Abstract
X-linked mental retardation (XLMR) is a complex human disease that causes intellectual disability1. Causal mutations have been found in approximately 90 X-linked genes2; however, molecular and biological functions of many of these genetically defined XLMR genes remain unknown. PHF8 (PHD Finger 8) is a JmjC domain-containing protein and its mutations have been found in patients with XLMR and craniofacial deformities. Here we provide multiple lines of evidence establishing PHF8 as the first mono-methyl histone H4 lysine 20 (H4K20me1) demethylase, with additional activities towards histone H3K9me1 and me2. PHF8 is located around the transcription start sites (TSS) of ~7,000 refseq genes and in gene bodies and intergenic regions (non-TSS). PHF8 depletion resulted in up-regulation of H4K20me1 and H3K9me1 at the TSS and H3K9me2 in the non-TSS sites, respectively, demonstrating differential substrate specificities at different target locations. PHF8 positively regulates gene expression, which is dependent on its H3K4me3-binding PHD and catalytic domains. Importantly, patient mutations significantly compromised PHF8 catalytic function. PHF8 regulates cell survival in the zebrafish developing brain and jaw development, thus providing a potentially relevant biological context for understanding the clinical symptoms associated with PHF8 patients. Lastly, genetic and molecular evidence supports a model whereby PHF8 regulates zebrafish neuronal cell survival and jaw development in part by directly regulating the expression of the homeodomain transcription factor MSX1/MSXB, which functions downstream of multiple signaling and developmental pathways3. Our findings suggest that an imbalance of histone methylation dynamics plays a critical role in XLMR.
XLMR affects 1–4 out of 2,000 males, causing intellectual disability (Intelligence Quotient (IQ) <70)1. Approximately 1/3 of them are syndromic patients in that they also have additional clinical symptoms including defects in craniofacial, muscular and hematopoietic development 1. About 25% of the XLMR genes identified by human genetic studies are predicted to encode nuclear proteins including transcriptional regulators and chromatin remodeling proteins 2,4. However, molecular mechanisms by which these proteins regulate biological processes relevant to the disease remain incompletely understood.
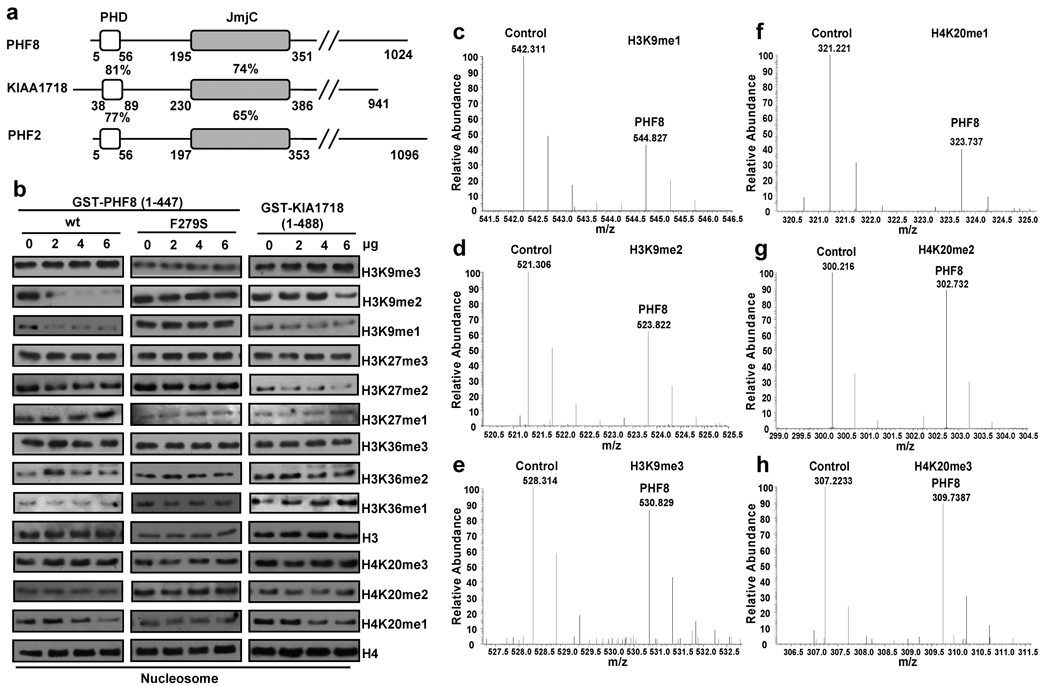
PHF8 belongs to a subfamily of JmjC domain-containing proteins that also includes KIAA1718 and PHF2. All three proteins share a similar domain structure with an N-terminus PHD finger adjacent to the catalytic JmjC domain (Figure 1a), predicting that they are candidate histone demethylases. We used nucleosomes as substrates and identified a novel H4K20me1-specific demethylase activity for PHF8 and KIAA1718, with additional activities towards H3K9me1/2 and H3K27me2, but not H4K20me2/3 or trimethylated H3K9, 27 and 36 (Figure 1b). However, when histone octomers were used as substrates, we were unable to detect the H4K20me1 demethylase activity (Supplemental Figure 2 and Supplemental Table 1), consistent with a number of recent reports that identified PHF8 as an H3K9me1/2 and H3K27me2 specific demethylase5–9. This discrepancy could be due to the use of nucleosomes versus histone octmers as substrates, and, as such, is not unprecedented. For instance, a number of histone methyltransferases have been shown to only methylate nucleosomal substrates or to methylate a specific lysine residue only in the nucleosomal context10,11. Importantly, the above in vitro demethylation reactions were also analyzed by mass spectrometry, which confirmed the Western blot results (Figure 1c to h and Supplemental Figure 1). Furthermore, a point mutation in the catalytic domain (phenylalanine to serine, F279S) abolished PHF8 demethylase activities (Figure 1b), suggesting that the reduction of the H3K9me1/2 and H4K20me1 signals by the wild type PHF8 was due to its demethylase activity. The F279S mutation has been found in several male XLMR patients from a Finnish family12, indicating the importance of the demethylase activity in PHF8-associated mental retardation.
Figure 1. PHF8 and KIA1718 demethylate multiple lysines of histone 3 and 4 in vitro.
a. Schematic diagram of human PHF8 subfamily proteins. Percentages indicate identical amino acid in the PHD and JmjC domains. b. In vitro demethylation assays. Mono-nucleosomes (1µg) were incubated with GST-fused wtPHF8, PHF8 (F279S) and KIAA1718 proteins followed by Western blot with indicated antibodies. c–h. Mass spectrometry analysis of Control (GST) and PHF8 reacted nucleosomes, which were isotopically labeled with D0-propionyl and D5-propionyl, respectively. This labeling induces a 5 Da mass shift between the two samples, which is observed as a 2.5 m/z shift for doubly charged peptides. The m/z ranges depicting peptides are indicated. Decreases in H3K9me1 (c), H3K9me2 (d) and H4K20me1 (f) were detected from the PHF8 reaction compared to GST only. No changes were observed in others.
We next investigated the impact of PHF8 on the global H4K20 and H3K9 methylation states in the cells. PHF8 over-expression in U2OS but not HeLa cells, which express a high level of PHF8 (Supplemental Figure 3), resulted in a significant reduction of H3K9me2 (but not H3K9me1), and only a slight reduction of the global level of H4K20me1 (~10%) (Supplemental Figure 4, and Table 2). Although over-expression or RNAi of PHF8 in HeLa cells did not affect the global levels of H4K20me1 and H3K9me1 assayed by Western blotting (Supplemental Figure 5a), we did observe an increase in the overall H4K20me1 level in the late G2/M and early G1 stages of the cell cycle when we inhibited PHF8 expression by RNAi (Supplemental Figure 5b). This is consistent with the previous reports that H4K20me1 is cell cycle-regulated13,14 and provides further support that PHF8 regulates H4K20me1 in vivo. Interestingly, inhibition of PHF8 also reduced cell proliferation (Supplemental Figure 6), suggesting a possible role for PHF8 in regulation of cell growth via histone demethylation.
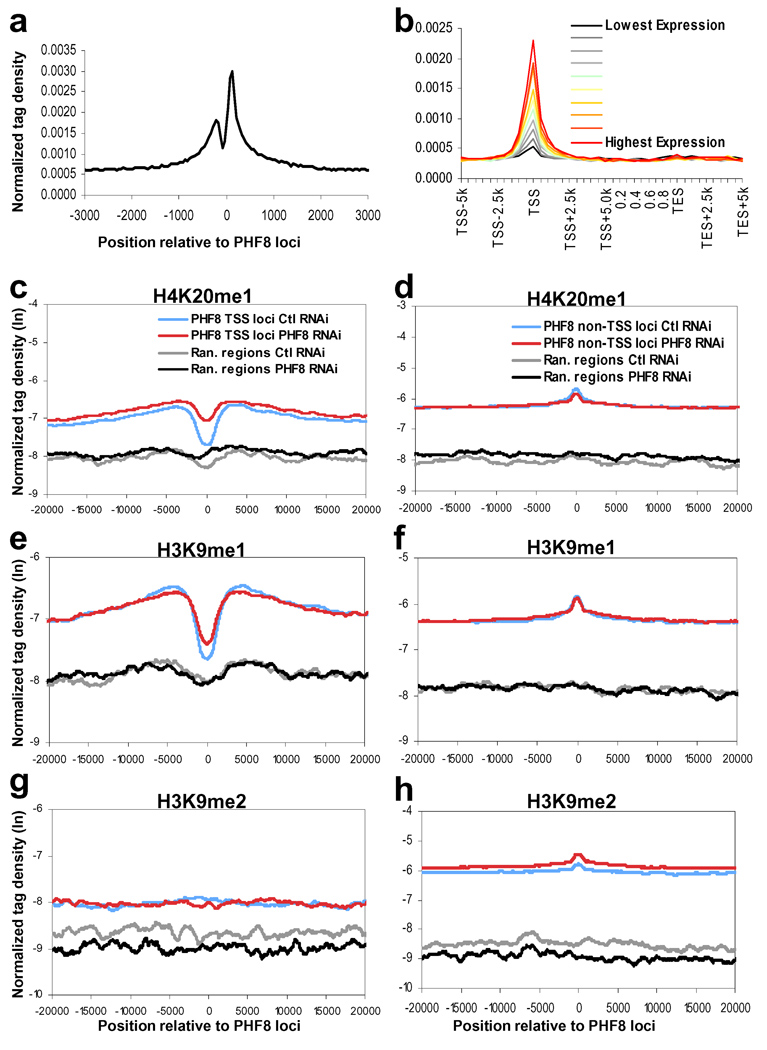
To identify the genomic locations of PHF8, we performed ChIP-seq (chromatin immunoprecipitation followed by DNA deep sequencing). We obtained 17,293 peaks in HeLa cells after IgG normalization. Of those, 8,077 peaks representing 7,469 refseq genes are mapped to within 2.5 kb of TSS (Supplemental Table 3) as well as 9,824 potential PHF8 binding events in the gene bodies (23.5%) and intergenic regions (33.3%) (Figure 2a). This pattern of PHF8 genomic distribution was corroborated by ChIP-chip analysis. Importantly, majority of the PHF8 binding signals identified by ChIP-chip were erased by PHF8 RNAi, supporting the specificity of the PHF8 binding events detected by the PHF8 antibody (Supplemental Figures 3, 7 and Table 4). Interestingly, the pattern of PHF8 binding around TSS (Figure 2a) resembles that of H3K4me315,16, consistent with the fact that the PHD domain of PHF8 binds H3K4me35,17. Point mutations (Y7A, W29A, Y7A/Y14A, W29A/Y14A) that abolished binding of PHF8 to H3K4me3 in vitro disrupted its localization in vivo (Supplemental Figure 8), suggesting that PHF8 TSS localization may be dependent on its PHD domain recognizing H3K4me3. Importantly, analysis of the ChIP-seq and gene expression microarray data (Supplemental Table 6) shows that PHF8 binding events are positively correlated with gene expression (Figure 2b), suggesting that PHF8 binds H3K4me3 and functions to promote gene transcription.
Figure 2. PHF8 differently regulates H4K20me1, H3K9me1 and H3K9me2.
a. Profile of PHF8 binding on all UCSC annotated genes. Normalized tag density of PHF8 ChIP-seq in regions of transcription start sites (TSS) at resolution of 50 bp. b. Correlation between PHF8 binding and gene expression. Normalized tag density of PHF8 across transcripted genes, which are separated into ten equal size groups based on expression levels (see Supplemental methods). c, e, g. Histone modification profiles of H4K20me1, H3K9me1, and H3K9me2 in control and PHF8 RNAi cells around PHF8 binding sites near TSS regions of 7,469 refseq genes. Randomly chosen sites from TSS regions not bound by PHF8 are shown for comparison. d, f, h. Histone modification profiles of H4K20me1, H3K9me1, and H3K9me2 in control and PHF8 RNAi cells around PHF8 binding sites in 9,824 non-TSS regions. Randomly chosen sites from non-TSS regions not bound by PHF8 are shown for comparison.
We next used ChIP-seq to investigate histone methylation regulation by PHF8 at its direct loci using HeLa cells stably expressing either control or PHF8 shRNA. We found that PHF8 depletion caused a modest albeit significant increase in H4K20me1 and H3K9me1 at the PHF8 TSS target loci (Figure 2c and e), compared with the regions not bound by PHF8 (p-value < 0.01), suggesting that PHF8 regulates transcription by demethylating H4K20me1 and H3K9me1. Unlike H4K20me1 and H3K9me1, H3K9me2 level remained unchanged (Figure 2g). Interestingly, for the 9824 non-TSS PHF8 sites, we found that reduction of PHF8 is instead correlated with an increase in H3K9me2 (p-value < 0.01) (Figure 2h), but not H3K9me1 or H4K20me1 (Figures 2d and f), indicating that PHF8 may regulate non-TSS regions via demethylation of H3K9me2. Consistently, a recent study reported PHF8 regulation of rDNA transcription via demethylation of H3K9me25. The finding that PHF8 may exhibit differential substrate specificities at different genomic locations (TSS versus non-TSS) implies that additional factors are involved in the regulation of PHF8 demethylase specificity in vivo.
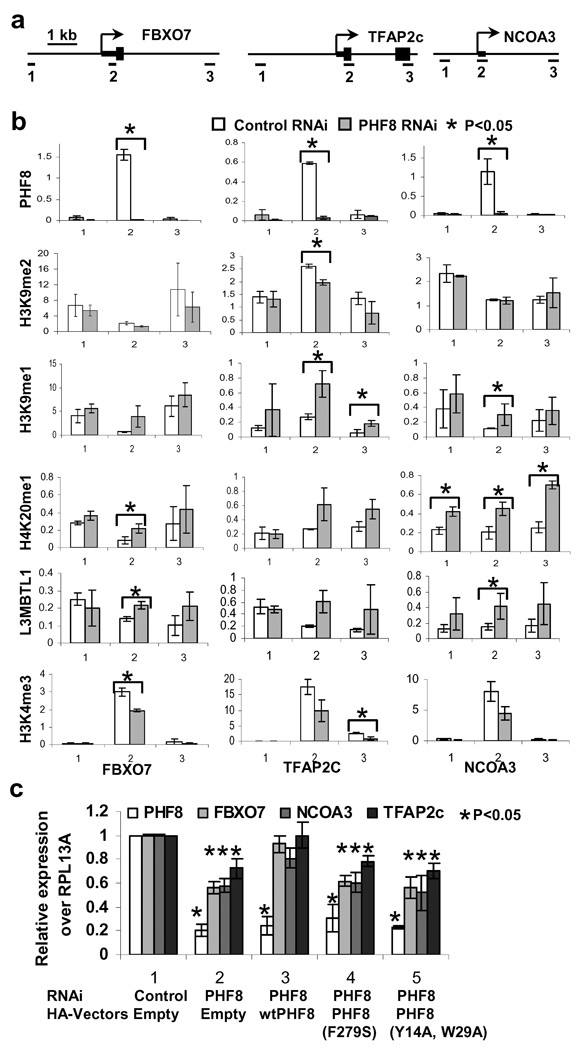
To confirm the ChIP-seq histone modification data, we initially focused on three TSS-bound PHF8 target genes, FBXO7, TFAP2C and NCOA3, and first confirmed PHF8 binding near TSS (amplicon 2) by conventional ChIP assays (Figures 3a and b). Inhibition of PHF8 resulted in up-regulation of H4K20me1 and H3K9me1 at amplicon 2 of the target genes (Figure 3b). In some instances, we also observed an increase in H4K20me1 and H3K9me1 in amplicons #1 and 3 for reasons that are currently unclear. In contrast, we found no appreciable changes in H3K9me2, H3K27me1/2 or H3K36me1/2 (Supplemental Figure 9), many of which are also substrates of PHF8 in vitro (refer to Figure 1 and Supplemental Figures 1 and 2). We also did not observe changes in histone H3, suggesting that the increase in H4K20me1/H3K9me1 is not due to an increase in nucleosome density (Supplemental Figure 9). We further investigated an additional seven PHF8 target genes and found H4K20me1 and H3K9me1 increase for some if not all of these targets (Supplemental Figure 11). We next investigated the impact of PHF8 inhibition on the expression of these three target genes. As shown in Figure 3c, Consistent with the bioinformatics analysis (Figure 2b) implicating PHF8 as a positive regulator of transcription, their expression was down-regulated in PHF8 RNAi cells, restored by wild-type PHF8 but not by the patient F279 mutation or the H3K4me3-binding defective PHD finger mutant (Y7A, W29A). The expression of the addition seven genes was similarly reduced upon PHF8 inhibition (Supplemental Figure 11). Taken together, these data indicate that PHF8 demethylates H4K20me1 and H3K9me1 both in vitro and in vivo, and that both H3K4me3-binding and catalytic functions of PHF8 are important for its ability to positively regulate gene expression.
Figure 3. Depletion of PHF8 increases H3K9me1, H4K20me1, L3MBTL1 at TSS-bound PHF8 target genes.
a. Three amplicons for real-time PCR are illustrated for the three direct PHF8 target genes. Amplicon 2 spans the PHF8 binding site. b. Conventional ChIP assays with indicated antibodies were performed using HeLa cells stably expressing control or PHF8 shRNA. Real time PCR was performed on the input and ChIP DNA. Data are presented as % of input. p values were obtained by T-Test analysis and those that are lower than 0.05 are indicated by asterisks. c. HeLa stable cell lines were established that co-express control or PHF8 shRNA and/or indicated HA-tagged PHF8 constructs. mRNA expression of the three selected genes and endogenous PHF8 was measured by RT-Real time PCR. RPL13A was used as an internal control. p values were obtained by T-Test by comparing the data from column 2, 3, 4 and 5 with those of the corresponding controls (column 1). Standard deviations from both b and c were obtained from three independent experiments
Although H4K20me1 has been found downstream of TSS on active genes16,18, it has also been associated with repression and recruitment of L3MBTL1, which preferentially binds mono- and dimethylated lysines including H4K20me1 and H3K9me119 and induces chromatin compaction to further negatively regulate gene expression20. Upon PHF8 knockdown, we also observed a corresponding increase of L3MBTL1 at amplicon 2 but not amplicons 1 and 3 (Figure 3b and Supplemental Figure 10). This is interesting in light of the fact that the H4K20me1 increase in PHF8 RNAi cells extends beyond amplicon 2, suggesting that factors, in addition to H4K20me1, are involved in determining L3MBTL1 recruitment. Additionally, we observed a reduction of H3K4me3, a mark that is associated with active genes, consistent with these genes being in a transcriptionally repressed state (Figure 3b and Supplemental Figure 10). Taken together, these findings support the model that PHF8, when bound to TSS of the target genes, positively regulates transcription by demethylating H3K9me1 and H4K20me1.
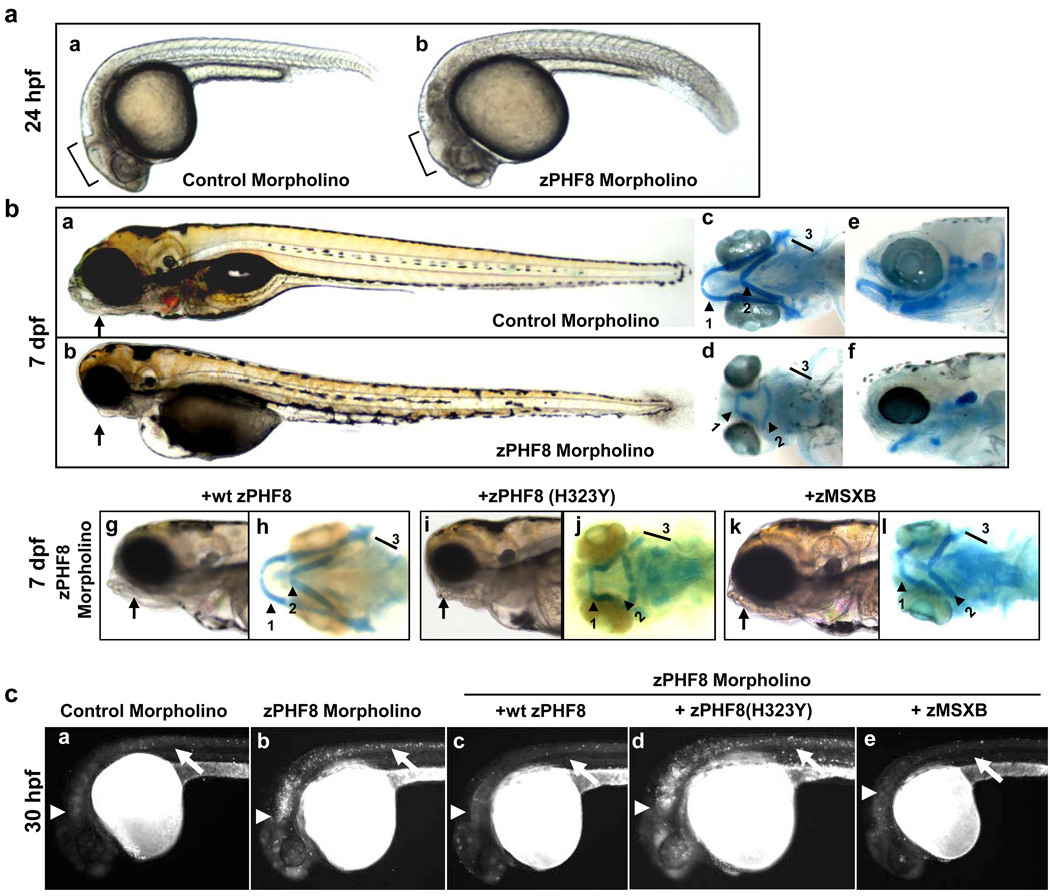
To address the biological function of PHF8, we turned to zebrafish where PHF8 is evolutionarily conserved (Supplemental Figure 11). zPHF8 expression was first detected at 14 hours post-fertilization (hpf) in the head and tail regions (Supplemental Figure 12), but starting at 1 day post-fertilization (dpf), zPHF8 was found mostly in the head region. zPHF8 expression was also detectable in the jaw of the embryo at 3 dpf. Injection of a zPHF8 morpholino caused delay in brain development at 24 hpf (Figure 4a, compare panel b with a), and apoptosis in the developing brain and the neural tube at 30 hpf (Figure 4c, compare panel b with a). Significantly, this apoptosis can be rescued by reintroduction of wild type but not catalytically inactive zPHF8 (Figure 4c, compare panels c, d with a, b). However, whether PHF8 regulates cell death by directly modulating the apoptotic machinery or indirectly through regulation of cell proliferation remains to be investigated. Furthermore, it remains to be seen whether PHF8 also plays a role in post-mitotic mature neurons. In addition to apoptosis in the developing brain, we also detected significant differences in the craniofacial development between control and zPHF8 morpholino injected embryos at 3, 4 and 7 dpf, with the most pronounced defect being the absence of a lower jaw (Figure 4b, compare panel a with b, Supplemental Figure 13). To better understand the craniofacial developmental abnormality, we used Alcian blue to visualize the extent of cartilage development in these embryos. We found that the 1st through 5th pharyngeal arches were all either developmentally affected or were absent upon zPHF8 inhibition (Figure 4b, compare panel d,f with c,e). Specifically, the 1st pharyngeal arch had not migrated to the full extent of the lower jaw and the 2nd pharyngeal was inverted. The 3rd through 5th pharyngeal arches were also significantly under-developed. Importantly, wild type but not catalytically inactive zPHF8 showed significant rescue of the craniofacial defects induced by the zPHF8 morpholino (Figure 4b, Supplemental Table 5). Collectively, these findings identified a critical role of zPHF8 in zebrafish brain and craniofacial development, in a manner that is dependent on its demethylase activity.
Figure 4. zPHF8 regulates brain and craniofacial development in part through regulation of MSXB.
a. Zebrafish embryos injected at the 1 cell stage with 250uM of control or zPHF8 morpholino (MO). At 24 hours post fertilization (hpf), brain development was delayed (compare panels a and b). b. At 7 days post fertilization (dpf) PHF8 MO embryos displayed craniofacial developmental abnormalities including stunted lower jaw (compare panels a and b). Alcian Blue staining of the embryos at the same stages shows stunted 1st pharyngeal arch (1), inverted 2nd pharyngeal arch (2), deformed or absent 3rd through 5th pharyngeal arches (3) (compare panels d,f with c,e). Zebrafish embryos at 1 cell stage were co-injected with 125uM zPHF8 MO with either 200ng of wtzPHF8 (g,h), zPHF8 (H323Y) (i,j) or zMSXB mRNA (k,l). At 7dpf, the embryos were monitored for their craniofacial phenotypes (panels g, i and k) and the pharyngeal arches (Alcian Blue staining) (panels h, j and l) are indicated. c. At 30 hpf, cell death in zebrafish embryos with similar treatments as in b was monitored by Acridine Orange staining. Developing brain and neural tubes are indicated with arrowheads and arrows, respectively.
The msh/Msx gene family encodes homeodomain transcription factors that act downstream of the TGFβ, BMP and WNT signaling pathways and play important roles in many developmental processes including craniofacial and neural development3. Zebrafish has five homologs, MSXA through MSXE, that constitute the zebrafish orthologs of the tetrapod MSX1, MSX2, and MSX3 genes21, of which, MSX1 was identified as a PHF8 direct target (Supplemental Figures 14a and b). Interestingly, a MSXB morpholino produced craniofacial deformities similar to those seen in a zPHF8 morphant22. We therefore investigated whether MSXB was regulated by zPHF8 and whether it functions to mediate the role of zPHF8 in zebrafish craniofacial development. We found that the msxB transcript level was reduced to 40% of wild-type level in embryos injected with a zPHF8 morpholino (Supplemental Figure 14c). The msxB transcript was largely restored to wild type level in the animals rescued with zPHF8 mRNA encoding wild type but not with a catalytically inactive form of the zPHF8 protein (Supplemental Figure 14c). Importantly, injection of msxB mRNA also significantly corrected the craniofacial defects induced by the zPHF8 morpholino (Figure 4b, compare panels k, l with a, c and g, h). Although the rescue was not complete, the lower jaw developed better and the 2nd pharyngeal arch was no longer inverted (Figure 4b, compare panels l with d and h, and data not shown). Similarly, the apoptosis seen in the developing brain and neural tube was also suppressed by forced expression of MSXB (Figure 4c, panel e). These findings raise the possibility that zPHF8 may control neuronal cell survival and craniofacial development at least in part by regulating MSXB expression. However, it is likely that MSXB is not the only PHF8 target gene that mediates PHF8 biology. Consistent with this idea, bioinformatics analysis of the expression microarray data from HeLa cells identified additional potential PHF8 target genes in a number of pathways including RA (Retinoic Acid) and Notch signaling pathways (Supplemental Table 6), both of which have been previously shown to play a role in neural and craniofacial development23,24 and therefore may also be involved in PHF8 biological functions. In addition, the PHF8 direct target genes (FBXO7, NCOA3 and TFAP2C) discussed earlier (Figure 3) also function in neural development25–27. Lastly, ChIP-seq/ChIP-chip identified many XLMR genes as potential PHF8 direct targets (Supplemental Figure 15), consistent with a recent report7. This finding raises the interesting possibility of a connection between PHF8 and other XLMR gene products in regulating cognitive functions. However, given that the ChIP-seq data were obtained from heterologous cells, whether PHF8 regulation of other XLMR genes is physiologically and pathologically relevant remains to be investigated.
In conclusion, we have identified the XLMR gene PHF8 to encode a novel H4K20me1 and H3K9me1/2 demethylase, and provided insights into molecular mechanisms by which PHF8 regulates histone methylation and gene transcription. The identification of a role for PHF8 in zebrafish brain apoptosis and craniofacial development suggests a potential biological basis for the involvement of PHF8 mutations in mental retardation and craniofacial deformities. Importantly, our finding of PHF8 as a histone demethylase whose mutations are correlated with XLMR supports the emerging theme of a critical link between histone methylation dynamics in X-linked mental retardation.
METHODS SUMMARY
In vitro demethylation assay
GST tagged PHF8 and KIAA1718 proteins and full length PHF8 protein were purified from bacteria and from insect Sf9 cells. Demethylation assay was carried out with 0.6µg of histone octamers or 1µg mono-nucleosomes in the demethylation buffer [50mM Hepes-KOH, pH7.9, 50mM KCl, 1mM MgCl2, 100µM (NH4) 2Fe(SO4) 2·6(H2O), 1mM α-ketoglutarate, and 2mM ascorbic acid] for 4 h at 37°C. Western blot was performed with antibodies recognizing specific modifications. ChIP-qPCR. Chromatin was prepared from HeLa stable cell lines expressing pBabe-control or PHF8 shRNAs and was sonicated to the range of 0.6 to 1.2kb. ChIP-chip and ChIP-qPCR experiment for PHF8 were carried out with PHF8 antibody (Ab36068) in the buffer (50mM Hepes, 500mM NaCl, 1mM EDTA, 1% triton, 0.1% Deoxylcholate-Na). For conventional ChIP of selected histone marks, 100–200µg of chromatin was used and the final ChIP DNA was dissolved in 100µl H2O. 2µl of ChIP DNA and input DNA were used for real time PCR. ChIP-chip and ChIP-seq were essentially described in the supplemental methods. Rescue experiments. Double stable HeLa cell lines were established with co-expressed pOZ-FH, or pOZ-FH-PHF8 (wt or mutant) and pBabe shRNAs (control or PHF8 RNAi1). RT was obtained from 200ng RNA followed by real-time PCR. Zebrafish experiments. Whole-mount in situ hybridization was performed using digoxigenin labeled antisense RNA probes. zPHF8 antisense morpholino was injected with at the one-cell stage. Pharyngeal arch and apoptosis were monitored by Alcian blue and acridine orange staining, respectively.
Methods
Vectors and Antibodies
Full length PHF8 (1–1024) and (60–1024) were amplified by RT-PCR and inserted into the XhoI/XbaI sites of pcDNA3-C-HA vector. Full length PHF8 was also inserted into XhoI/NotI sites of pOZ-N-Flag-HA vector. Full length KIAA1718 was also PCR-amplified and cloned into Gateway entry vector and pcDNA3-C-HA vector. GST-PHF8 (1–447), GST-PHF8(1–76), GST-KIAA1718 (1– 488) were constructed by inserting the PCR-amplified fragments into pGEX-4T-3 vector. F279S mutation was introduced into pcDNA3-PHF8-HA, GST-PHF8 (1–447), and pOZ-N-Flag-HA-PHF8 vectors by mutagenesis. Single (Y5A, Y14A, W29A) and double mutations (Y14A,W29A and Y5A,Y14A) were generated in GST-PHF8 (1–76), pcDNA3-PHF8-HA and pOZ-N-Flag-HA-PHF8 vectors. Control28 and PHF8 shRNAs were constructed in pBabe-U6-puro vector 28 with the passenger strand (5’ to 3’) sequences for PHF8 shRNA1 (ORF): GCTTCATGATCGAGTGTGACA, PHF8 shRNA2 (3’UTR): GGCCTAGAAATGCCAACTTCA. PHF8 silent mutations that confer shRNA1 resistant were introduced into pOZ -N-Flag-HA-PHF8wt, F278S, and Y5W,W29A vectors. To generate BB-Flag insect cell expression vectors, full length PHF8 was first cloned into the Gateway Entry vector (Invitrogen) and subcloned into Gateway Flag-baculoviral expression vector. Antibodies: All antibodies are listed in Supplemental Table 7. The Abcam PHF8 antibody (ab36068) recognizes only the HA-tagged PHF8 at about 145 kDa, but not HA-KIAA1718 which is at 110 kDa (Supplemental Figure 4). The Active Motif PHF8 antibody was generated using His-tagged PHF8 (539–689) as an immunogen. Both antibodies detect a single band at about 145 kDa, which was abolished by PHF8 RNAi, suggesting that they recognize endogenous PHF8 (Supplemental Figure 4).
PHF8 RNAi in HeLa cells
PHF8 stable knockdown and control HeLa cell lines were established by transfecting HeLa cells with pBabe-PHF8 shRNA1 or pBabe-control shRNA28 vectors. The transfectants were selected with puromycin (1µg/ml) till the cells are stabilized. The stable cell lines were transfected again with the corresponding shRNA for an additional 3 days for ChIP chromatin preparation, gene expression (microarray and RT-qPCR) and cell cycle analysis. The knockdown efficiency was monitored by Western blot with an anti-PHF8 antibody (Active Motif).
Mass Spectrometry
In vitro demethylation assay using GST and GST-PHF8 (1–447) was performed on nucleosomes (1µg per reaction), which were isolated from HeLa cells. To compare histone PTMs from those reacted nucleosomes, we used a chemical isotopic labeling approach recently described29 In brief, equal amounts of control and PHF8 treated total nucleosome samples were derivatizated using D0-proionic anhydride followed by trypsin digestion at a substrate enzyme ratio of 20:1 for 5 hours at 37°C. Samples were quenched by acidification with acetic acid and then labeled on the peptide N-termini using either D0- or D5-propionic anhydride followed by desalting on homemade STAGE tips as reported earlier30. Desalted peptides were loaded onto a fused silica capillary column (75um) packed with C18 resin constructed with an ESI tip by an Eksigent AS-2 autosampler (Eksigent Technologies Inc., Dublin, CA). Peptides were eluted from the C18 material at a flow rate of ~200 nL/min using a 5–40% solvent B in 60 minute gradient (solvent A= 0.1 M acetic acid, solvent B = 70% MeCN in 0.1 M acetic acid) into an Orbitrap mass spectrometer (ThermoFisher Scientific, San Jose, CA) as previously described29.
Gene expression experiments and microarray analysis
For target gene expression and rescue experiments, PHF8 shRNA1-resistant mutant pOZ-FH-wtPHF8, PHF8 (F278S), PHF8 (Y5A, W29A) and empty vectors were packed into retrovirus as described previously 28. HeLa cells were infected with the virus and selected with anti-IL-2α receptor, which was conjugated with dynal beads. The stable cell lines were transfected with control or PHF8 shRNA1 and subjected to puromycin selection. RT was performed using 100ng RNA and poly(dT) primer, real time PCR with Light cycler 480 were done with the primers listed in Supplemental Table 8. Microarray analysis was performed on Human Whole Genome OneArray™ by Phalanx Biotech Group. RNA was prepared from three independent control and PHF8 RNAi HeLa cells. Each RNA sample was subjected to replicate hybridizations to the array. Based on RT-qPCR analysis of 10 PHF8 target genes, which were down-regulated for −30% to −70% in PHF8 RNAi cells, we chose −30% and p<0.05 as cutoff for down-regulated genes. For up-regulated genes, we chose 2 fold and p<0.05.
Purification of GST fusion proteins and Flag-PHF8
GST fusion proteins were produced from Rosetta bacteria strain and purified with Glutathione Sepharose 4B (GE healthcare). Briefly, Rosetta bacteria transformed with the corresponding plasmids were cultured till OD reached 0.8 and induced with 500mM IPTG for 5 hours at RT (room temperature). The bacteria were harvested and lysed with 10mM Tris-HCl (pH8.0), 1mM EDTA, 150mM NaCl and 0.2% Triton. GST-fused proteins were eluted with 50mM Tris.HCl and 20mM Glutathione. The Flag-PHF8 was expressed in Sf9 cells using BAC-N-BLUE baculoviral expression system (Invitrogen). Cells were lysed in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40, 0.2% Triton X-100, 1 mM DTT, 1 mM PMSF, and Protease Inhibitor Cocktail (Roche). Recombinant proteins were immobilized on Flag M2 affinity gel (Sigma), washed with buffer W (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 8.0% glycerol, 1 mM DTT, 0.01% NP40), and eluted in buffer W containing 100 µg/ml of 3× Flag peptide (Sigma).
Histone peptide binding assay
GST-fused PHF8 (1–76) and corresponding mutants were purified as described above. Five micrograms of purified proteins were incubated with 0.2 µg of biotinylated histone peptide in 100 µl of the binding buffer (50 mM Tris-HCl (pH7.5), 150 mM NaCl, 0.05% NP-40, 0.3 mg/ml BSA) overnight at 4°C. Protein-peptide complexes were pulled down with streptavidin beads (Upstate), washed five times with binding buffer and subjected to SDS-PAGE gel and Commassie blue staining.
Immunofluorescence and antibodies
HA-tagged PHF8 constructs were transfected into U2OS or HeLa cells for 24 hours and plated onto coverslips in 6-well plates at 3 × 105 cells/well. 24 hours later, cells were fixed with 4% paraformaldehyde, permeabilized (0.2% Triton X-100 in PBS), and blocked with 1% BSA. The coverslips were incubated at 4° overnight with corresponding primary antibodies, washed, and incubated with the corresponding secondary antibodies for 1 hr at RT. The coverslips were then washed, mounted with Vectashield (Vector Laboratories), and analyzed by a fluorescence microscopy (Nikon E600) using a 60× objective. Images were acquired and processed with Openlab 3.1.5 software. Primary antibodies are detailed in the antibody table. Alexa 594-conjgated donkey-anti-rabbit and Alexa 488-conjugated goat-anti-mouse secondary antibodies (Molecular probes) were used at 1:10,000. Hoechst 33342 was used to stain the nucleus.
qChIP (Conventional ChIP followed by real time PCR) and ChIP-chip
Hela stable cells (Control or PHF8 RNAi) from a p150cm plate were cross-linked with 1% formaldehyde and quenched with 0.125M glycine. Cells were washed with cold PBS and then lysed with lysis buffer (50mM Hepes, 500mM NaCl, 1mM EDTA, 1% triton, 0.1% Deoxylcholate-Na) complemented with protease inhibitor Cocktail (Roche). DNA fragmentation was performed with Misonix 3000 sonicator using 1ml cell lysate with 4 cycles of 25 second ON and 45 second OFF. In qChIP, 200µg of chromatin were beads (Protein A or G beads) cleared and incubated with 4µg of PHF8 or 2µg the histone antibodies ON at 4°C. 25µl of Protein A or G beads was added for 1 hour and the complex were washed five times with the lysis buffer and once with RIPA buffer (50mM Hepes pH7.6, 300mM LiCl, 1mM EDTA, 0.5% NP40 and 0.7% NA-dexycholate). The immunoprecipitants were de-cross-linked at 65°C overnight. The immunoprecipitated DNA were dissolved in 50µl H2O. 1% of input DNA and 2µl of ChIP DNA were used for real time PCR. The primers for qChIP is shown in Supplemental Table 9.
ChIP-chip
Chromatin from HeLa cells, control RNAi and two PHF8 RNAi HeLa cells were prepared as described above. ChIP-chip was performed on an Agilent 244K array (AMADID 14841; G4495A) that tiles across the genome from chr20:045004127 to chrX: 025615738 according to the Agilent Mammalian ChIP-on-chip protocol (Version 9.2) (http://www.chem.agilent.com/Library/usermanuals/Public/ChIP-on-chip_Mammalian_9.2.pdf). Briefly, input and the immunoprecipitated DNA using PHF8 specific antibody (Ab36068) undergo linker ligations and two rounds of LM-PCR. The LM-PCR product from input and ChIP DNA were labeled with Cy3 and Cy5, respectively and hybridized to Agilent 244K array. Enrichments were obtained by extracting the Cy5 (ChIP DNA) signals over Cy3 (input DNA) signals. To identify the significant binding events among the ChIP-chip data, we first performed lowess normalization, and then applied the “Pre-defined Peak Shape Detection 2.0” algorithm provided within the ChIP Analytics software package from Agilent Technologies, using the default algorithm parameters, a score threshold of 2.0, and applying the option to repeat the peak-finding calculation after optimizing the peak-shape model. Associations of the resulting binding events with genes were determined by comparing predicted peak centers to transcription start sites as defined by the “refFlat” table of RefSeq genes31 for Hg18 provided at the UCSC Genome Browser Database32.
ChIP-seq
(Chromatin immunoprecipitation followed by DNA sequencing). Control and PHF8 shRNA knockdown HeLa cells were described above. For PHF8 ChIP-seq, cells were cross-linked with 1% formaldehyde, and quenched with 0.125M glycine. Cross-linked chromatin templates were generated as described in33. 20×107 cells were immunoprecipitated with 8 µg of anti-PHF8 antibody (ab36068). For H3K9me2,1 and H4K20me1, we generated native chromatin from 107 cells with Micrococcal nuclease to gain mainly mononucleosomes for each ChIP34. ChIP-seq library construction, Illumina sequencing, and data analyses were performed as described previously 33,34. Briefly, sequence reads of PHF8, H3K9me1, H3K9me2 and H4K20me1 were mapped to the human genome (hg18) using Solexa Analysis Pipeline. All uniquely matching reads were retained. PHF8 enriched regions were predicted with SICER using IgG as control (SICER program, E value=1,000, FDR = 0.001) 35. In brief, it first identifies islands whose tag counts are above a threshold determined by an E-value and then uses IgG control to reduce false positives (if provided) 35. Here, the E value refers to the expected number of islands whose tag counts are above the threshold under a background model of random tags. Each PHF8 site was assigned with a p-value and a false discovery rate, which measure to what extend the tag enrichment of the sites is significant compared to the IgG control 35.
To examine the correlation of PHF8 density with gene expression levels (Figure 2b), the 16,639 UCSC known genes were classified into 10 equal-size groups based on mRNA expression levels, obtained from the microarray analysis in HeLa cells as described above. The density in the promoter regions was profiled with 500-bp windows after aligning to TSS (±5 kb). The same window size was applied to the 5-kb downstream regions of transcription ending sites (TES). Profiles in gene body were generated by aligning genes in each bin relative to TSS + 5 kb and TES, and with a window size of 10% of the region length. The densities were normalized against the size of Chip-Seq library and the size of sliding windows.
To detect changes of histone modifications in PHF8 RNAi cells, we aligned ChIP-seq tags according to the centers of PHF8 sites and manipulated the modification densities with a 50-bp window. As a control, we randomly chose 3,000 non-PHF8 sites from promoter regions [±2.5 kb of TSS] and non-TSS regions, respectively. To reduce noise from the background, following the procedures described in 36, we filtered the tags outsides of the modification enriched regions determined by SICER before calculating the densities (window =200, gap =1000, E-value =1000). To test the significance of modification changes, we averaged the fold change of densities within a region 2.5 kb around the PHF8 site for each modification, and compared it with an expectation to calculate a p-value (t-test). The expectation was estimated by the ratio of tag number from control RNAi to PHF8 RNAi (normalized by library sizes) in the promoter and non-promoter regions.
Cell growth and cell cycle analysis
HeLa stable knockdown and control cells were seeded at 1×104 cells in triplicate 60mm plates. Cells were counted from day 2 till day 5. For cell cycle analysis, HeLa stable cells were seeded at 4×104 cells in 60mm plate and synchronized in S phase by a double thymidine or in mitosis with a thymidine-nocodazole block as essentially described previously37. Cells were then released from the block, stained with propidium iodide (PI) and analyzed with FACS following the protocol of Current Protocol of Cell Biology.
Zebrafish PHF8 vectors, in situ hybridization, zPHF8 knockdown and phenotypic analysis
Zebrafish PHF8 (zPHF8) was cloned by RT-PCR. The coding protein, as shown in Supplemental Figure 11, is shorter at the C-terminus than human PHF8 and the predicted zPHF8, and differences in the coding region were also noticed. pCS-N-HA- zPHF8 was cloned and H323Y mutation was introduced into this vector. pBSKS-zPHF (corresponding to PHF8 cDNA nucleotides 1868–2505) was used to generate a cRNA probe. Whole-mount zebrafish embryo in situ hybridizations using digoxigenin-labeled antisense and sense zPHF8 RNA probes were carried out following the method described previously 38. A morpholino (MO) targeting −2 to +23 of zPHF8 (5’-CAGTAAACCGGAACAGATGCCATTC-3’) and control Morpholino (5’CCTCTTACCTCAGTTACAATTTATA 3’) were obtained from Gene-Tools, LLC.
Zebrafish embryos injected at the 1 cell stage with 2nl of 250uM control or PHF8 morpholino with or without 200ng of GFP, zPHF8, or zMSXb mRNA were injected using a gas-driven microinjector (Medical Systems Corp). For in vivo detection of cell death, live 30 hpf embryos were incubated in 2 µg/ml acridine orange (Sigma) in embryo media in the dark for 30 min and washed three times in fresh embryo media, and fluorescence was observed under 488 nm wavelength excitation. Pharyngeal arches were stained with Alcian blue.
Supplementary Material
Acknowledgements
We thank Shi Lab members for helpful discussions and Drs. Nima Mosammaparast and Shuzhen Chen for providing the nucleosomes. We thank Geoff Rosenfeld for sharing unpublished PHF8 results, and Geoff Rosenfeld, Alex Schier and Phil Hinds for helpful discussions. Hank H. Qi is a recipient of Ruth L. Kirschstein-National Service Research Award (T32 NS007473, and T32 CA09031-32). Nasser Yaghi was supported by Harvard SHARP (Summer Honors Undergraduate Research Program). Huijun Lim is a recipient of the Agency for Science, Technology and Research (A*STAR) National Science Scholarship. This work was supported by grants from the National Science Foundation (CBET-0941143) to B.A.G.; NIH (GM 071004 and NCI118487) to Y.S.; a Senior Scholar Grant from the Ellison Foundation, and an NIH PO1 grant (CA50661) to T.M.R.
Footnotes
Full methods and any associated references are available in the online version of the paper at www.nature.con/nature
Supplemental Information is linked to the online version of the paper at www.nature.com/nature
Author contributions
Y.S and H.H.Q conceived, designed the study, and co-wrote the manuscript. H.H.Q performed cloning, demethylation, PHF8 knockdown, conventional ChIP, gene expression, cell cycle analysis and histone profiling experiments. T.M.R directed the zebrafish work and M.S performed zebrafish experiments; both T.M.R and M.S contributed to the writing of the manuscript. Z.W. performed the ChIP-Seq experiments; G.-Q.H. analyzed the ChIP-Seq data; K.Z. directed the ChIP-Seq analysis. A.B, B.G and L.B contributed to the ChIP-chip experiments and data analysis. M.G and B.A.G performed Mass spectrometry experiments and data analysis. F.L. identified that the PHD domain of PHF8 and KIAA1718 bind to H3K4me3. P.P.O designed and helped in the cell cycle work. M.H cloned the full length KIAA1718 cDNA. N.K.Y generated mutations in the PHD domain of PHF8 and H.L helped in the demethylation assays.
Accession Numbers. Genome-wide mapping data of PHF8, H4K20me1, H3K9me1, 2 and gene expression microarray analysis in HeLa cells reported in this paper are available at the Gene Expression Omnibus server (http://www.ncbi.nlm.nih.gov/geo/) with the accession numbers GSE21108 and GSE21555, respectively.
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Ropers HH. X-linked mental retardation: many genes for a complex disorder. Curr Opin Genet Dev. 2006;16:260–269. doi: 10.1016/j.gde.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 2.Gecz J, Shoubridge C, Corbett M. The genetic landscape of intellectual disability arising from chromosome X. Trends Genet. 2009;25:308–316. doi: 10.1016/j.tig.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Ramos C, Robert B. msh/Msx gene family in neural development. Trends Genet. 2005;21:624–632. doi: 10.1016/j.tig.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Chiurazzi P, Schwartz CE, Gecz J, Neri G. XLMR genes: update 2007. Eur J Hum Genet. 2008;16:422–434. doi: 10.1038/sj.ejhg.5201994. [DOI] [PubMed] [Google Scholar]
- 5.Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol. 2010;17:445–450. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- 6.Fortschegger K, et al. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol Cell Biol. 2010 doi: 10.1128/MCB.01520-09. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleine-Kohlbrecher D, et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38:165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loenarz C, et al. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nepsilon-dimethyl lysine demethylase. Hum Mol Genet. 2010;19:217–222. doi: 10.1093/hmg/ddp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu L, et al. Structural insights into a novel histone demethylase PHF8. Cell Res. 2010;20:166–173. doi: 10.1038/cr.2010.8. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem. 2009;284:34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishioka K, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 12.Koivisto AM, et al. Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate. Clin Genet. 2007;72:145–149. doi: 10.1111/j.1399-0004.2007.00836.x. [DOI] [PubMed] [Google Scholar]
- 13.Houston SI, et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem. 2008;283:19478–19488. doi: 10.1074/jbc.M710579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol. 2008;28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Horton JR, et al. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalakonda N, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene. 2008;27:4293–4304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trojer P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 21.Postlethwait JH. The zebrafish genome: a review and msx gene case study. Genome Dyn. 2006;2:183–197. doi: 10.1159/000095104. [DOI] [PubMed] [Google Scholar]
- 22.Phillips BT, et al. Zebrafish msxB, msxC and msxE function together to refine the neural-nonneural border and regulate cranial placodes and neural crest development. Dev Biol. 2006;294:376–390. doi: 10.1016/j.ydbio.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Cau E, Blader P. Notch activity in the nervous system: to switch or not switch? Neural Dev. 2009;4:36. doi: 10.1186/1749-8104-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupo G, Harris WA, Lewis KE. Mechanisms of ventral patterning in the vertebrate nervous system. Nat Rev Neurosci. 2006;7:103–114. doi: 10.1038/nrn1843. [DOI] [PubMed] [Google Scholar]
- 25.Di Fonzo A, et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72:240–245. doi: 10.1212/01.wnl.0000338144.10967.2b. [DOI] [PubMed] [Google Scholar]
- 26.Eckert D, Buhl S, Weber S, Jager R, Schorle H. The AP-2 family of transcription factors. Genome Biol. 2005;6:246. doi: 10.1186/gb-2005-6-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tetel MJ. Nuclear receptor coactivators: essential players for steroid hormone action in the brain and in behaviour. J Neuroendocrinol. 2009;21:229–237. doi: 10.1111/j.1365-2826.2009.01827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi HH, et al. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–424. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plazas-Mayorca MD, et al. One-pot shotgun quantitative mass spectrometry characterization of histones. J Proteome Res. 2009;8:5367–5374. doi: 10.1021/pr900777e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rappsilber J, Friesen WJ, Paushkin S, Dreyfuss G, Mann M. Detection of arginine dimethylated peptides by parallel precursor ion scanning mass spectrometry in positive ion mode. Anal Chem. 2003;75:3107–3114. doi: 10.1021/ac026283q. [DOI] [PubMed] [Google Scholar]
- 31.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karolchik D, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui K, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwase S, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.