Abstract
MicroRNAs (miRNAs) post-transcriptionally repress complementary target gene expression and can contribute to cell differentiation. The coordinate expression of miRNA-183 family members (miR-183, miR-96, and miR-182) has been demonstrated in sensory cells of the mouse inner ear and other vertebrate sensory organs. To further examine hair cell miRNA expression in the mouse inner ear, we have analyzed miR-183 family expression in wild type animals and various mutants with defects in neurosensory development. miR-183 family member expression follows neurosensory cell specification, exhibits longitudinal (basal-apical) gradients in maturating cochlear hair cells, and is maintained in sensory neurons and most hair cells into adulthood. Depletion of hair cell miRNAs resulting from Dicer1 conditional knockout (CKO) in Atoh1-Cre transgenic mice leads to more disparate basal-apical gene expression profiles and eventual hair cell loss. Results suggest that hair cell miRNAs subdue cochlear gradient gene expression and are required for hair cell maintenance and survival.
Keywords: microRNA, Dicer, conditional knockout, inner ear, cochlea, sensory epithelium, hair cells, sensory neurons, development, maturation, maintenance
Introduction
The mammalian inner ear is a complex organ comprised of sensory and non-sensory tissues within the cochlea and vestibule that are organized to facilitate hearing and balance, respectively. The cochlea contains a single sensory epithelium called the organ of Corti within which highly organized rows of hair cells and supporting cells run from the base to the apex (Barald & Kelley, 2004) and detect high to low frequency stimuli, respectively (Carey & Amin, 2006). The vestibule contains five sensory epithelia comprised of mechanosensory hair cells and supporting cells (Barald & Kelley, 2004). The anterior crista, horizontal crista, and posterior crista function to detect linear or angular acceleration, whereas the saccular macula and utricular macula detect horizontal or vertical motion (Fritzsch et al., 2007). The mouse inner ear has been well studied as a model system for determining the molecular mechanisms that dictate sensory epithelia development. While considerable progress has been made in identifying the molecular pathways required for inner ear morphohistogenesis, maturation, and homeostasis (Eatock & Hurley, 2003; Barald & Kelley, 2004; Fritzsch et al., 2006), less is known about the degree to which microRNAs influence inner ear gene expression and affect cell specification and differentiation.
MicroRNAs are small (~21 nucleotide) RNAs processed from endogenous transcripts that function to mediate post-transcriptional silencing of complementary target genes (Ambros, 2004). Mammalian species possess as many as one thousand miRNA genes that might each influence expression of hundreds of target genes (Griffiths-Jones et al., 2006; Griffiths-Jones et al., 2008). It has been recently estimated that 60% of human genes contain conserved miRNA binding sites (Friedman et al., 2009a). Many miRNAs display distinct expression patterns in specific tissues and cell types (Wienholds et al., 2005; Landgraf et al., 2007) that can affect cell specification and differentiation (Bushati & Cohen, 2007; Papagiannakopoulos & Kosik, 2008; Lu & Liston, 2009; Zorio et al., 2009). For example, Drosophila miR-9 overexpression or deletion effects loss or ectopic formation of sensory organ precursors, respectively (Li et al., 2006). Additionally, mammalian miR-124 overexpression leads to decreased proliferation and promotes neuronal differentiation, whereas miR-124 repression leads to increased proliferation and increased levels of non-neuronal transcripts (Lim et al., 2005; Cheng et al., 2009). These studies demonstrate that such highly conserved miRNAs are critical for neurosensory development.
In mice, approximately one hundred microRNAs were shown to be expressed in the newborn and adult inner ear (Weston et al., 2006) or vestibular and cochlear tissues (Friedman et al., 2009b), suggesting miRNAs make considerable contributions to inner ear development and maintenance. Indeed, depletion of mature miRNAs by conditional knockout (CKO) of Dicer1 in embryonic mouse inner ear results in severe defects in neurogenesis and morphohistogenesis (Soukup et al., 2009; Kersigo et al., 2011). Moreover, the extent of hair cell development in miRNA-depleted sensory epithelia appears to correlate with residual hair cell miRNA expression. In another study, CKO of Dicer1 in mouse inner ear sensory epithelia effects hair cell degeneration and sensorineural hearing loss (Friedman et al., 2009b). Interestingly, the morphology of surviving, miRNA-depleted hair cells is highly similar between these Dicer1 CKO models, suggesting that sensory epithelial miRNAs are crucial for both hair cell development and maintenance.
In vertebrates, the miR-183 family (miR-183, miR-96, and miR-182) is expressed in neurosensory cells including zebrafish inner ear hair cells and neuromast hair cells (Wienholds et al., 2005), chicken and mouse cranial and spinal ganglia (Darnell et al., 2006; Kloosterman et al., 2006), and mouse eye photoreceptors and inner ear hair cells (Weston et al., 2006; Xu et al., 2007). The three miRNAs appear to be strictly co-expressed and processed from the same primary transcript (Weston et al., 2006; Saini et al., 2008). Moreover, miR-183-related miRNAs including miR-228 and miR-263b demonstrate a wider taxonomic distribution and expression in ciliated neurosensory cells and organs (Pierce et al., 2008), suggesting that these highly conserved miRNAs are important for neurosensory cell development and function. Indeed, recent studies demonstrate that mutations in miR-96 effect hereditary deafness in humans and mice (Lewis et al., 2009; Mencia et al., 2009; Weston & Soukup, 2009), and that overabundance or inhibition of miR-183 family members in zebrafish development, respectively, effect ectopic hair cell formation or reduction in hair cell number (Li et al., 2010).
In this report, we examine the pattern of miR-183 family expression in the mouse inner ear from early development through functional maturation. Mouse models that fail to develop mature hair cells and/or sensory neurons demonstrate that miR-183 family expression follows sensory neuron and hair cell specification, and is concomitant with hair cell differentiation. Upon functional maturation of cochlear hair cells, miR-183 family members demonstrate a marked basal-apical expression gradient. To determine whether such hair cell miRNAs affect basal-apical gene expression, we performed microarray analyses from basal and apical organ of Corti from control versus hair cell-specific Dicer1 CKO mice. Results from these studies reveal the general impact of hair cell miRNAs on cochlear gene expression and on hair cell maintenance and survival.
Results
Embryonic miRNA expression in neurosensory cells
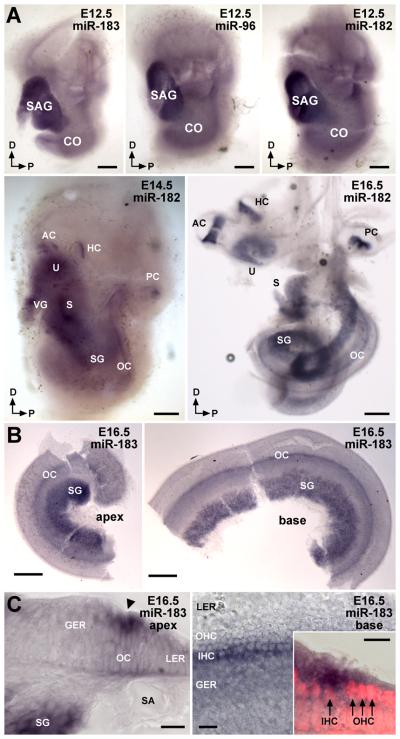
To examine the developmental expression pattern of neurosensory miR-183 family members in mouse inner ear, whole-mount in situ hybridization (ISH) using locked nucleic acid (LNA) probes was performed over the course of hair cell development from embryonic day (E) 12.5 to E16.5 (Fig. 1A). Robust expression is observed only in statoacoustic ganglia (SAG) at E12.5, which is the earliest time of hair cell development in the vestibular end organs (Kelley, 2006). At E14.5, miRNA-183 family expression is most apparent in inner ear ganglia and vestibular sensory epithelia, and faint detection is observed in the developing cochlea (Fig 1A). At E16.5, miR-183 family expression is observed in all sensory ganglia and epithelia throughout the inner ear (Fig. 1A). In the cochlea, expression appears greatest in spiral ganglia (SG), and a distinct strip of expression is seen from base to apex within the organ of Corti (Fig. 1A,B). Frozen sections from the base and apex of the cochlea (Fig. 1C) show that the miRNAs are first detected in inner hair cells (IHCs), the differentiation of which precedes that of outer hair cells (IHCs) (Kelley, 2007; Dabdoub et al., 2008). Some diffuse staining is observed in transient cells of the greater epithelial ridge (Fig. 1B,C). At E18.5 (data not shown) and postnatal day (P) 0 (Weston et al., 2006; see also Supplementary Fig. S1), miR-183 family member expression is limited to ganglia and hair cells throughout the inner ear. Identical expression patterns are observed for miR-96, miR-182 and miR-183 (Fig. 1A, Supplementary Fig. S1), consistent with the miRNAs being processed from a common primary transcript (Weston et al., 2006; Saini et al., 2008). miR-183 family members thus serve as excellent markers for neurosensory cell differentiation within the developing inner ear.
Figure 1.
Embryonic expression of miR-183 family members in hair cells and peripheral sensory neurons of mouse inner ear by in situ hybridization using LNA probes. A: Detection of miR-183 family members in E12.5, E14.5, and E16.5 whole mount inner ears. Expression is indicated in statoacoustic ganglia (SAG), which segregate to form vestibular ganglia (VG) and spiral ganglia (SG) in the cochlea (CO), and in all sensory epithelia (AC, anterior crista; HC, horizontal crista; PC, posterior crista; U, utricular macula; S, saccular macula; OC, organ of Corti). Bars represent 200 μm. B: Detection of miR-183 in the apex and base of E16.5 cochlea. Bars represent 200 μm. C: Detection of miR-183 in inner hair cells (IHC) of E16.5 cochlea. Depicted are frozen cross-sections of apical and basal segments of cochlea, and the basal organ of Corti in whole mount (GER, greater epithelial ridge; LER, lesser epithelial ridge; SA, spiral artery; OHC, outer hair cells). An arrowhead denotes the approximate position of IHC along the boundary between the GER and OC in the apex, whereas the position of IHC in the base is more evident by DAPI staining of nuclei (red). Bars represent 20 μm.
Postnatal miRNA expression gradients
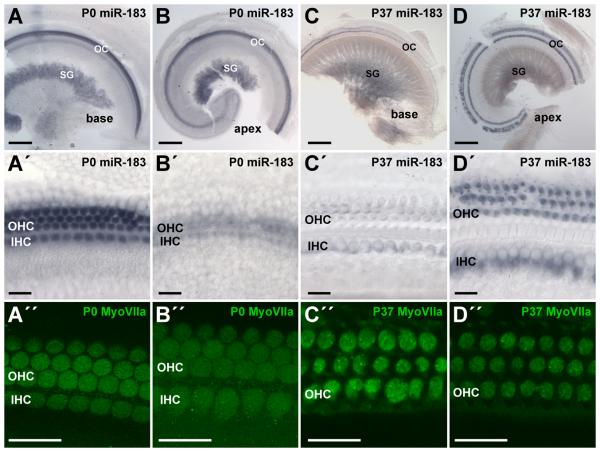
Postnatal miR-183 family member expression in cochlear hair cells demonstrates dynamic changes in intensity as the organ attains functional maturity. At P0, miR-183 family members show robust expression in hair cells that gradually diminishes from base to apex (Fig 2A,B,A,′B′), which is a pattern consistent with the progression of hair cell differentiation from base to apex (Kelley, 2007). Additionally, expression of the miRNAs generally appears greater in OHCs than IHCs. However, in the functionally mature cochlea at P37, miR-183 family members exhibit robust expression in apical hair cells that gradually diminishes toward the base (Fig. 2C,D,C,′D′). Moreover, OHC and IHC miRNA expression ranges from being similar in apical hair cells to greater in IHCs than OHCs approaching the base, whereas the relatively similar expression of the differentiated hair cell marker myosin VIIa suggests that miRNA gradient expression is not an artifact of histological differences in hair cells (Fig. 2A″-D″). Basal hair cells show relatively less miR-183 family member expression, especially in OHCs. These longitudinal and radial gradients in cochlear miR-183 family member hair cell expression are established around the time of functional maturation (2-3 weeks of age; data not shown) and persist well into adulthood (Supplementary Fig. S2), where similar patterns are observed at P100 while vestibular miR-183 family member hair cell expression remains relatively stable. These data suggest that miR-183 family member function extends beyond developmental effects and might contribute to gradients in hair cell gene expression that affect differentiated cell functionality and maintenance.
Figure 2.
Expression of miR-183 exhibits temporal changes in longitudinal and radial gradients in postnatal mouse cochlea by in situ hybridization. A-D: Detection of miR-183 in the apex and base of P0 and P37 cochleae. Scale bars represent 200 μm. A′-D′: Corresponding segments of OC in the apex and base. Scale bars represent 20 μm. Intensity of miR-183 detection in hair cells appears as basal>apical and OHC>IHC at P0 versus an apical>basal and IHC>OHC at P37. A″-D″: Immunohistochemical detection of hair cell myosin VIIa in apex and base of P0 and P37 cochleae. Scale bars represent 20 μm. Labels are as described in the legend to Fig. 1.
Dependence of miRNA expression on neurosensory transcription factors
Neurosensory ganglia and hair cells are both derived from proneuronal precursors that are respectively dependent on the basic helix-loop-helix (bHLH) transcription factors Neurog1 and Atoh1 (Fritzsch et al., 2010). Lack of Neurog1 expression prevents sensory neuron specification and development and results in a reduction of sensory epithelia in both the cochlea and the vestibule (Ma et al., 2000). miR-183 expression in Neurog1 null inner ear demonstrates that hair cell miRNA expression is unaffected in the cochlea and vestibular endorgans whereas sensory neurons are altogether absent (Fig. 3A). Conversely, lack of Atoh1 prevents hair cell specification and leads to loss of sensory neurons primarily due to absence of hair cell neurotrophic support (Matei et al., 2005). Atoh1 null inner ear shows no miR-183 hair cell expression, whereas surviving ganglia in the cochlea demonstrate weak miRNA expression (Fig. 3B). Pou4f3 is a POU domain factor essential for the differentiation and maintenance of hair cells in the cochlea (Xiang et al., 1997). Pou4f3 mutant (Dreidel mouse) inner ear develops immature hair cells that rapidly undergo apoptosis especially in the cochlea (Hertzano et al., 2004), but retains some sensory neurons (Xiang et al., 2003). Pou4f3 mutant inner ear demonstrates that miR-183 is weakly expressed in surviving cochlear hair cells, but that expression of the miRNA is absent in vestibular hair cells (Fig. 3C). The data demonstrate that miR-183 expression is downstream of Neurog1 and Atoh1 specification of sensory neurons and hair cells, respectively. Moreover, miR-183 expression is likely dependent on downstream factors for differentiation and maintenance.
Figure 3.
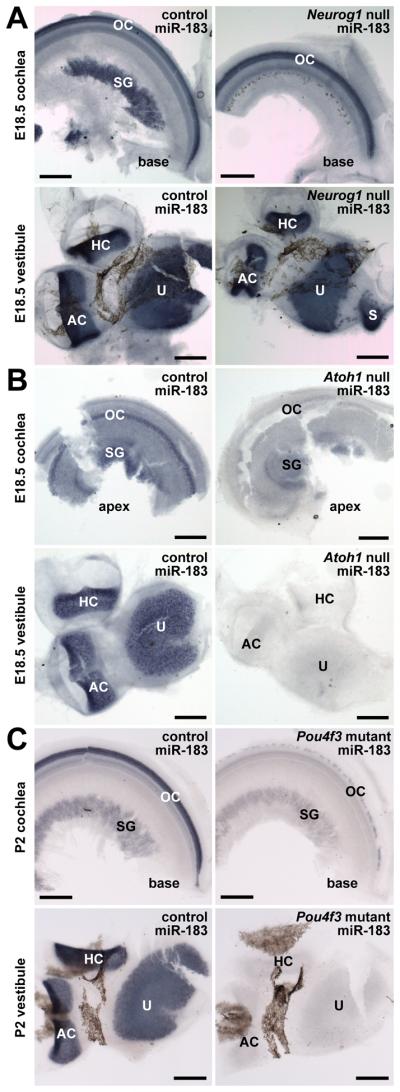
Expression of miR-183 in inner ear of Neurog1 null, Atoh1 null and Pou4f3 mutant mice and controls by in situ hybridization follows neurosensory cell fate specification and differentiation. A: Hair cell expression of miR-183 is unaffected in Neurog1 null inner ear lacking in sensory neurons. B: Expression of miR-183 is evident in remaining sensory neurons of Atoh1 null inner ear lacking hair cells. C: Expression of miR-183 is unaffected in sensory neurons of Pou4f3 mutant inner ear, but absent in vestibular hair cells and weak in remaining cochlear hair cells. Scale bars represent 200 μm. Labels are as described in the legend to Fig. 1.
Hair cell miRNA depletion and hair cell loss in Dicer1 CKO mice
To investigate the function of small RNAs in hair cell development and maintenance, we generated Atoh1-Cre conditional Dicer1 knockout (CKO) mice (Atoh1-Cre;Dicer1flox/flox). Atoh1 and the Atoh1-Cre transgene are expressed in all hair cells beginning approximately E12.5 to E14.5 in addition to other tissues, most notably including the cerebellum and intestinal epithelium (Ben-Aire et. al., 1997; Yang et al., 2001; Matei et al., 2005). Dicer1 CKO mice thus exhibit a general failure to thrive, increasing ataxia, and seizures that contribute to lethality around 4 weeks of age (data not shown).
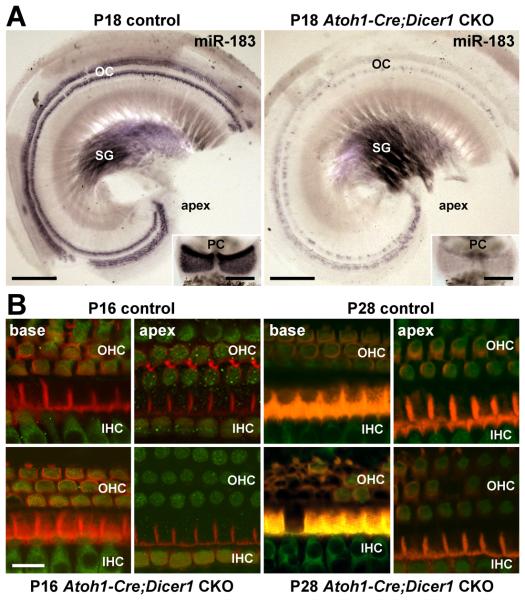
To confirm the loss of functional Dicer1 and depletion of mature miRNAs in hair cells, miR-183 expression was examined in P18 Dicer1 CKO mice and littermate controls (Atoh1-Cre;Dicer1flox/wt). In situ hybridization demonstrates that miR-183 expression is largely depleted in Dicer1 CKO inner ear hair cells by P18, whereas expression in an equally reacted littermate control inner ear appears normal despite hemizygousity of Dicer1 (Fig. 4A). Notably, miR-183 expression in spiral ganglia is equivalent between the Dicer1 CKO and control cochlea, and an analysis of supporting cell expression of an epithelial miR-200 family member (miR-141; Weinholds et al., 2005; Darnell et al., 2006) shows little difference in expression within the organ of Corti (Supplementary Fig. 3). These data demonstrate the intended hair cell-specificity of Dicer1 CKO and subsequent depletion of hair cell miRNAs.
Figure 4.
Hair cell miRNA depletion and eventual hair cell loss in Atoh1-Cre;Dicer1 CKO inner ears versus controls. A: Hair cell expression of miR-183 is largely depleted at P18 in Atoh1-Cre;Dicer1 CKO inner ear by in situ hybridization. Depicted are the posterior cristae and apical turns of cochleae from control and Dicer1 CKO mouse inner ears that are equally reacted. Scale bars represent 200 μm. B: Hair cell loss in the cochlea of Atoh1-Cre;Dicer1 CKO inner ear compared to control. Depicted is immunohistochemical detection of myosin VIIa in hair cells (green) and phalloidin staining of F-actin (red) in organ of Corti near the base and apex of P16 and P28 control and Dicer1 CKO cochleae. Scale bar represents 10 μm. Labels are as described in the legend to Fig. 1.
Residual expression of miR-183 in the P18 Dicer1 CKO cochlea (Fig. 4A; Supplementary Fig. S4) follows the expression pattern observed for the functionally mature mouse cochlea (Fig. 2C,D; Supplementary Fig. S4), where there is a rapidly declining apical to basal gradient and IHC to OHC radial gradient of residual expression. These data suggest that beyond the half-life of Dicer1, the depletion of hair cell miRNAs is largely dependent on miRNA half-life from recent miRNA expression levels. Moreover, the model demonstrates that mature miRNAs are extremely long-lived in post-mitotic, differentiating hair cells, where detection is evident upwards of three weeks from the time of Dicer1 deletion.
Considering the nuances of the model, Atoh1-Cre knockout of Dicer1 mainly provides an opportunity to examine the effect of slow depletion of miRNAs on hair cell maturation and maintenance. With regard to the latter, hair cell viability in Dicer1 CKO and control cochleae was examined by phalloidin staining of F-actin and immunostaining of myosin VIIa at P16 and P28 (Fig. 4B). At P16, both Dicer1 CKO and control cochleae demonstrate a normal histological organization and presence of hair cells in apical and basal segments of the organ of Corti, with minor loss of OHCs in the base of the Dicer1 CKO cochlea. At P28, however, the Dicer1 CKO cochlea shows a marked reduction in basal OHCs and some loss of apical OHCs, whereas IHCs are substantially less affected. These data demonstrate that hair cell miRNA depletion results in a progressive loss of OHCs from base to apex, and that miRNAs are thus required for cochlear hair cell maintenance and survival.
Response of gene expression profiles to hair cell miRNA depletion
Given that hair cell miR-183 family member expression exhibits a longitudinal gradient in the mature cochlea, it is plausible that these apparently abundant miRNAs might function to establish gradient expression of other genes. To address this hypothesis, microarray analysis was used to examine the gene expression profiles of apical versus basal organ of Corti dissected from P16 Dicer1 CKO and control cochleae, where hair cell miRNAs have been largely depleted without affecting hair cell viability. The expectation is that apical-basal gradient gene expression might be reduced when hair cell miRNAs are depleted.
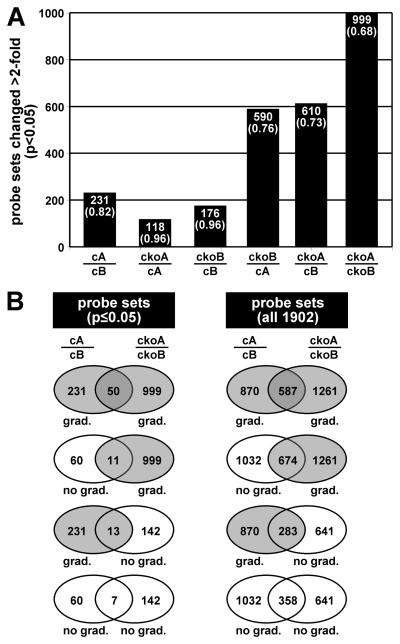
Analysis and comparisons of expression profiles from control apex (cA), control base (cB), Dicer1 CKO apex (ckoA), and Dicer1 CKO base (ckoB) yielded a data set of 1,902 probe sets for which there is determinable presence of expression in any one of the profiles and significant changes in gene expression >2-fold (Fig 5A; Supplementary Table S1). Expectedly, comparisons of cA to ckoA and cB to ckoB show the highest degree of correlation, suggesting that neither apical gene expression nor basal gene expression has been drastically altered in the Dicer1 CKO organ of Corti relative to the control organ of Corti. However, whereas comparison of cA to cB establishes the number of probe sets exhibiting >2-fold changes representing apical-basal gradient gene expression, comparisons of cA to ckoB, ckoA to cB, and finally ckoA to ckoB show increasing gradient gene expression that is more than quadrupled from control to Dicer1 CKO organ of Corti. These data effectively demonstrate that depletion of hair cell miRNAs has a relatively nominal effect on apical or basal gene expression per se that is nevertheless accentuated upon consideration of apical-basal gradient gene expression, contrary to the hypothesis.
Figure 5.
Microarray analysis of gradient gene expression in control and Atoh1-Cre;Dicer1 CKO organ of Corti. A: Significant changes in gene expression comparing apical and basal organ of Corti. Shown for each comparison of expression profiles from control apex (cA), control base (cB), Dicer1 CKO apex (ckoA), and Dicer1 CKO base (ckoB) are the number of probe sets out of 1,902 expressed that exhibit significant changes greater than 2-fold. Numbers in parentheses indicate the correlation coefficient for each comparison. B: Comparison of gradient and non-gradient expression profiles between control and Dicer1 CKO organ of Corti. Venn diagrams show the overlap among probe sets that show gradient expression (grad.; >2-fold change) or non-gradient expression (no grad.; <2-fold change) considering those with significance (p<0.05) and all 1,902 probe sets.
Venn diagrams illustrate the overlap between Dicer1 CKO and control apical/basal probe set data in consideration of significant >2-fold changes (gradient expression) or significant <2-fold changes (no gradient expression) versus consideration of >2-fold or <2-fold changes for all 1,902 probe sets (Fig. 5B). Analysis of this presentation reveals that probe sets exhibiting significant gradient expression in the Dicer1 CKO model do not result from a conversion of probe sets exhibiting significant non-gradient expression in the control, but instead are largely derived from probe sets that did not show significant gradient or non-gradient expression in the control. The data suggest that depletion of hair cell miRNAs mainly allows significant gradient gene expression to arise from gene expression levels that already show some variability. Results from the microarray analysis thus suggest that hair cell miRNAs, in so far as gradient miR-183 family member expression is representative, are not the cause of cochlear gradient gene expression, but rather function to suppress it.
Perturbation of cochlear gradient gene expression in the Dicer1 CKO mouse represents the combined direct and indirect effects of miRNA regulation of target gene expression that ultimately contribute to hair cell degeneration in the model. Although it is tempting to correlate expectant increases in the level of predicted miRNA target gene expression with the loss of miRNA function, heterogeneity of tissue cell types represented in the analysis and the fact that miRNAs rarely elicit >2-fold changes in mRNA expression level (Baek et al., 2008; Selbach et al., 2008) likely preclude the observation of strong correlations (data not shown). Nevertheless, it is useful to consider predicted miR-183 family member target genes represented in the data set as candidates to scrutinize potential miRNA functions in hair cell development and maintenance. A gene ontological listing of predicted miR-183 family member target genes within the data set (Supplementary Table S2) suggests a number of targets with established roles in inner ear biology. Notable among these is Sox2, an SRY-box containing transcription factor required for the establishment of prosensory domains and development of hair cells (Kiernan et al., 2005; Oesterle et al., 2008). Sox2 expression is nevertheless downregulated upon differentiation and maturation of hair cells, whereas neighboring supporting cells retain its expression (Kiernan et al., 2005). We therefore investigated whether hair cell miR-183 family member expression can directly effect Sox2 downregulation.
Sox2 is a miR-183 family target
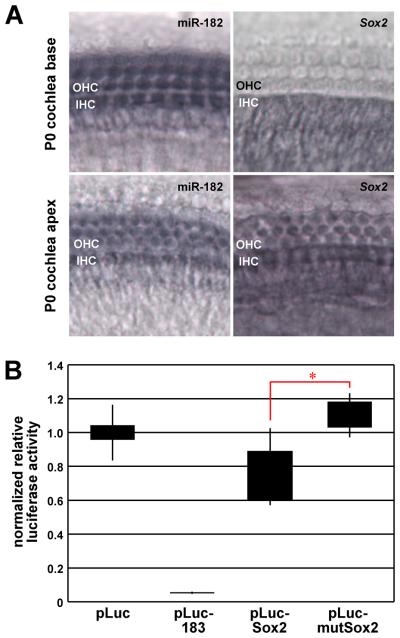
Sox2 contains a single predicted and highly conserved miR-182 binding site within its 1.1 Kb 3′ untranslated region (3′UTR). To address whether Sox2 mRNA might be a miR-182 target, coexpression of the RNAs in cochlear hair cells was examined by in situ hybridization (Fig. 6A). At P0, apical and basal IHCs show miR-182 and Sox2 mRNA coexpression, whereas the RNAs are coexpressed in OHC only in apical OHCs by this time point. These data demonstrate the opportunity for interaction between the RNAs in developing hair cells. To determine whether miR-182 binding to the site within Sox2 mRNA can inhibit translation, the entire Sox2 3′UTR was cloned downstream of the open reading frame encoding Photinus luciferase (pLuc-Sox2), and the miR-182 binding site was mutated to disrupt miRNA seed pairing (pLuc-mutSox2). Negative and positive control vectors, respectively, contained no insert (pLuc) or a synthetic 3′UTR with tandem miR-183 family member binding sites that are perfectly complementary to each miRNA (pLuc-183). Dual luciferase assays were performed using HEK293 cells co-transfected with reporter vector and siRNA mimic for miR-182 or scrambled control siRNA to determine normalized relative luciferase activity for each construct (Fig. 6B). Expectedly, pLuc is unaffected by miR-182 while pLuc-183 is effectively reduced ~95% where the miRNA is designed to act as an siRNA. pLuc-Sox2 is repressed ~25% by miR-182, whereas pLuc-mutSox2 is significantly and fully derepressed. The specificity and significance of pLuc-Sox2 repression demonstrates that the miR-182 binding site is functionally accessible and suggest that Sox2 is a bona fide hair cell miR-183 family member target gene.
Figure 6.
Coexpression and validation of Sox2 as a target of miR-182. A: In situ hybridization detecting miR-182 and Sox2 mRNA at P0. Depicted are apical and basal segments of OC, where coexpression of the RNAs is evident in OHCs at the apex and in IHCs. B: Dual luciferase assay demonstrating specificity of Sox2 silencing by miR-182. The parental Photinus luciferase reporter vector (pLuc) was modified to contain tandem complementary sites to miR-183 family members (pLuc-183), the Sox2 3′UTR (pLuc-Sox2), or the Sox2 3′UTR with a mutated miR-182 binding site (pLuc-mutSox2). The box and whisker plot represents the distribution of relative luciferase activity for each reporter construct with miR-182 normalized to control siRNA. Bars span the 10th to 90th percentiles, and boxes span the 25th to 75th percentiles. The asterisk denotes a significant and specific reduction of Photinus luciferase expression with miR-182 (p<0.01; Mann-Whitney-Wilcoxon test).
Discussion
Several recent studies have demonstrated the functional relevance of miR-183 family members in the development and maintenance of hair cells in the inner ear. The first Mendelian disease associated with microRNAs results from a point mutation in the seed region of miR-96, which causes progressive hearing loss in humans (Mencia et al., 2009). A similar ENU-induced mutation in miR-96 is associated with rapid hair cell degeneration and deafness in the Diminuendo mouse (Lewis et al., 2009). Interestingly, each of these miR-96 mutations is different, suggesting that the hearing loss phenotype results from loss of function rather than gain of function. In zebrafish embryos, injection of miR-96 or miR-182 causes the formation of ectopic hair cells, whereas knockdown of miR-96, miR-182 or miR-183 results in significantly fewer hair cells formed from apparently normal sensory maculae (Li et al., 2010).
Considering the importance of miR-183 family members in hair cell development and maintenance, we sought to better define their patterns with regard to hair cell commitment, differentiation, maturation, and maintenance of hair cells in the mouse inner ear. We show that the miRNAs exhibit coordinated expression in sensory neurons from E12.5 and in hair cells from E14.5. Moreover, expression in cochlear hair cells is first detectable in inner hair cells, and subsequently in all hair cells by P0. The data demonstrate that expression of the miRNAs specifically follows the timing of specification of sensory neurons and hair cells throughout the inner ear (Kelley, 2006; Kelley, 2007; Fritzsch et al., 2010). Moreover, we show that expression of the miRNAs is dependent upon Neurog1 and Atoh1 required for the specification of sensory neurons and hair cells, respectively. The results show that miR-183 family expression is a property of committed sensory cell types and is thus likely to exert the greatest influence upon sensory cell differentiation in early development. Moreover, it is interesting to note the differential dependence of miR-183 family expression on Pou4f3 in surviving vestibular versus cochlear hair cells. The data suggest that miR-183 family expression is differentially regulated like other downstream hair cell factors such as Lhx3 (Hertzano et al., 2004), further highlighting nuances in the genetic programs that contribute to differentiation of distinct hair cell types.
Previous microarray analysis and quantitative RT-PCR of mouse inner ear miRNA expression indicated the continued postnatal expression of miR-183 family members (Weston et al., 2006). We now demonstrate that miR-183 family expression in sensory neurons and hair cells persists well into adulthood, where cochlear hair cells exhibit distinct longitudinal and radial gradient patterns that develop about the time the organ reaches functional maturity. Importantly, the observed expression pattern of miR-183 family members in the mouse inner ear is highly consistent with previously observed expression patterns in mouse, chicken, and zebrafish sensory cells and ganglia (Wienholds et al., 2005; Darnell et al., 2006; Kloosterman et al., 2006; Weston et al., 2006; Xu et al., 2007; Pierce et al., 2008; Friedman et al., 2009b; Lewis et al., 2009; Soukup et al., 2009; Li et al., 2010). Collectively, the data stand in stark contrast to two recent publications (Sacheli et al., 2009; Wang et al., 2010) that show non-coordinated and widespread miR-183 family expression in non-sensory cells of the mouse embryo and inner ear, and fail to detect hair cell expression in the juvenile and adult inner ear. Although there are points of consistency in miR-183 family expression in sensory neurons and hair cells, particularly at late embryonic and early postnatal time points, discrepancies in miRNA detection likely reflect differences in hybridization protocols using sectioned versus intact tissues. Results suggest that whole-mount in situ hybridization more accurately reflects the coordinated and sensory cell-specific expression pattern of miR-183 family members in the mouse inner ear.
Based on the overall apical-basal gradient expression pattern of miR-183 family members in hair cells of the mature cochlea, we hypothesized that hair cell miRNAs function to promote gradient gene expression. However, apical and basal organ of Corti gene expression profiles for control and Atoh1-Cre;Dicer1 CKO mice show that depletion of hair cell miRNAs results in relatively pronounced gradient gene expression. Importantly, these analyses were performed at a time when there is substantial miR-183 depletion, but little or no apparent hair cell loss. While analysis of mRNA levels does not rule out the possibility that miR-183 family members might effect gradient expression of some target genes, the model demonstrates that hair cell miRNAs significantly influence organ of Corti gene expression and dampen apical-basal gradients. Moreover, the depletion of hair cell miRNAs causes eventual hair cell death, particularly for basal OHCs.
It is interesting to note the range of effects observed in various Dicer1 CKO models and the Diminuendo mouse (reviewed in Weston & Soukup, 2009). Previous work has demonstrated that Pax2-Cre; Dicer1 CKO in the otic placode leads to severe morphological and histological defects of the inner ear (Soukup et al., 2009), whereas Pou4f3-Cre;Dicer1 CKO occurring later in the organ of Corti and hair cells throughout the inner ear causes severe hair cell degeneration and deafness by P38 (Friedman et al., 2009b). The morphological characteristics of immature and malformed hair cells in the two Dicer1 CKO models are strikingly similar. Depletion of miRNAs in the Atoh1-Cre;Dicer1 CKO model described here is hair-cell specific and relatively slow, where the long half-life of mature miRNAs in post-mitotic Dicer1 CKO models appears to be a feature consistent with other Dicer1 CKO models (Schaefer et al., 2007; Cuellar et al., 2008). Hair cells appear to develop normally and show moderate degeneration and loss at P28 compared to the Pou4f3-Cre;Dicer1 CKO model at P38. Together these models demonstrate that hair cell miRNAs play critical roles in both hair cell development and maintenance. Considering the effects of miRNA depletion in Dicer1 CKO models, it is pertinent to note that the loss of miR-96 in homozygous Diminuendo mice results in a relatively severe hair cell phenotype (Lewis et al., 2009). The developmental absence of this single miR-183 family member is thus more detrimental than the maturational depletion of all hair cell miRNAs in the Atoh1-Cre;Dicer1 CKO model, lending further credence to the notion that miR-183 family members play critical roles in early hair cell development. Among the undoubtedly numerous target genes through which miR-183 family members exert their influence, we have validated that Sox2, which remains expressed in supporting cells, is repressed by miR-182. These data are consistent with the hypothesis that miR-183 family members promote hair cell fate and differentiation in part by repressing supporting cell genetic programs (Soukup, 2009).
In summary, this study details the onset and persistent expression of miR-183 family members in sensory cells of the mammalian inner ear, where the regulatory functions of such hair cell miRNAs contribute to development and maintenance. As therapeutic strategies for stimulating hair cell regeneration and hearing restoration continue to advance (Beisel et al., 2008; Brigande & Heller, 2009; Groves, 2010), they might benefit substantially from miRNA-based therapies (Hammond, 2006) designed to guide developmental or maturational processes that contribute to hair cell maintenance and function.
Experimental Procedures
Animals
Animal care and handling complied with protocols approved by the Creighton University Institutional Animal Care and Use Committee and employed measures to minimize pain and discomfort. FVB/N mice were purchased from Charles River Laboratories. Various mutant and littermate control mouse ears were derived from Neurog1 null mice (Neurog1tm1And; Ma et al., 1998; Ma et al., 2000), Atoh1 null mice (Atoh1tm2Hzo; Ben-Aire et al., 1997; Matei et al., 2005), and Pou4f3 mutant (Dreidel) mice (Pou4f3ddl; Frankel et al., personal communication; Hertzano et al., 2004). Mouse embryos were harvested from timed pregnant females counting the day a vaginal plug was present as E0.5. Embryos and postnatal mice were transcardially perfused with 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (pH 7.4) to preserve tissues for in situ hybridization and immunohistochemistry.
Mice carrying floxed Dicer1 (Dicer1tm1Bdh; Harfe et al., 2005) alleles (Dicer1flox/flox) were mated to mice carrying an Atoh1-Cre transgene (Tg(Atoh1-cre)1Bfri; Matei et al., 2005) to generate Atoh1-Cre;Dicer1flox/wt mice. Atoh1-Cre;Dicer1flox/wt mice were subsequently mated to Dicer1flox/flox mice to generate Atoh1-Cre;Dicer1flox/flox CKO animals and Atoh1-Cre;Dicer1flox/wt littermate controls. Offspring were genotyped by PCR analysis of tail DNA using Cre-specific primers (5′-GCCTGCATTACCGGTCGATGCAACGA and 5′-GTGGCAGATGGCGCGGCAACACCATT) that produce a 726 bp product, and Dicer1-specific primers (5′-CCTGACAGTGACGGTCCAAAG and 5′-CATGACTCTTCAACTCAAACT) that produce a 420 bp product from the Dicer1flox allele and a 351 bp product from the Dicer1wt allele (Harfe et al., 2005).
Whole mount in situ hybridization (ISH)
Whole-mount ISH was performed as previously described (Weston et al., 2006; Pierce et al., 2008) using locked nucleic acid (LNA) probes for miRNAs labeled with digoxigenin (DIG Oligonucleotide 3′-End Labeling Kit; Roche) or riboprobe for Sox2 mRNA labeled with DIG by transcription using T7 RNA polymerase (DIG RNA Labeling Kit; Roche). LNA probes were either purchased (miRCURY LNA probes; Exiqon) or custom synthesized (Integrated DNA Technologies) by incorporating LNA modifications at every third nucleotide position from the 5′ end. LNA probes are antisense to miR-183 (5′-CAGTGAATTCTACCAGTGCCATA), miR-96 (5′-AGCAAAAATGTGCTAGTGCCAAA), miR-182 (5′-TGTGAGTTCTACCATTGCCAAA), or miR-141 (5′-CCATCTTTACCAGACAGTGTTA). Riboprobe antisense to Sox2 mRNA (659 nt) was transcribed and purified by denaturing polyacrylamide gel electrophoresis from template DNA generated by PCR amplification of a sequence-verified clonal isolate of Sox2 derived from mouse genomic DNA using primers (5′-GCTCTGCACATGAAGGAGCAC and 5′-TAATACGACTCACTATAGGGCATGTGCGACAGGGGCAG). Briefly, fixed tissues were defatted with ethanol, digested with proteinase K, hybridized with 12 pmol labeled LNA probe or 100 ng labeled riboprobe, and washed and digested with RNase A. Labeled LNA probe was detected using alkaline phosphatase (AP) conjugated sheep anti-DIG Fab fragment and BM Purple AP Substrate (Roche). Tissues were whole mounted in glycerol and imaged by light microscopy using a Nikon Eclipse 800 microscope. A minimum of two samples was prepared for each genotype and time point described.
Immunohistochemistry
FVB/N mouse cochleae were microdissected and stained with a rabbit anti-mouse myosin VIIa antibody (Affinity Bioreagents) as previously described (Matei et al., 2005; Pauley et al., 2006). Briefly, fixed tissues were defatted, blocked with normal goat serum, incubated with 1:50 dilution of primary antibody, rinsed, incubated with Alexa Fluor 568-conjugated goat anti-rabbit antibody (Invitrogen), rinsed, mounted and imaged using a Zeiss LSM 510 META LNO confocal microscope. The organ of Corti was optically sectioned at 2 μm intervals and images are the composite of sections including hair cell bodies.
Inner ears from Atoh1-Cre;Dicer1 CKO and control mice were embedded in 2.5% agarose, and sagittal sections (300 ± 50 μm) containing the apical, middle, and basal cochlear turns were obtained using a vibratome. Sections were similarly stained with rabbit anti-mouse myosin VIIa antibody (Abcam) detected with Alexa Fluor 488-conjugated goat anti-rabbit antibody (Molecular Probes), and Alexa Fluor 568-conjugated Phalloidin (Molecular Probes) to detect F-actin. Sections were imaged by confocal microscopy as described above.
RNA extraction
Total RNA from the organ of Corti (OC) was isolated using the mirVana miRNA Isolation Kit (Ambion). Each biological replicate included the combined apical or basal OC from each ear of a Dicer1 CKO mouse or littermate control mouse. Tissues were disrupted by rotor-stator homogenization in lysis buffer to facilitate RNA isolation. The quality and quantity of each RNA preparation were determined using a Model 2100 Agilent BioAnalyzer.
Microarray Analysis
Two biological replicate samples of total RNA (300 ng each) from apical and basal OC from Dicer1 CKO and littermate control mice were analyzed using Affymetrix Mouse Genome 430 2.0 Arrays (8 total microarrays). Sample preparation, hybridization, and analysis were performed by the Microarray Core Facility at the University of Nebraska Medical Center. The mas5 function from the affy software package (Bioconductor project; http://www.bioconductor.org/) was used to calculate probe set expression values. Expression values from replicate microarrays were used to determine statistically significant changes in gene expression (p<0.05, fold changes >2.0 or <0.5) using the Student's t-test. Microarray data have been deposited in the NCBI Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE26822 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26822).
Dual Luciferase Assays
Tandem sites complementary to miR-183, miR-96 and miR-182 were generated from synthetic oligonucleotides and inserted into the 3′UTR of the parental Photinus luciferase reporter vector pMIR-REPORT (Ambion) to produce pLuc-183. The ~1.1 Kb 3′UTR of mouse Sox2 was amplified from genomic DNA and similarly inserted into pMIR-REPORT to produce pLuc-Sox2. The miR-182 complementary seed match of pLuc-Sox2 (5′-TTGCCAA) was mutated to contain two single base changes (5′-TTCCGAA) using the QuikChange II Site Directed Mutagenesis Kit (Stratagene) to produce pLuc-mutSox2. HEK293 cells (~2×105 cell/well; 24-well plate) were co-transfected with 200 ng Photinus luciferase reporter vector, 50 ng Renilla luciferase reporter vector phRL-TK (Promega), and 25 pmol synthetic RNA duplex representing miR-182 or scrambled control siRNA (Integrated DNA Technologies) using Lipofectamine 2000 (Invitrogen). Cells were cultured post-transfection for 48-72 h and harvested to perform dual-luciferase assays using the Dual-Glo Luciferase Assay System (Promega) on a Modulus Microplate Luminometer with dual injectors (Turner Biosystems). Two replicate readings from 3-6 independent transfections were performed. The ratio of Photinus and Renilla luciferase activity for each Photinus luciferase reporter vector with miR-182 was normalized to that for each with scrambled control siRNA. The Mann-Whitney-Wilcoxon test was performed to determine the statistical significance of differences between samples.
Supplementary Material
Acknowledgments
Grant Information: This research was supported by NIH–NIDCD:R01DC009025, NIH–NIDCD:F32DC008253, NIH–NCRR:P20RR018788, and Nebraska State LB692, and conducted in facilities supported by NIH–NCRR:G20RR024001 and NIH–NCRR:C06RR017417 including the Creighton University Integrative Biological Imaging Facility. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SF, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:44–45. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barald KF, Kelley MW. From placode to polarization: new tunes in inner ear development. Development. 2004;131:4119–4130. doi: 10.1242/dev.01339. [DOI] [PubMed] [Google Scholar]
- Ben-Aire N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granular neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Beisel K, Hansen L, Soukup G, Fritzsch B. Regenerating cochlear hair cells: quo vadis stem cell. Cell Tissue Res. 2008;333:373–379. doi: 10.1007/s00441-008-0639-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigande JV, Heller S. Quo vadis, hair cell regeneration? Nat Neurosci. 2009;12:679–685. doi: 10.1038/nn.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Carey J, Amin N. Evolutionary changes in the cochlea and labyrinth: Solving the problem of sound transmission to balance organs in the inner ear. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:482–489. doi: 10.1002/ar.a.20306. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci USA. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KSE, Pevny LH, Kelley MW. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci USA. 2008;105:18396–18401. doi: 10.1073/pnas.0808175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell DK, Kaur S, Stanislaw S, Konieczka JH, Yatskievych TA, Antin PB. MicroRNA expression during chick embryo development. Dev Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- Eatock RA, Hurley KM. Functional development of hair cells. Curr Top Dev Biol. 2003;57:389–448. doi: 10.1016/s0070-2153(03)57013-2. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of miRNAs. Genome Res. 2009a;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, Biesemeier DJ, Shomron N, Fekete DM, Hornstein E, Avraham KB. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci USA. 2009b;106:7915–7920. doi: 10.1073/pnas.0812446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Eberl DF, Beisel KW. The role of bHLH genes in ear development and evolution: revisiting a 10-year-old hypothesis. Cell Mol Life Sci. 2010 doi: 10.1007/s00018-010-0403-x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Pauley S, Soukup G. Molecular evolution of the vertebrate mechanosensory cell and ear. Int J Dev Biol. 2007;51:663–678. doi: 10.1387/ijdb.072367bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Pauley S, Beisel KW. Cells, molecules and morphogenesis: the making of the vertebrate ear. Brain Res. 2006;1091:151–171. doi: 10.1016/j.brainres.2006.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK. The challenge of hair cell regeneration. Exp Biol Med. 2010;235:434–446. doi: 10.1258/ebm.2009.009281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. MicroRNA therapeutics: a new niche for antisense nucleic acids. Trends Mol Med. 2006;12:99–101. doi: 10.1016/j.molmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzano R, Montcouquiol M, Rashi-Elkeles S, Elkon R, Yucel R, Frankel WN, Rechavi G, Moroy T, Friedman TB, Kelley MW, Avraham KB. Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet. 2004;13:2143–2153. doi: 10.1093/hmg/ddh218. [DOI] [PubMed] [Google Scholar]
- Kelley MW. Cellular commitment and differentiation in the organ of Corti. Int J Dev Biol. 2007;51:571–583. doi: 10.1387/ijdb.072388mk. [DOI] [PubMed] [Google Scholar]
- Kelley MW. Hair cell development: commitment through differentiation. Brain Res. 2006;1091:172–185. doi: 10.1016/j.brainres.2006.02.062. [DOI] [PubMed] [Google Scholar]
- Kersigo J, D'Angelo A, Gray B, Soukup GA, Fritzsch B. The role of sensory organs and the forebrain for the development of the craniofacial shape as revealed by Foxg1-cre mediated microRNA loss. Genesis. 2011 doi: 10.1002/dvg.20714. [Epub ahead of print. PMID: 21225654] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KS. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MA, Quint E, Glazier AM, Fuchs H, De Agnelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, Dalmay T, Moreno-Pelyao MA, Entright AJ, Steel KP. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li H, Kloosterman W, Fekete DM. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J Neurosci. 2010;30:3254–3263. doi: 10.1523/JNEUROSCI.4948-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lu LF, Liston A. MicroRNA in the immune system, microRNA as an immune system. Immunology. 2009;127:291–298. doi: 10.1111/j.1365-2567.2009.03092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J Assoc Res Otolaryngol. 2000;1:129–143. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, Morris K, Feng F, Jones K, Lee J, Fritzsch B. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MA. Mutations in the seed region of miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- Oesterle EC, Campbell S, Taylor RR, Forge A, Hume CR. Sox2 and JAGGED1 expression in normal and drug-damaged adult mouse inner ear. J Assoc Res Otolaryngol. 2008;9:65–89. doi: 10.1007/s10162-007-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannakopoulos T, Kosik KS. MicroRNAs: regulators of oncogenesis and stemness. BMC Med. 2008;6:15. doi: 10.1186/1741-7015-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev Dyn. 2006;235:2470–2482. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol Dev. 2008;10:106–113. doi: 10.1111/j.1525-142X.2007.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacheli R, Nguyen L, Borgs L, Vandenbosch R, Bodson M, Lefebvre P, Malgrange B. Expression patterns of miR-96, miR-182, and miR-183 in the development of the inner ear. Gene Expr Patterns. 2009;9:364–370. doi: 10.1016/j.gep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Saini HK, Enright AJ, Griffiths-Jones S. Annotation of mammalian primary microRNAs. BMC Genomics. 2008;9:564. doi: 10.1186/1471-2164-9-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, O'Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhauser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Soukup GA. Little but loud: small RNAs have a resounding affect on ear development. Brain Res. 2009;1277:104–114. doi: 10.1016/j.brainres.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009;328:328–341. doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XR, Zhang XM, Zhen J, Zhang PX, Xu G, Jiang H. MicroRNA expression in the embryonic mouse inner ear. Neuroreport. 2010;21:611–617. doi: 10.1097/WNR.0b013e328338864b. [DOI] [PubMed] [Google Scholar]
- Weston MD, Soukup GA. MicroRNAs sound off. Genome Med. 2009;1:59. doi: 10.1186/gm59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Xiang M, Gan L, Li D, Chen ZY, Zhou L, O'Malley BW, Jr, Klein W, Nathans J. Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc Natl Acad Sci USA. 1997;94:9445–9450. doi: 10.1073/pnas.94.17.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Makland A, Pirvola U, Fritzsch B. Brn3c null mutant mice show long-term incomplete retention of some afferent inner ear innervation. BMC Neurosci. 2003;4:2. doi: 10.1186/1471-2202-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- Zorio E, Medina P, Rueda J, Millán JM, Arnau MA, Beneyto M, Marín F, Gimeno JR, Osca J, Salvador A, España F, Estellés A. Insights into the role of microRNAs in cardiac diseases: from biological signaling to therapeutic targets. Cardiovasc Hematol Agents Med Chem. 2009;7:82–90. doi: 10.2174/187152509787047676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.