Abstract
We validate specific binding activity of a fluorescence-labeled peptide to colorectal dysplasia in living mice using a miniature, flexible, fiber microendoscope that passes through the instrument channel of an endoscope. The microendoscope delivers excitation light at 473 nm through a fiber-optic bundle with outer diameter of 680 µm to collect en face images at 10 Hz with 4 µm lateral resolution. We applied the FITC-labeled peptide QPIHPNNM topically to colonic mucosa in genetically engineered mice that spontaneously develop adenomas. More than two-fold greater fluorescence intensity was measured from adenomas compared to adjacent normal-appearing mucosa. Images of adenomas showed irregular morphology characteristic of dysplasia.
OCIS codes: (110.2350) Fiber optics imaging, (170.2150) Endoscopic imaging, (170.2680) Gastrointestinal, (170.4580) Optical diagnostics for medicine
1. Introduction
Although use of colonoscopy for early detection of colorectal cancer has led to a decreased incidence over the past two decades, this disease is still one of the most common cancers in the U.S [1]. Traditional white-light endoscopy relies on gross architectural changes and is not sensitive to molecular changes that develop as normal colonic epithelium transforms into a pre-malignant condition (dysplasia) prior to evolving into adenocarcinoma [2]. Furthermore, white-light endoscopy cannot distinguish between dysplasia and hyperplasia, a benign epithelial proliferation. Intra-vital microscopes can be used to validate the unique expression pattern of molecular targets in diseased tissues with use of highly specific exogenous probes [3,4]. Additionally, these probes can be used to guide therapy [5], stratify risk, and localize margins, aiding the clinician’s decision-making ability on a patient-to-patient basis [6].
Genetically engineered small animal models can be used to study the molecular progression of cancer that develops spontaneously over time [7]. While non-invasive techniques such as PET, bioluminescence, and MRI can serially image an animal, they cannot provide cellular detail in real time due to limitations in spatial resolution [8]. On the other hand, conventional intra-vital microscopes can achieve sub-cellular resolution, but their large dimensions in general require either an invasive incision or use of optical windows, limiting serial studies or working distance, respectively [9]. We have recently demonstrated a miniature microendoscope that can directly and repetitively image the epithelium of deep tissues with sub-cellular resolution in living mice [10].
Our laboratory has previously discovered a fluorescent peptide probe that specifically binds to murine colorectal dysplasia. This peptide (QPIHPNNM) was FITC-tagged and used to localize colonic dysplasia in vivo on wide-field fluorescence endoscopy [11]. Here, we aim to demonstrate the use of microendoscopy to validate selective binding by this peptide on a sub-cellular scale. This microendoscope is sufficiently small to pass through the instrument channel of the endoscope for direct placement onto the mucosal surface. Its fast frame rate can overcome motion artifact from the live animal introduced by organ peristalsis, heart beating, and breathing. We use this instrument to validate specific binding activity of QPIHPNNM to dysplasia in comparison to adjacent normal mucosa and to hyperplasia. Additionally, we validate the in vivo images with conventional confocal images collected with a bench top instrument. This study demonstrates a rigorous methodology for the validation of novel molecular probes being developed as a targeting agent for disease in hollow organs.
2. Methods
2.1. System design
A schematic of the imaging system is shown in Fig. 1 . A diode pumped solid state laser (CNI MBL-III-473, Changchun, China) produces emission centered at 473 nm. The power is adjusted to achieve 1 mW at the distal end of the microendoscope. The beam is collimated by an aspheric lens (L1, f = 18 mm), reflected by a 495 nm dichroic beamsplitter (Semrock FF495-Di02-25x36), and focused into a coherent fiber bundle (Sumitomo IGN-06/17, Osaka, Japan) by an infinity corrected objective lens (Olympus UIS2 PLN 20X, Tokyo, Japan). The bundle has ~17,000 fibers with core-to-core spacing of ~4 µm. The fluorescence image is magnified by the objective, passes through the dichroic, and is focused by a condenser lens (L2, f = 100 mm) onto a CCD camera (Hamamatsu Orca R2, Hamamatsu City, Japan). A 500 nm long-pass filter (LPF, Semrock BLP01-488R-25) blocks the reflected excitation light.
Fig. 1.
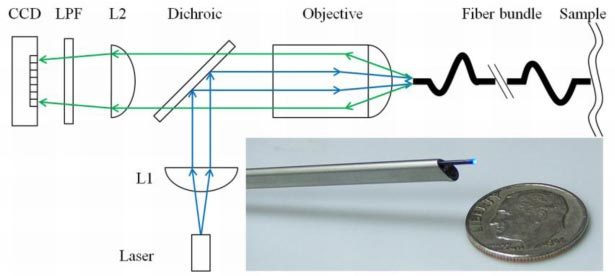
Schematic. Excitation from the laser diode at 473 nm is collimated by an asphere (L1), reflected by a dichroic beamsplitter, and focused into a 680 µm (O.D.) coherent fiber bundle. Fluorescence is transmitted to the CCD. The bundle is sufficiently small in dimension to pass through the instrument channel of a small animal endoscope (inset).
The 680 µm outer diameter fiber bundle is sufficiently small to pass through the 3 Fr (~1 mm diameter) instrument channel of the rigid endoscope (Karl Storz, Tuttlingen, Germany) (Fig. 1, inset). This endoscope has outer dimensions of 2.7 mm x 3.5 mm, and is used to localize the adenomas with white light illumination [12]. The instrument channel is also used to administer the peptides and to provide insufflation to distend the lumen of the colon for imaging.
2.2. Animal models
All animal experiments were approved by the University of Michigan Committee for Use and Care of Animals (UCUCA). We used the CPC;Apc mouse model of spontaneous colorectal dysplasia, developed by Hinoi et al. [13], to validate the peptide probe. APC is a tumor suppressor gene that is mutated in over 75% in human colorectal tumors [14]. In this mouse, the Apc gene locus is deleted by a Cre recombinase, resulting in the development of adenomas in the distal colon and rectum beginning at 10 weeks of age. Cre( + ) mice develop adenomas; Cre(-) mice and the Kras mouse model of colorectal hyperplasia are used as controls [15]. The CPC;Apc mice ranged in age from 5 to 6 months, and the Kras mice from 1 to 2 months.
2.3. In vivo microendoscopy
The target (QPIHPNNM) and control (GGGAGGGA) peptides were synthesized with Fmoc chemistry, conjugated with 5′-fluorescein isothiocyanate (FITC) via amino hexanoic acid linker, and purified with HPLC, as previously described [11]. These peptides were diluted to 100 µM in 1X phosphate buffered saline (PBS). Mice were placed prone and anesthetized with isoflurane. After rinsing the colon with water to remove debris and mucous, approximately 1 mL of the peptide solution was administered intra-luminally to the mucosa. The peptide was allowed to incubate for 5 minutes, and subsequently, the unbound peptide was rinsed 3X with water. The endoscope was used to localize adenomas with white light illumination. The microendoscope was then passed through the instrument channel, and the distal end was positioned for complete contact with the mucosal surface. The white light source was turned off, and fluorescence images were collected with an exposure of 100 ms (frame rate of 10 Hz). All microendoscopy images are presented as single frames extracted from video recordings. A representative video is provided for target peptide on the surface of an adenoma.
The target and control peptides are individually applied to the mucosa of Cre( + ) mice (n = 6 and n = 4, respectively). Microendoscopic images were acquired from the surface of the one adenoma and the adjacent normal-appearing mucosa in each mouse. The specificity of QPIHPNNM for dysplasia was further validated by applying the peptide to normal mucosa of Cre(-) mice (n = 1) that do not develop adenomas, and to hyperplastic mucosa of Kras mice (n = 2). Autofluorescence images were taken from Cre( + ) mice (n = 1) and Kras mice (n = 2) for purposes of comparison.
2.4. Ex vivo confocal microscopy
To validate the in vivo images collected with the microendoscope, we collected fluorescence images from freshly excised tissues with a bench top confocal microscope (Olympus FluoView 500, Tokyo, Japan). The mice were euthanized, and a pair of adenomas were excised (n = 3 mice, n = 5 adenoma pairs) from each mouse. Adenomas were incubated in 100 µM solution of either QPIHPNNM or GGGAGGGA for 5 minutes, and then rinsed 3X with PBS. The adenomas were then placed with the mucosal surface facing the objective lens and imaged using 488 nm excitation.
2.5. Data analysis
Microendoscopic images were collected at 12-bit resolution (0 – 4095 AU). All images were acquired as videos of 2 second duration, and contained 21 frames. Criteria for frame selection include: 1) entire image being in focus (flat contact of microendoscope with the mucosal surface), and 2) no saturated pixels. The mean fluorescence intensity ± one standard deviation error was then determined for each image. Those mean intensities were then averaged for all adenomas and normal tissues from each mouse. Additionally, we measured autofluorescence from the adenomas and hyperplastic mucosa. Statistical significance was determined by 2-sample t-test, α<0.05 (Minitab 16, State College, PA, USA).
The target-to-background ratios (T/B) of peptide binding to adenomas versus normal-appearing adjacent mucosa for each peptide were calculated by dividing the average mean intensity from adenomas by that from normal mucosa for each mouse. The results from all mice were then averaged to determine the overall T/B for each peptide.
3. Results
3.1. System design
The microendoscope achieves a field-of-view with a diameter of 540 µm and has a lateral resolution of 4 µm, verified by a standard (USAF) resolution target.
3.2. In vivo microendoscopy
The microendoscopy images revealed that the target peptide QPIHPNNM demonstrates greater binding to adenomas than to the normal-appearing surrounding mucosa, as demonstrated by the greater fluorescence intensity (Fig. 2a-b ). In addition, the binding pattern reflects cellular features specific to the mucosal surface. In contrast, the control peptide (GGGAGGGA) shows minimal fluorescence and does not reveal cellular features for either dysplastic or normal mucosa (Fig. 2c-d). Furthermore, QPIHPNNM shows minimal fluorescence signal from the normal mucosa, demonstrating little non-specific binding (Fig. 2e). The autofluorescence signal from adenomas is also minimal (Fig. 2f). Fluorescence intensity from QPIHPNNM applied to the hyperplastic mucosa of the Kras mouse model (Fig. 2g) is comparable to the autofluorescence signal from the same tissue (Fig. 2h). The full acquired video for Fig. 2a is provided (Media 1 (775.7KB, MOV) ).
Fig. 2.
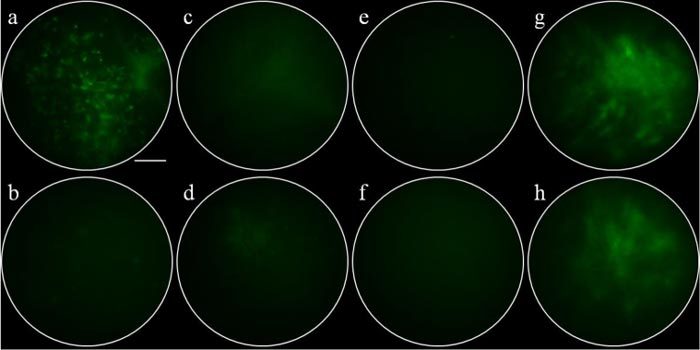
In vivo microendoscopy images. QPIHPNNM applied to the surface of a) an adenoma (Media 1 (775.7KB, MOV) ) and b) normal-appearing adjacent mucosa. GGGAGGGA (control peptide) applied to the surface of c) an adenoma and d) normal-appearing adjacent mucosa. e) QPIHPNNM applied to normal colonic mucosa in Cre(-) mice. f) Autofluorescence from adenoma in Cre(+) mice. g) QPIHPNNM applied to hyperplastic mucosa of Kras mouse. h) Autofluorescence signal from Kras mouse. Scale bar = 100 µm.
The mean fluorescence intensity from adenomas averaged across all mice, 773 ± 99, is significantly higher than that from the normal-appearing surrounding mucosa, 384 ± 93, (p<0.001). The T/B of QPIHPNNM from adenomas to the adjacent normal mucosa, 2.11 ± 0.61, is significantly higher than that of the control peptide GGGAGGGA, 1.16 ± 0.10 (p = 0.016) (Fig. 3a ). The autofluorescence, measured from adenomas on the Cre( + ) mice and hyperplasia on the Kras mice, are 297 ± 48 and 471 ± 36, respectively. After subtraction of autofluorescence, the mean intensity from QPIHPNNM applied to adenomas, normal-surrounding mucosa of Cre( + ) mice, hyperplasia, and normal mucosa of Cre(-) mice are 476 ± 110, 87 ± 105, 46 ± 64, and 81 ± 62, respectively (Fig. 3b), resulting in a T/B for dysplasia to normal (Cre + ), hyperplasia, and normal (Cre-) of 5.5, 10.3, and 5.9, respectively.
Fig. 3.
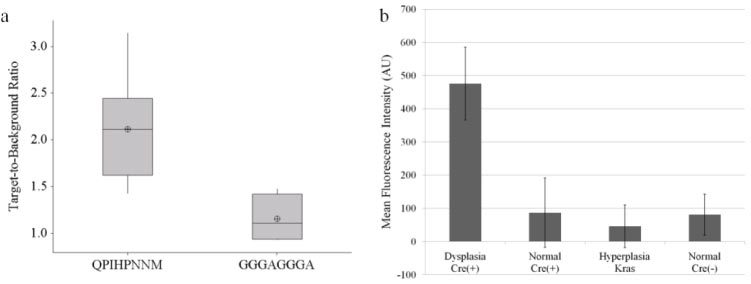
Quantification of average fluorescence intensities. (a) Boxplot of T/B of peptides from adenomas to adjacent normal mucosa. T/B of QPIHPNNM is significantly higher than that of GGGAGGGA. Lower, middle, and top lines of boxes indicate lower quartile, median, and upper quartile, respectively. Whiskers indicate minima and maxima, and crosshairs indicate means. (b) Mean fluorescence intensity of QPIHPNNM, after autofluorescence subtraction. Fluorescence signal from dysplasia is at least five-fold greater than from all other tissues.
3.3. Comparison of in vivo microendoscopy to ex vivo confocal microscopy
Fluorescence images of freshly excised adenomas incubated with QPIHPNNM and GGGAGGGA collected with a bench top confocal microscope are shown in Fig. 4 . The image of QPIHPNNM (Fig. 4a) demonstrates binding pattern of the target peptide to single epithelial cells, representing similar morphological features to that seen on microendoscopy. Comparatively, the ex vivo confocal microscopy (Fig. 4b) image shows minimal fluorescence signal and no resolvable cellular features using the GGGAGGGA (control) peptide. Hematoxylin and eosin stained biopsy specimens of adenomas reveal enlarged nuclei and distorted crypts characteristic of dysplasia (Fig. 4c), as previously shown [11].
Fig. 4.

Ex vivo confocal microscopy images. (a) QPIHPNNM shows binding to single epithelial cells (arrow). (b) Minimal binding was revealed on using the GGGAGGGA peptide. (c) Histology confirms dysplastic crypts in adenoma biopsy specimens. Scale bar = 20 µm.
4. Discussion
Here, we demonstrate a methodology for validation of selective binding of a fluorescence-labeled peptide to colonic dysplasia with sub-cellular resolution in living mice using a microendoscope. The experiments confirm specific binding activity of the target peptide (QPIHPNNM) to dysplasia in comparison to hyperplastic and normal colonic mucosa and to the control peptide (GGGAGGGA) on a microscopic scale. The T/B of both the target and control peptides for adenomas and to adjacent normal mucosa on microendoscopy, shown in Fig. 3a, are consistent with those found for the same peptides on wide-field fluorescence endoscopy [11]. This result is expected because the microendoscope images the mucosal surface, as does the wide-field fluorescence endoscope. Moreover, The T/B of the target peptide improves with subtraction of mucosal autofluorescence. This integrated strategy combines the use of a wide-area endoscope to guide placement of the microendoscope to achieve both a large field of view for localization and sub-cellular resolution for validation in real time.
In addition, a comparison of the microendoscope to bench top confocal images of peptide binding to adenomas reveals similar spatial patterns. This result demonstrates the ability of the microendoscope to acquire sub-cellular images in living mice, in real time, allowing for the study of molecular expression patterns of pre-malignant epithelial cells in vivo, using each animal as its own control. This enabling technology can greatly reduce the number, hence cost, of animals needed to perform validation studies with robust statistics. In addition, the process of euthanizing animals and excising tissue may introduce artifacts in the data. Given the time needed to dissect the animal, resect the specimen, and bring the tissue to the microscope, the fluorescent probes may undergo enzymatic degradation or photobleaching, and the cells may undergo apoptosis or desiccation. Moreover, surgical incisions may introduce trauma, hypoxia, and bleeding that can alter the molecular expression patterns.
5. Conclusion
These results demonstrate a rigorous methodology for multi-scale validation of a novel targeting agent being developed to localize disease that can be generalized to hollow organs.
Acknowledgments
Research was supported in part by NIH grants P30 DK034993, P50 CA93990, and R01 CA142750. The bench top confocal microscopy work was performed in the Microscopy & Image Analysis Laboratory (MIL) at the University of Michigan.
References and links
- 1.American Cancer Society, Cancer Facts & Figures 2010 (American Cancer Society, Atlanta, Ga., 2010). [Google Scholar]
- 2.R. S. Cotran, Robbins and Cotran Pathalogic Basis of Disease, 7th ed., V. Kumar, A. K. Abbas, and N. Fausto, eds. (Saunders, Philadelphia, Pa., 2005). [Google Scholar]
- 3.Goetz M., Ziebart A., Foersch S., Vieth M., Waldner M. J., Delaney P., Galle P. R., Neurath M. F., Kiesslich R., “In vivo molecular imaging of colorectal cancer with confocal endomicroscopy by targeting epidermal growth factor receptor,” Gastroenterology 138(2), 435–446 (2010). 10.1053/j.gastro.2009.10.032 [DOI] [PubMed] [Google Scholar]
- 4.Hsiung P. L., Hardy J., Friedland S., Soetikno R., Du C. B., Wu A. P., Sahbaie P., Crawford J. M., Lowe A. W., Contag C. H., Wang T. D., “Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy,” Nat. Med. 14(4), 454–458 (2008). 10.1038/nm1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciardiello F., Tortora G., “EGFR antagonists in cancer treatment,” N. Engl. J. Med. 358(11), 1160–1174 (2008). 10.1056/NEJMra0707704 [DOI] [PubMed] [Google Scholar]
- 6.De Roock W., Biesmans B., De Schutter J., Tejpar S., “Clinical biomarkers in oncology: focus on colorectal cancer,” Mol. Diagn. Ther. 13(2), 103–114 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Sharpless N. E., Depinho R. A., “The mighty mouse: genetically engineered mouse models in cancer drug development,” Nat. Rev. Drug Discov. 5(9), 741–754 (2006). 10.1038/nrd2110 [DOI] [PubMed] [Google Scholar]
- 8.Sandhu G. S., Solorio L., Broome A.-M., Salem N., Kolthammer J., Shah T., Flask C., Duerk J. L., “Whole animal imaging,” Wiley Interdiscip Rev. Syst. Biol. Med. 2(4), 398–421 (2010). 10.1002/wsbm.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goetz M., Fottner C., Schirrmacher E., Delaney P., Gregor S., Schneider C., Strand D., Kanzler S., Memadathil B., Weyand E., Holtmann M., Schirrmacher R., Weber M. M., Anlauf M., Klöppel G., Vieth M., Galle P. R., Bartenstein P., Neurath M. F., Kiesslich R., “In-vivo confocal real-time mini-microscopy in animal models of human inflammatory and neoplastic diseases,” Endoscopy 39(4), 350–356 (2007). 10.1055/s-2007-966262 [DOI] [PubMed] [Google Scholar]
- 10.Elahi S. F., Liu Z., Luker K. E., Kwon R. S., Luker G. D., Wang T. D., “Longitudinal molecular imaging with single cell resolution of disseminated ovarian cancer in mice with a LED-based confocal microendoscope,” Mol. Imaging Biol. (2010), doi:. 10.1007/s11307-010-0455-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller S. J., Joshi B. P., Feng Y., Gaustad A., Fearon E. R., Wang T. D., “In vivo fluorescence-based endoscopic detection of colon dysplasia in the mouse using a novel Peptide probe,” PLoS ONE 6(3), e17384 (2011). 10.1371/journal.pone.0017384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker C., Fantini M. C., Neurath M. F., “High resolution colonoscopy in live mice,” Nat. Protoc. 1(6), 2900–2904 (2007). 10.1038/nprot.2006.446 [DOI] [PubMed] [Google Scholar]
- 13.Hinoi T., Akyol A., Theisen B. K., Ferguson D. O., Greenson J. K., Williams B. O., Cho K. R., Fearon E. R., “Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation,” Cancer Res. 67(20), 9721–9730 (2007). 10.1158/0008-5472.CAN-07-2735 [DOI] [PubMed] [Google Scholar]
- 14.Arnold C. N., Goel A., Blum H. E., Boland C. R., “Molecular pathogenesis of colorectal cancer: implications for molecular diagnosis,” Cancer 104(10), 2035–2047 (2005). 10.1002/cncr.21462 [DOI] [PubMed] [Google Scholar]
- 15.Trobridge P., Knoblaugh S., Washington M. K., Munoz N. M., Tsuchiya K. D., Rojas A., Song X., Ulrich C. M., Sasazuki T., Shirasawa S., Grady W. M., “TGF-β receptor inactivation and mutant Kras induce intestinal neoplasms in mice via a β-catenin-independent pathway,” Gastroenterology 136(5), 1680–1688e7 (2009). 10.1053/j.gastro.2009.01.066 [DOI] [PMC free article] [PubMed] [Google Scholar]


