not change significantly with increasing protein concentration, consistent with a lack of oligomerization. The Rg–values of ~ 17 and 28 Å correlate well with the hydrodynamic radius/molecular mass of the monomeric states of UBCUbe2S and Ube2S, respectively, as derived by gel filtration and multi-angle light scattering (data not shown). C. Comparison of experimental and simulated scattering curves generated with CRYSOL (Svergun et al., 1995) based on a UBCUbe2S monomer (PDB ID: 1ZDN, chain A) and a crystallographic dimer (PDB ID: 1ZDN, both chains). I(q) is plotted as a function of the momentum transfer q=(4π*sin(θ))/λ), where 2θ is the scattering angle and λ is the wavelength of the incident X-ray beam. D. The hydrophobic patch of ubiquitin is required for Ube2S- and APC/C-dependent chain formation over a wide range of ubiquitin concentrations. Ub-L-cycA was produced by IVT/T and incubated with APC/CCdh1, Ube2S, E1, ATP, and the indicated concentrations of either wt-ubiquitin or ubiI44A. The formation of K11-linked ubiquitin chains on Ub-L-cycA was monitored by SDS-PAGE and autoradiography. As a comparison, the concentration of ubiquitin in HeLa cells has been estimated at 90 μM (Ryu et al., 2006). E. The hydrophobic patch in ubiquitin is required for APC/C-dependent ubiquitin chain elongation. 35S-labeled APC/C-substrate cyclin A was incubated with APC/CCdh1, E1, low concentrations of Ube2C (for chain initiation) and Ube2S (for chain elongation). Reaction products were separated by SDS-PAGE and analyzed by autoradiography. Due to the presence of Ube2S, the majority of modified cyclin A is decorated with long ubiquitin chains. F. The hydrophobic patch of ubiquitin is not required for charging of Ube2S by the E1. Ube2S was incubated with E1 and ubiquitin or the indicated ubiquitin mutants in the absence of reducing agents. When indicated, β-mercaptoethanol was added to gel-loading buffer to reduce thioester linkages. Charging of Ube2S with ubiquitin results in a βME-sensitive conjugate representing the thioester (Ube2S-Cys95~ubi) and in a βME-insensitive conjugate, most likely a covalent modification of a lysine residue in the UBC-domain of Ube2S (Ube2S-ubi). G. A functional hydrophobic patch is not required in the acceptor ubiquitin. The L8A and I44A/V70A-mutations were introduced into the acceptor ubiΔGG. ubiΔGG and indicated mutants were mixed with ubiquitin, E1, ATP, and Ube2S, and the formation of ubiΔGG-ubi and ubi-ubi dimers was monitored by SDS-PAGE and Silver staining.
Figure S2: Characterization of donor ubiquitin binding to Ube2S. A. Ubiquitin causes similar chemical shift perturbations on UBCUbe2S regardless of whether it is added in trans or is covalently linked to the active site. To obtain a more stable oxy-ester complex, the C95S mutant of UBCUbe2S was used instead of wt. Weighted combined chemical shift perturbations, Δδ(1H15N), are plotted over the residue number. 1H-15N HSQC spectra were recorded of 22μM 15N-enriched UBCUbe2S C95S ester-linked to unlabelled ubiquitin (yellow) and 140μM 15N-enriched UBCUbe2S C95S in the presence of an 11-fold molar excess of unlabelled ubiquitin (red) and were referenced to the spectrum of 140μM 15N-enriched UBCUbe2S C95S in the absence of ubiquitin. Gaps are due to proline residues or missing assignments. B. Residues with significant binding-induced chemical shift perturbations and significant surface accessibility (see Table S1) are mapped onto the surface of UBCUbe2S (PDB ID: 1ZDN) and ubiquitin (PDB ID: 1UBQ), respectively. C. Mutations in the UBCUbe2S-donor ubiquitin interface interfere with the interaction detected by NMR. Weighted combined chemical shift perturbations, ⊗™(1H15N), are plotted over residue number. The data are based on 1H-15N HSQC spectra of mixtures of 200μM 15N-enriched ubiquitin and a 6-fold molar excess of UBCUbe2S. Gaps are due to proline residues or missing assignments. As justified under Materials and Methods, we interpret the amplitude of ⊗™(1H15N) as a measure of binding affinity. D. Determination of the dissociation constants, Kd, for the interaction between Ube2S and ubiquitin in solution. NMR-derived isotherms for the binding of ubiquitin to Ube2S (left panel) and UBCUbe2S (right panel) were fitted globally to a single-site model. Only those resonances were included that show a weighted combined chemical shift perturbation, ⊗™(1H15N), of at least 0.5ppm at the highest excess of ubiquitin used. The concentration of Ube2S and UBCUbe2S was 240μM. E. Mutations in the non-covalent donor ubiquitin binding interface of Ube2S do not inhibit charging of Ube2S by E1. Ube2S or indicated mutants were incubated with ubiquitin E1 and ATP, resulting in formation of a βME-sensitive thioester (Ube2S-Cys95~ubi) and a βME-insensitive conjugate (Ube2S-ubi). Reactions were analyzed by αUbe2S-Western. F. Mutations of Ube2S do not interfere with binding of Ube2S to the APC/C. APC/C-subunits were synthesized by in vitro-transcription/translation. The radiolabeled proteins are incorporated into full APC/C present in reticulocyte lysate (our unpublished observations). The 35S-labeled proteins were then incubated with MBPUbe2S, which was immobilized on amylose-resin; MBP was used as a control. After extensive washing, binding reactions were analyzed by SDS-PAGE and Coomassie staining (for inputs; bottom panel) or autoradiography (for binding; top panel).
Figure S3: Characterization of the NMR-based model of the Ube2S-donor ubiquitin interface. A. Comparison of docked models generated by two different programs, HADDOCK, including NMR-based restraints (green), and ClusPro without any restraints (blue). Major differences are only seen for the C-terminal tail of ubiquitin, which is highlighted red in the ClusPro model. The chosen ClusPro model represents 9 out of 965 models generated, 38 of which have the Sγ atom of Cys95 of Ube2S within a distance of 7.5Å from the C-terminal carbon atom of donor ubqiutin. B. Surface electrostatic potentials of the donor interface, as calculated using APBS (Baker et al., 2001; Dolinsky et al., 2004, 2007). Intermolecular salt bridges, as predicted by the PISA server at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html; Krissinel et al., 2007), are illustrated by solid lines. C. Illustration of the ionic contact between Arg74 of donor ubiquitin and Asp102 on Ube2S, as seen in our model of the Ube2S-donor ubiquitin complex. D. Residues in ubiquitin that are at the Ube2S-donor ubiquitin interface are not required on the acceptor ubiquitin. Mutations were introduced into the acceptor ubiΔGG. ubiΔGG and indicated mutants were incubated with E1, Ube2S, ubiquitin, and ATP, and formation of ubiΔGG-ubi and ubi-ubi dimers was monitored by SDS-PAGE and Silver staining. Except for Lys6, no residue was required in the acceptor ubiquitin. E. Lys6 is required in the donor ubiquitin. Ubiquitin and the respective mutants K6A and K6E were incubated with the acceptor ubiΔGG, E1, Ube2S, and ATP. Reaction products were analyzed by SDS-PAGE and Coomassie staining. Mutation of Lys6 to Glu affects both acceptor and donor ubiquitin, and thus, neither ubiΔGG-ubi nor ubi-ubi dimers are formed. Mutation of Lys6 to Ala only affects acceptor ubiquitin function, and thus, ubiΔGG-ubi dimers are formed with this mutant. Both ubiK6E and ubiK6A showed higher mobility in SDS-PAGE compared to wt-ubiquitin. F. With exception of Arg72, all ubiquitin residues at the Ube2S-donor ubiquitin interface can be mutated without affecting Ube2S-charging by E1. Ube2S was incubated with ubiquitin or indicated mutants, E1, and ATP, resulting in formation of a βME-sensitive thioester (Ube2S-Cys95~ubi) and a βME-insensitive conjugate (Ube2S-ubi). Reaction products were analyzed by SDS-PAGE and αUbe2S-Western. G. Ube2S-residues at the binding interface with donor ubiquitin are not required for charging of Ube2S by E1. Ube2S and indicated mutants were incubated with E1, ubiquitin, and ATP. Reaction products were analyzed by SDS-PAGE and αUbe2S-Western. H. The E51K-mutant of Ube2S does not rescue defective ubiquitin dimer formation in the presence of the I44A-mutation on ubiquitin. Ubiquitin or ubiI44A were incubated with acceptor ubiΔGG, Ube2S or Ube2SE51K, E1, and ATP. Reaction products were analyzed by SDS-PAGE and Silver staining.
Figure S4: Characterization of donor ubiquitin recognition by Ube2R1 and Ube2G2. A. The hydrophobic patch of ubiquitin is required for ubiquitin-dimer formation by the K48-specific E2s Ube2G2 (top panel) and Ube2R1 (bottom panel). Ubiquitin and indicated mutants were incubated with Ube2R1 or Ube2G2/gp78, E1, and ATP. Reaction products were separated by SDS-PAGE and analyzed by Coomassie staining. B. The hydrophobic patch of ubiquitin is not required for charging of Ube2G2 (top panel) or Ube2R1 (bottom panel) by E1. Ubiquitin and indicated mutants were incubated with E1, ATP, and Ube2G2 or Ube2R1 in the absence of reducing agents. Where indicated, βME was added to gel loading buffer to show thioester formation. Reaction products were monitored by αHis-Western, detecting a His-epitope used to purify the E2 proteins. C. The same surface used by Ube2S for donor ubiquitin-binding is required on Ube2R1 for E2-activity. Ube2R1 or indicated mutants were incubated with E1, ubiquitin, and ATP. Formation of K48-linked ubiquitin dimers (ubi~ubi) was monitored by SDS-PAGE and Coomassie staining. D. Ube2R1-residues at the donor ubiquitin binding interface are not required for charging of Ube2R1 by the E1. Ube2R1 or indicated mutants were incubated with E1, ATP, and ubiquitin, and analyzed for charging as described above.
Figure S5: HADDOCK output for the docking of acceptor ubiquitin onto the UBCUbe2S-donor ubiquitin complex. Cartoon representation of the top-scoring models of the two clusters of run 1 without experimental restraints (see Table S2).
Figure S6: Characterization of acceptor ubiquitin recognition by the Ube2S-donor ubiquitin complex. A. The TEK-box of ubiquitin is required for APC/C-dependent chain formation catalyzed by the E2s Ube2C and Ube2S. Ubiquitin or indicated mutants were incubated with 35S-labeled cyclin A, APC/CCdh1, Ube2C, Ube2S, E1, and ATP. Reaction products were separated by SDS-PAGE and analyzed by autoradiography. B. The TEK-box residues of ubiquitin are not required for charging of Ube2S by E1. Ubiquitin or indicated mutants were incubated with E1, Ube2S, and ATP, and analyzed for charging as described above. C. Mutations in the Ube2S-acceptor ubiquitin interface do not influence the interaction between Ube2S and donor ubiquitin, as detected by NMR. Weighted combined chemical shift perturbations, ⊗™(1H15N), are plotted over residue number. The data are based 1H-15N HSQC spectra of mixtures of 200μM 15N-enriched ubiquitin and a 6-fold molar excess of UBCUbe2S. Gaps are due to proline residues or missing assignments. D. Surface electrostatic potentials of the acceptor interface for the selected HADDOCK complex between Ube2S, donor, and acceptor ubiquitin (cluster 1, no. 1; Table S2), as calculated using APBS. Intermolecular salt bridges and hydrogen bonds, as predicted by the PISA server are illustrated by solid and dashed lines, respectively. E. Ube2S-residues at the acceptor binding interface are required for APC/C-dependent ubiquitin chain formation. Ube2S or indicated mutants were incubated with 35S-labeled cyclin A, APC/CCdh1, E1, Ube2C, and ubiquitin, and reaction products were analyzed by SDS-PAGE and autoradiography. F. Ube2S-residues at the acceptor binding interface are not required for charging of Ube2S by E1. Ube2S or indicated mutants were incubated with E1, ATP, and ubiquitin, and analyzed for charging as described above.
Figure S7: Characterization of acceptor ubiquitin recognition by Ube2S. A. The E131A mutant of Ube2S rescues the phenotype of the K6E-mutation in acceptor ubiquitin. ubiΔGG or ubiΔGG/K6E were incubated with ubiquitin, E1, ATP, and Ube2S or indicated Ube2S-mutants. Formation of ubiΔGG-ubi and ubi-ubi dimers was analyzed by SDS-PAGE and Coomassie staining. In contrast to Ube2SE131K, Ube2SE131A does not interfere with recognition of Lys6 in wt-ubiquitin, explaining the formation of ubi-ubi dimers in the presence of Ube2SE131A, but not Ube2SE131K. B. Specific rescue of the K6E-mutation in acceptor ubiquitin by Ube2SE131K, but not other mutants at the acceptor ubiquitin binding interface. Ube2S or indicated mutants were incubated with E1, ATP, ubiΔGG or ubiΔGG/K6E, and ubiquitin. Formation of ubiΔGG-ubi and ubi-ubi dimers was analyzed by SDS-PAGE and Coomassie staining. C. Complete rescue of the ubiK6E-phenotype by Ube2SE51K/E131K. Ubiquitin or ubiK6E were incubated with Ube2S or Ube2SE51K/E131K, E1, and ATP, and formation of ubiquitin dimers was monitored by SDS-PAGE and Silver staining. D. Validation of the Ube2S-acceptor ubiquitin interaction by charge swap analysis between Glu64 of ubiquitin and Arg135 of Ube2S. The indicated mutants of ubiquitin and Ube2S were tested for their ability to produce ubiquitin dimers (ubi~ubi). Reaction products were analyzed by SDS-PAGE and Coomassie staining. E. Specific rescue of the deleterious effects of a R135E-mutant of Ube2S by a E64K-mutant of ubiquitin. Ube2S or Ube2SR135E were incubated with ubiquitin or indicated TEK-box mutants, and formation of ubiquitin dimers (ubi~ubi) was monitored by SDS-PAGE and Coomassie staining. F. Rescue of impaired ubiquitin dimer formation by a K63E-mutant of ubiquitin by Ube2SE139K. Ube2S or Ube2SE139K were incubated with ubiquitin or ubiK63E, E1, and ATP, and formation of ubiquitin-dimers was analyzed by SDS-PAGE and Coomassie staining. G. Ube2S-residues required for ubiquitin-linkage formation are not required for charging of Ube2S by the E1. Ube2S or indicated mutants were incubated with E1, ATP, and ubiquitin, and analyzed for charging as described above. H. Reduced substrate-specificity of Ube2S at higher pH. A peptide of 26 C-terminal residues of Ube2S tagged with biotin (BCTP) was incubated with Ube2S, E1, ATP, and ubiquitin at either pH7.5 or at pH9. Modification of lysine residues in BCTP was detected by Western-blotting using HRP-coupled streptavidin. I. Increasing the pH allows Ube2S to modify ubiquitin lysine residues other than K11. The indicated single lysine ubiquitin mutants were incubated with Ube2S, E1, and ATP at either pH7.5 or pH9. Formation of ubiquitin-dimers (ubi~ubi) was detected by Western blotting using an α-ubiquitin antibody. J. Increasing the pH does not completely obliterate the K11-specificity of Ube2S. Ubiquitin or the indicated mutants were incubated with Ube2S, E1, and ATP at pH 9. The formation of ubiquitin-dimers (ubi~ubi) was monitored by SDS-PAGE and Silver staining.
Figure S8: HADDOCK analysis of Ube2S-donor-acceptor complexes exposing each of the seven lysine residues of the acceptor ubiquitin to the active site of Ube2S. Details of the active site are shown for the top-scoring model of each of 7 HADDOCK runs in stereo representation with relevant side chains rendered as ball-and-stick. Only the C-terminal tail (residues 71–76) of donor ubiquitin is displayed (blue). Ribbons for acceptor ubiquitin and Ube2S are shown in pink and grey, respectively. While structures docked around K11 show a favorable active site constellation (see Figure 7D), complexes exposing other lysine residues have features incompatible with efficient catalysis. In the following we describe these features for the top representatives of the most populated clusters for each of 7 HADDOCK runs; note, however, that these conclusions also hold for the lower-ranked clusters. Around K6 and K48 no acidic groups are found on the acceptor within a radius of ~ 8 Å and ~ 10 Å, respectively. K33 is neighbored by an acidic residue, E34; in this case, however, the acidic side chain points away from the active site, which puts K33 in an unfavorable orientation for the nucleophilic attack. Structures docked around K27 display productive active site geometry including a proximal acidic residue, D52. However, these complexes contain an additional acidic residue, D39, near the active site cysteine of Ube2S, which might interfere with catalysis by destabilizing the thiolate intermediate formed during the nucleophilic substitution reaction. Complexes docked around K29 contain D21 of the acceptor ubiquitin in a distance of ~ 5 Å from K29, but their overall geometry appears sterically unfavorable. K63 can be docked in a reasonable orientation, but the position of the neighboring acidic side chain, E64, appears less optimal than in the case of K11.
Acknowledgments
We thank HJ Meyer for Ub-L-cycA; A Williamson for golden extracts; members of the Rape and Kuriyan labs, J Winger, S Kassube, and J Kirsch for discussions; J Schaletzky for reading the manuscript and suggestions; J Pelton for help with NMR experiments; D King and T Iavarone for mass spectrometry; and thank the staff at beamline 12.3.1 at LBNL for technical support. NMR instrumentation and operation were supported by NIH-GM 68933, NIH GM68933, NSF BBS 0119304 and NIH RR15756, and NSF BBS 8720134. SL is a fellow of The Leukemia & Lymphoma Society. MR is a Pew fellow, supported by NIH GM83064 and an NIH New Innovator Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–47. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau SR, Kozakov D, Brenke R, Shen Y, Beglov D, Vajda S. ClusPro: Performance in CAPRI rounds 6–11 and the new server. Proteins. 2007;69:781–5. doi: 10.1002/prot.21795. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING-domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- de Vries, et al. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–71. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–20. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–9. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton KS, et al. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Wagner S, Rogov V, Dötsch V, Dikic I. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–8. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Huang DT, Zhuang M, Ayrault O, Schulman BA. Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nat Struct Mol Biol. 2008;15:280–7. doi: 10.1038/nsmb.1387. [DOI] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Phillip I, Rape M. Mechanism of ubiquitin chain formation by the human Anaphase-Promoting Complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CR, Hwang KS, Yoo J, Cho WK, Kim JM, Kim WH, Im DS. E2-EPF UCP targets pVHL for degradation and associates with tumor growth and metastasis. Nat Med. 2006;12:809–16. doi: 10.1038/nm1440. [DOI] [PubMed] [Google Scholar]
- Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–73. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci U S A. 2009;106:3722–7. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto ML, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11-linkage specific antibody. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.07.001. in press. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–20. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- Pierce NW, Kleiger G, Shan SO, Deshaies RJ. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–9. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–92. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KY, Baker RT, Kopito RR. Ubiquitin-specific protease 2 as a tool for quantification of total ubiquitin levels in biological specimens. Anal Biochem. 2006;353:153–5. doi: 10.1016/j.ab.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–31. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinerman FB, Honig B. On the role of electrostatic interactions in the design of protein-protein interfaces. J Mol Biol. 2002;318:161–77. doi: 10.1016/S0022-2836(02)00030-X. [DOI] [PubMed] [Google Scholar]
- Song L, Rape M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol Cell. 2010;38:369–82. doi: 10.1016/j.molcel.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell. 2001;105:711–20. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- Wagner KW, et al. Overexpression, genomic amplification and therapeutic potential of inhibiting the UbcH10 ubiquitin conjugase in human carcinomas of diverse anatomic origin. Oncogene. 2004;3:6621–9. doi: 10.1038/sj.onc.1207861. [DOI] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci USA. 2009;106:18213–8. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci U S A. 2010;107:1355–60. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–45. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–64. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–9. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Characterization of Ube2S and its interaction with donor ubiquitin. A. The UBC-domain of Ube2S (UBCUbe2S) promotes formation of K11-linkages between ubiquitin molecules. UBCUbe2S or Ube2S were incubated with E1, ubiquitin or ubiK11R, and ATP for the indicated times. Reaction products were analyzed by SDS-PAGE and Coomassie staining. B. Radius of gyration, Rg, of Ube2S and UBCUbe2S at various protein concentrations, as derived from Guinier analysis in PRIMUS (Konarev et al., 2003). Analysis with the indirect transform package GNOM (Svergun et al., 1988) yielded similar Rg values (data not shown). For both proteins, Rg does not change significantly with increasing protein concentration, consistent with a lack of oligomerization. The Rg–values of ~ 17 and 28 Å correlate well with the hydrodynamic radius/molecular mass of the monomeric states of UBCUbe2S and Ube2S, respectively, as derived by gel filtration and multi-angle light scattering (data not shown). C. Comparison of experimental and simulated scattering curves generated with CRYSOL (Svergun et al., 1995) based on a UBCUbe2S monomer (PDB ID: 1ZDN, chain A) and a crystallographic dimer (PDB ID: 1ZDN, both chains). I(q) is plotted as a function of the momentum transfer q=(4π*sin(θ))/λ), where 2θ is the scattering angle and λ is the wavelength of the incident X-ray beam. D. The hydrophobic patch of ubiquitin is required for Ube2S- and APC/C-dependent chain formation over a wide range of ubiquitin concentrations. Ub-L-cycA was produced by IVT/T and incubated with APC/CCdh1, Ube2S, E1, ATP, and the indicated concentrations of either wt-ubiquitin or ubiI44A. The formation of K11-linked ubiquitin chains on Ub-L-cycA was monitored by SDS-PAGE and autoradiography. As a comparison, the concentration of ubiquitin in HeLa cells has been estimated at 90 μM (Ryu et al., 2006). E. The hydrophobic patch in ubiquitin is required for APC/C-dependent ubiquitin chain elongation. 35S-labeled APC/C-substrate cyclin A was incubated with APC/CCdh1, E1, low concentrations of Ube2C (for chain initiation) and Ube2S (for chain elongation). Reaction products were separated by SDS-PAGE and analyzed by autoradiography. Due to the presence of Ube2S, the majority of modified cyclin A is decorated with long ubiquitin chains. F. The hydrophobic patch of ubiquitin is not required for charging of Ube2S by the E1. Ube2S was incubated with E1 and ubiquitin or the indicated ubiquitin mutants in the absence of reducing agents. When indicated, β-mercaptoethanol was added to gel-loading buffer to reduce thioester linkages. Charging of Ube2S with ubiquitin results in a βME-sensitive conjugate representing the thioester (Ube2S-Cys95~ubi) and in a βME-insensitive conjugate, most likely a covalent modification of a lysine residue in the UBC-domain of Ube2S (Ube2S-ubi). G. A functional hydrophobic patch is not required in the acceptor ubiquitin. The L8A and I44A/V70A-mutations were introduced into the acceptor ubiΔGG. ubiΔGG and indicated mutants were mixed with ubiquitin, E1, ATP, and Ube2S, and the formation of ubiΔGG-ubi and ubi-ubi dimers was monitored by SDS-PAGE and Silver staining.
Figure S2: Characterization of donor ubiquitin binding to Ube2S. A. Ubiquitin causes similar chemical shift perturbations on UBCUbe2S regardless of whether it is added in trans or is covalently linked to the active site. To obtain a more stable oxy-ester complex, the C95S mutant of UBCUbe2S was used instead of wt. Weighted combined chemical shift perturbations, Δδ(1H15N), are plotted over the residue number. 1H-15N HSQC spectra were recorded of 22μM 15N-enriched UBCUbe2S C95S ester-linked to unlabelled ubiquitin (yellow) and 140μM 15N-enriched UBCUbe2S C95S in the presence of an 11-fold molar excess of unlabelled ubiquitin (red) and were referenced to the spectrum of 140μM 15N-enriched UBCUbe2S C95S in the absence of ubiquitin. Gaps are due to proline residues or missing assignments. B. Residues with significant binding-induced chemical shift perturbations and significant surface accessibility (see Table S1) are mapped onto the surface of UBCUbe2S (PDB ID: 1ZDN) and ubiquitin (PDB ID: 1UBQ), respectively. C. Mutations in the UBCUbe2S-donor ubiquitin interface interfere with the interaction detected by NMR. Weighted combined chemical shift perturbations, ⊗™(1H15N), are plotted over residue number. The data are based on 1H-15N HSQC spectra of mixtures of 200μM 15N-enriched ubiquitin and a 6-fold molar excess of UBCUbe2S. Gaps are due to proline residues or missing assignments. As justified under Materials and Methods, we interpret the amplitude of ⊗™(1H15N) as a measure of binding affinity. D. Determination of the dissociation constants, Kd, for the interaction between Ube2S and ubiquitin in solution. NMR-derived isotherms for the binding of ubiquitin to Ube2S (left panel) and UBCUbe2S (right panel) were fitted globally to a single-site model. Only those resonances were included that show a weighted combined chemical shift perturbation, ⊗™(1H15N), of at least 0.5ppm at the highest excess of ubiquitin used. The concentration of Ube2S and UBCUbe2S was 240μM. E. Mutations in the non-covalent donor ubiquitin binding interface of Ube2S do not inhibit charging of Ube2S by E1. Ube2S or indicated mutants were incubated with ubiquitin E1 and ATP, resulting in formation of a βME-sensitive thioester (Ube2S-Cys95~ubi) and a βME-insensitive conjugate (Ube2S-ubi). Reactions were analyzed by αUbe2S-Western. F. Mutations of Ube2S do not interfere with binding of Ube2S to the APC/C. APC/C-subunits were synthesized by in vitro-transcription/translation. The radiolabeled proteins are incorporated into full APC/C present in reticulocyte lysate (our unpublished observations). The 35S-labeled proteins were then incubated with MBPUbe2S, which was immobilized on amylose-resin; MBP was used as a control. After extensive washing, binding reactions were analyzed by SDS-PAGE and Coomassie staining (for inputs; bottom panel) or autoradiography (for binding; top panel).
Figure S3: Characterization of the NMR-based model of the Ube2S-donor ubiquitin interface. A. Comparison of docked models generated by two different programs, HADDOCK, including NMR-based restraints (green), and ClusPro without any restraints (blue). Major differences are only seen for the C-terminal tail of ubiquitin, which is highlighted red in the ClusPro model. The chosen ClusPro model represents 9 out of 965 models generated, 38 of which have the Sγ atom of Cys95 of Ube2S within a distance of 7.5Å from the C-terminal carbon atom of donor ubqiutin. B. Surface electrostatic potentials of the donor interface, as calculated using APBS (Baker et al., 2001; Dolinsky et al., 2004, 2007). Intermolecular salt bridges, as predicted by the PISA server at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html; Krissinel et al., 2007), are illustrated by solid lines. C. Illustration of the ionic contact between Arg74 of donor ubiquitin and Asp102 on Ube2S, as seen in our model of the Ube2S-donor ubiquitin complex. D. Residues in ubiquitin that are at the Ube2S-donor ubiquitin interface are not required on the acceptor ubiquitin. Mutations were introduced into the acceptor ubiΔGG. ubiΔGG and indicated mutants were incubated with E1, Ube2S, ubiquitin, and ATP, and formation of ubiΔGG-ubi and ubi-ubi dimers was monitored by SDS-PAGE and Silver staining. Except for Lys6, no residue was required in the acceptor ubiquitin. E. Lys6 is required in the donor ubiquitin. Ubiquitin and the respective mutants K6A and K6E were incubated with the acceptor ubiΔGG, E1, Ube2S, and ATP. Reaction products were analyzed by SDS-PAGE and Coomassie staining. Mutation of Lys6 to Glu affects both acceptor and donor ubiquitin, and thus, neither ubiΔGG-ubi nor ubi-ubi dimers are formed. Mutation of Lys6 to Ala only affects acceptor ubiquitin function, and thus, ubiΔGG-ubi dimers are formed with this mutant. Both ubiK6E and ubiK6A showed higher mobility in SDS-PAGE compared to wt-ubiquitin. F. With exception of Arg72, all ubiquitin residues at the Ube2S-donor ubiquitin interface can be mutated without affecting Ube2S-charging by E1. Ube2S was incubated with ubiquitin or indicated mutants, E1, and ATP, resulting in formation of a βME-sensitive thioester (Ube2S-Cys95~ubi) and a βME-insensitive conjugate (Ube2S-ubi). Reaction products were analyzed by SDS-PAGE and αUbe2S-Western. G. Ube2S-residues at the binding interface with donor ubiquitin are not required for charging of Ube2S by E1. Ube2S and indicated mutants were incubated with E1, ubiquitin, and ATP. Reaction products were analyzed by SDS-PAGE and αUbe2S-Western. H. The E51K-mutant of Ube2S does not rescue defective ubiquitin dimer formation in the presence of the I44A-mutation on ubiquitin. Ubiquitin or ubiI44A were incubated with acceptor ubiΔGG, Ube2S or Ube2SE51K, E1, and ATP. Reaction products were analyzed by SDS-PAGE and Silver staining.
Figure S4: Characterization of donor ubiquitin recognition by Ube2R1 and Ube2G2. A. The hydrophobic patch of ubiquitin is required for ubiquitin-dimer formation by the K48-specific E2s Ube2G2 (top panel) and Ube2R1 (bottom panel). Ubiquitin and indicated mutants were incubated with Ube2R1 or Ube2G2/gp78, E1, and ATP. Reaction products were separated by SDS-PAGE and analyzed by Coomassie staining. B. The hydrophobic patch of ubiquitin is not required for charging of Ube2G2 (top panel) or Ube2R1 (bottom panel) by E1. Ubiquitin and indicated mutants were incubated with E1, ATP, and Ube2G2 or Ube2R1 in the absence of reducing agents. Where indicated, βME was added to gel loading buffer to show thioester formation. Reaction products were monitored by αHis-Western, detecting a His-epitope used to purify the E2 proteins. C. The same surface used by Ube2S for donor ubiquitin-binding is required on Ube2R1 for E2-activity. Ube2R1 or indicated mutants were incubated with E1, ubiquitin, and ATP. Formation of K48-linked ubiquitin dimers (ubi~ubi) was monitored by SDS-PAGE and Coomassie staining. D. Ube2R1-residues at the donor ubiquitin binding interface are not required for charging of Ube2R1 by the E1. Ube2R1 or indicated mutants were incubated with E1, ATP, and ubiquitin, and analyzed for charging as described above.
Figure S5: HADDOCK output for the docking of acceptor ubiquitin onto the UBCUbe2S-donor ubiquitin complex. Cartoon representation of the top-scoring models of the two clusters of run 1 without experimental restraints (see Table S2).
Figure S6: Characterization of acceptor ubiquitin recognition by the Ube2S-donor ubiquitin complex. A. The TEK-box of ubiquitin is required for APC/C-dependent chain formation catalyzed by the E2s Ube2C and Ube2S. Ubiquitin or indicated mutants were incubated with 35S-labeled cyclin A, APC/CCdh1, Ube2C, Ube2S, E1, and ATP. Reaction products were separated by SDS-PAGE and analyzed by autoradiography. B. The TEK-box residues of ubiquitin are not required for charging of Ube2S by E1. Ubiquitin or indicated mutants were incubated with E1, Ube2S, and ATP, and analyzed for charging as described above. C. Mutations in the Ube2S-acceptor ubiquitin interface do not influence the interaction between Ube2S and donor ubiquitin, as detected by NMR. Weighted combined chemical shift perturbations, ⊗™(1H15N), are plotted over residue number. The data are based 1H-15N HSQC spectra of mixtures of 200μM 15N-enriched ubiquitin and a 6-fold molar excess of UBCUbe2S. Gaps are due to proline residues or missing assignments. D. Surface electrostatic potentials of the acceptor interface for the selected HADDOCK complex between Ube2S, donor, and acceptor ubiquitin (cluster 1, no. 1; Table S2), as calculated using APBS. Intermolecular salt bridges and hydrogen bonds, as predicted by the PISA server are illustrated by solid and dashed lines, respectively. E. Ube2S-residues at the acceptor binding interface are required for APC/C-dependent ubiquitin chain formation. Ube2S or indicated mutants were incubated with 35S-labeled cyclin A, APC/CCdh1, E1, Ube2C, and ubiquitin, and reaction products were analyzed by SDS-PAGE and autoradiography. F. Ube2S-residues at the acceptor binding interface are not required for charging of Ube2S by E1. Ube2S or indicated mutants were incubated with E1, ATP, and ubiquitin, and analyzed for charging as described above.
Figure S7: Characterization of acceptor ubiquitin recognition by Ube2S. A. The E131A mutant of Ube2S rescues the phenotype of the K6E-mutation in acceptor ubiquitin. ubiΔGG or ubiΔGG/K6E were incubated with ubiquitin, E1, ATP, and Ube2S or indicated Ube2S-mutants. Formation of ubiΔGG-ubi and ubi-ubi dimers was analyzed by SDS-PAGE and Coomassie staining. In contrast to Ube2SE131K, Ube2SE131A does not interfere with recognition of Lys6 in wt-ubiquitin, explaining the formation of ubi-ubi dimers in the presence of Ube2SE131A, but not Ube2SE131K. B. Specific rescue of the K6E-mutation in acceptor ubiquitin by Ube2SE131K, but not other mutants at the acceptor ubiquitin binding interface. Ube2S or indicated mutants were incubated with E1, ATP, ubiΔGG or ubiΔGG/K6E, and ubiquitin. Formation of ubiΔGG-ubi and ubi-ubi dimers was analyzed by SDS-PAGE and Coomassie staining. C. Complete rescue of the ubiK6E-phenotype by Ube2SE51K/E131K. Ubiquitin or ubiK6E were incubated with Ube2S or Ube2SE51K/E131K, E1, and ATP, and formation of ubiquitin dimers was monitored by SDS-PAGE and Silver staining. D. Validation of the Ube2S-acceptor ubiquitin interaction by charge swap analysis between Glu64 of ubiquitin and Arg135 of Ube2S. The indicated mutants of ubiquitin and Ube2S were tested for their ability to produce ubiquitin dimers (ubi~ubi). Reaction products were analyzed by SDS-PAGE and Coomassie staining. E. Specific rescue of the deleterious effects of a R135E-mutant of Ube2S by a E64K-mutant of ubiquitin. Ube2S or Ube2SR135E were incubated with ubiquitin or indicated TEK-box mutants, and formation of ubiquitin dimers (ubi~ubi) was monitored by SDS-PAGE and Coomassie staining. F. Rescue of impaired ubiquitin dimer formation by a K63E-mutant of ubiquitin by Ube2SE139K. Ube2S or Ube2SE139K were incubated with ubiquitin or ubiK63E, E1, and ATP, and formation of ubiquitin-dimers was analyzed by SDS-PAGE and Coomassie staining. G. Ube2S-residues required for ubiquitin-linkage formation are not required for charging of Ube2S by the E1. Ube2S or indicated mutants were incubated with E1, ATP, and ubiquitin, and analyzed for charging as described above. H. Reduced substrate-specificity of Ube2S at higher pH. A peptide of 26 C-terminal residues of Ube2S tagged with biotin (BCTP) was incubated with Ube2S, E1, ATP, and ubiquitin at either pH7.5 or at pH9. Modification of lysine residues in BCTP was detected by Western-blotting using HRP-coupled streptavidin. I. Increasing the pH allows Ube2S to modify ubiquitin lysine residues other than K11. The indicated single lysine ubiquitin mutants were incubated with Ube2S, E1, and ATP at either pH7.5 or pH9. Formation of ubiquitin-dimers (ubi~ubi) was detected by Western blotting using an α-ubiquitin antibody. J. Increasing the pH does not completely obliterate the K11-specificity of Ube2S. Ubiquitin or the indicated mutants were incubated with Ube2S, E1, and ATP at pH 9. The formation of ubiquitin-dimers (ubi~ubi) was monitored by SDS-PAGE and Silver staining.
Figure S8: HADDOCK analysis of Ube2S-donor-acceptor complexes exposing each of the seven lysine residues of the acceptor ubiquitin to the active site of Ube2S. Details of the active site are shown for the top-scoring model of each of 7 HADDOCK runs in stereo representation with relevant side chains rendered as ball-and-stick. Only the C-terminal tail (residues 71–76) of donor ubiquitin is displayed (blue). Ribbons for acceptor ubiquitin and Ube2S are shown in pink and grey, respectively. While structures docked around K11 show a favorable active site constellation (see Figure 7D), complexes exposing other lysine residues have features incompatible with efficient catalysis. In the following we describe these features for the top representatives of the most populated clusters for each of 7 HADDOCK runs; note, however, that these conclusions also hold for the lower-ranked clusters. Around K6 and K48 no acidic groups are found on the acceptor within a radius of ~ 8 Å and ~ 10 Å, respectively. K33 is neighbored by an acidic residue, E34; in this case, however, the acidic side chain points away from the active site, which puts K33 in an unfavorable orientation for the nucleophilic attack. Structures docked around K27 display productive active site geometry including a proximal acidic residue, D52. However, these complexes contain an additional acidic residue, D39, near the active site cysteine of Ube2S, which might interfere with catalysis by destabilizing the thiolate intermediate formed during the nucleophilic substitution reaction. Complexes docked around K29 contain D21 of the acceptor ubiquitin in a distance of ~ 5 Å from K29, but their overall geometry appears sterically unfavorable. K63 can be docked in a reasonable orientation, but the position of the neighboring acidic side chain, E64, appears less optimal than in the case of K11.