Abstract
Paclitaxel is one of the most important chemotherapeutic drugs in the fight against cancer. This minireview covers the recent advances in the study of the bioactive conformation of paclitaxel in tubulin/microtubules. The tubulin-bound structure of paclitaxel has been studied by means of photoaffinity labeling, cryo-electron microscopy, solid-state NMR, molecular modeling, MD simulations and the synthesis of conformationally restrained analogues and paclitaxel mimics. The bioactive conformation of paclitaxel is important since it could provide critical information that would allow the design of novel analogues with simpler structures and/or increased potency against cancer.
Keywords: microtubules, molecular modeling, paclitaxel, REDOR NMR, REDOR-taxol, T-taxol, tubulin
Introduction
Cancer has become the leading cause of death among people under the age of 85 in the US, exceeding cardiovascular diseases.[1,2] Paclitaxel (taxol) and docetaxel (taxotere) are two of the most important antitumor drugs currently in use against various cancers, such as metastatic breast cancer, advanced ovarian cancer, nonsmall-cell lung cancer, and Kaposi's sarcoma.[3,4] Further clinical applications are ongoing against different types of cancers as well as for combination therapies with other anti-tumor drugs.
Paclitaxel binds to the β-tubulin component of the α,β-tubulin dimer, promotes the polymerization of tubulin, stabilizes microtubules, and blocks microtubular dynamics, which eventually leads to apoptosis.[5,6] Paclitaxel has a rigid baccatin core with four flexible side chains. Even though the mechanism of microtubule stabilization by paclitaxel was discovered almost 30 years ago, the binding conformation of paclitaxel to β-tubulin is still not fully understood. Investigation into the possible bioactive conformation of paclitaxel has attracted much interest among organic and medicinal chemists, as well as molecular pharmacologists and structural biologists.[7,8] This is not only because of the natural curiosity for the elucidation of microtubule-bound paclitaxel structure, but also because of a genuine possibility that novel drugs with much simpler structures than paclitaxel could be designed and developed based on the bioactive structure of paclitaxel.
The aim of the article is to review the recent advances in the study of the bioactive conformation of paclitaxel in β-tubulin, including molecular modeling studies, MD simulations, synthetic studies involving structurally restrained taxoids and paclitaxel mimics, and to envision future directions in the field.
Polar conformation and nonpolar conformation
The conformational analysis of paclitaxel in solution and in solid state was initially performed by NMR and X-ray crystallography, affording the “polar” (hydrophobic collapse) conformation and the “nonpolar” conformation, involving hydrophobic collapse between C3′ phenyl or C3′ N-benzoyl and the C-2 phenyl groups.[9–13] An exploration of potential common pharmacophores based on known microtubule-stabilizing agents, paclitaxel, epothilone,[14,15] eleutherobin[16] and discodermolide,[17] was also attempted.[18]
Various conformationally constrained taxoids were prepared to mimic the “polar“ and “nonpolar“ conformations. Compounds 1 and 2 were the first attempts to orient the C13 side chain to mimic the “polar“ conformation, by Heck reaction, esterification[19] and olefin metathesis,[18,20] but none of the reported taxoids were as active as paclitaxel. Later, several series of C2–C3′ N-linked macrocyclic taxoids (3–6) were reported to mimic the “nonpolar“ conformation, and again none of the compounds showed similar or higher activity compared with paclitaxel.[20–24] It was concluded that neither the “polar“ conformation nor the “nonploar“ conformation were the correct binding structure of paclitaxel. Nevertheless, through those endeavors, the olefin metathesis strategy proved to be a powerful method to synthesize conformationally restrained macrocyclic paclitaxel analogues.[7] These early efforts on the design, synthesis and biological evaluation of macrocyclic taxoids have been previously reviewed.[7]
The electron crystallographic structures and the T-taxol conformation
The structural biology study of paclitaxel did not start until the first cryo-electron microscopy (cryo-EM) (or “electron crystal-lography”) structure of a microtubule model, i.e. Zn2+-stablized α,β-tubulin dimer with paclitaxel bound, was reported in 1998 at 3.7 Å resolution (1TUB structure, Figure 1 a).[25] In the 1TUB structure, the drug-binding site was identified, which was consistent with the previous photolabeling studies using docetaxel.[26–28] This poorly resolved crystal structure was further refined to 3.5 Å resolution (1JFF structure, Figure 1 b) in 2001 using paclitaxel as the substrate.[29] However, the conformation of the crucial N-benzoylphenylisoserine moiety at C13 was still difficult to determine with confidence due to the low diffraction level in the electron density map for this moiety, especially, the 2-benzoate and 3′N-phenyl groups.[29]
Figure 1.
a) 1TUB and b) 1JFF.
In 2004, Downing et al. reported the structure of epothilone A bound to α,β-tubulin in zinc-stabilized sheets (1TVK); the structure was resolved at 2.89 Å by a combination of electron crystallography and NMR-based conformational analysis.[30] The overlay of epothilone A (1TVK) and paclitaxel (1JFF) revealed a common binding site in β-tubulin (Figure 2). In 2008, the binding structures of discodermolide and its cyclic analogue dicytostatin[31] were proposed by NMR and molecular modeling studies.[32] Similar to epothilone A, the binding structures roughly overlap with the taxane skeleton but do not make the additional contacts provided by the paclitaxel side chain, specifically with helix 1 and the other side of His 229.[32] The combined binding structures of these microtubule-stabilizing agents might be used to design hybrid compounds with much higher binding affinities.[18]
Figure 2.
a) Epothilone A (green) in β-tubulin (1TVK) and b) overlay of epothilone and paclitaxel by superimposing the backbones of 1TVK and 1JFF.
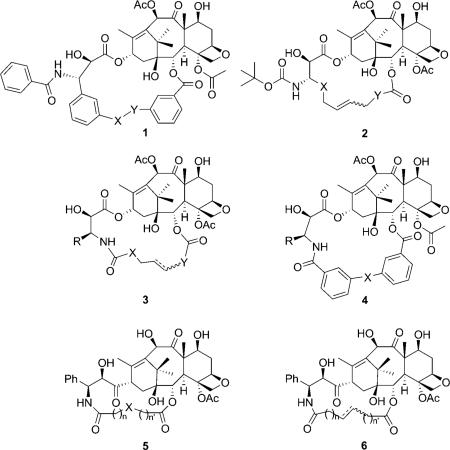
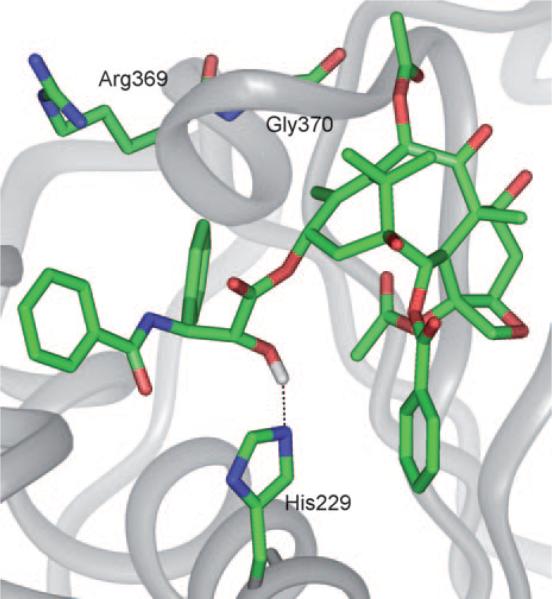
Based on further computational analysis of the solution structure of paclitaxel, and docking studies on the 1TUB structure, the “T-taxol” structure was proposed in 2001.[33] Unlike the polar and nonpolar conformations, which experience intramolecular hydrophobic collapse, T-taxol makes intermolecular hydrophobic associations, as seen for the irregularly stacked C3′-benzamido, His 229, and C2-benzoyl moieties. This model is consistent with three photoaffinity labeling studies performed on β-tubulin.[26–28] Sharing the same electron density map, the T-taxol structure is very similar to the conformation seen in the 1JFF co-crystal structure, except for torsional rotations of the side chain phenyl rings.[29] The critical C2′-OH group,[34,35] which forms an intramolecular H bond with C1′-O in the polar or nonpolar conformations, was claimed to form a H bond with the backbone carbonyl of Arg 369, however, a H bond was eventually observed between the NH of Gly 370 and C2′-O. This H bond change was probably caused by a change in the protein backbone from 1TUB to 1JFF. The new H bond is not only weaker than the original, but also inconsistent with well-known structure–activity (SAR) data that shows the C2′-OH acts as a H bond donor, but not as an acceptor (see Figure 3).[36]
Figure 3.
T-taxol in 1JFF.[37]
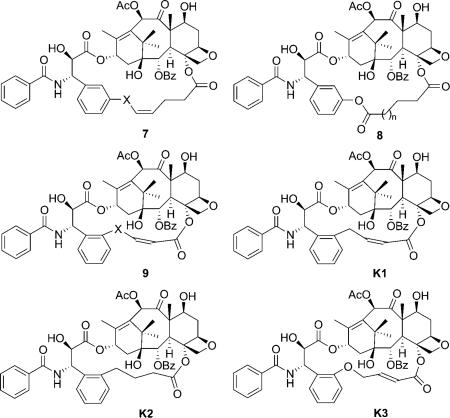
To prove the validity of the T-taxol structure, rigidified paclitaxel congeners were designed, synthesized and assayed for their tubulin polymerization activity and cytotoxicity.[7] Kingston et al. first synthesized the C4-meta-C3′ linked macrocyclic taxoids, which showed ten times weaker potency than paclitaxel due to the steric conflict between the long bridge and the Phe 270.[38] The macrolactone derivatives of the taxoids (8) showed even lower activity.[39] Later, the C4-ortho-C3′-linked macrocyclic taxoids with shorter linker (9) were synthesized.[40,41] Compound K1 is the most active compound in the series, which shows ~ 20 times higher activity than paclitaxel against A2780 human ovarian cancer cell line and twice higher activity in the tubulin polymerization experiment. The saturated congener K2 showed the same activity as paclitaxel.
The analogue with a longer linker (K3), which exhibited the T-taxol form in a population of 83 % in CDCl3 according to the NAMFIS (NMR analysis of molecular flexibility in solution) analysis,[42] is twice as effective in the tubulin polymerization assay and equally potent against both PC3 and A2780 cell lines. In 2007, the apparent affinity of the C4-ortho-C3′ linked macrocyclic taxoids for guanylyl-(α,β)-methylene-diphosphonate (GMPCPP)-stabilized microtubules was assessed by a competition assay. K1 and K2 bind to the GMPCPP-stabilized microtubules with two- to threefold greater affinity than paclitaxel, and decrease the critical concentration of guanosine diphosphate (GDP)-bound tubulin by two- to fourfold compared to paclitaxel.[43]
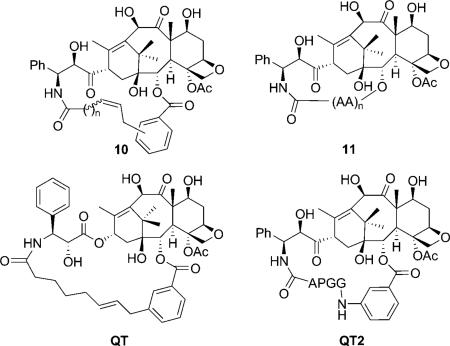
In 2004, Dubois et al. reported two new series of C2–C3′ N-linked macrocyclic taxoids with aliphatic and amino acid linkers to mimic the T-form docetaxel.[44–46] Most taxoids showed much lower activity compared with docetaxel, but one derivative (QT) was equipotent to paclitaxel in the microtubule depolymerization inhibitory assay, but ~ 10 times less active in the in vitro cytotoxicity assay.[44] The higher activity of 10 compared with 6 indicated that a hydrophobic group at C2 plays a major role in the interaction with binding site residues. It should be noted that the crystal structure of QT2 was resolved, which has a close similarity to the T-taxol structure, although it is almost inactive in both cytotoxicity and microtubule depolymerization inhibitory assays.[46]
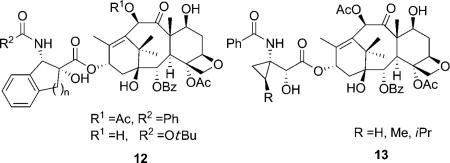
Another attempt to mimic the T-taxol structure was the introduction of a cyclic structure in the C13 side chain. In 2001, Barboni et al. reported the conformationally constrained taxoids by insertion of a carbon linker between the C2′ and the ortho position of the 3′-phenyl ring (12).[47] The taxoids with five-membered rings showed a two- to ninefold decrease in activity compared with paclitaxel against MCF-7 cell lines, and slightly lower activity compared with paclitaxel in the tubulin assembly assay. The compounds were claimed to be in the T-taxol conformation in a population of 47 % in DMSO as determined by 2D NMR/NAMFIS experiments. The taxoids with six-membered rings, however, are 240–340 times less active than paclitaxel due to their interaction with Phe 270 in the binding site. In 2005, Kingston et al. reported the C3′-cyclopropyl paclitaxel analogues (13) as T-taxol mimics.[48] These taxoids showed 13-to 1000-fold lower activity against A2780 and KB cell lines relative to paclitaxel. The interaction of the methylene group of a cyclopropyl taxane with the C4-acetyl group produced structures incompatible with the T-taxol structure, which was considered to be the reason for the weak activity of these compounds.
The REDOR NMR experiment and the REDOR-taxol conformation
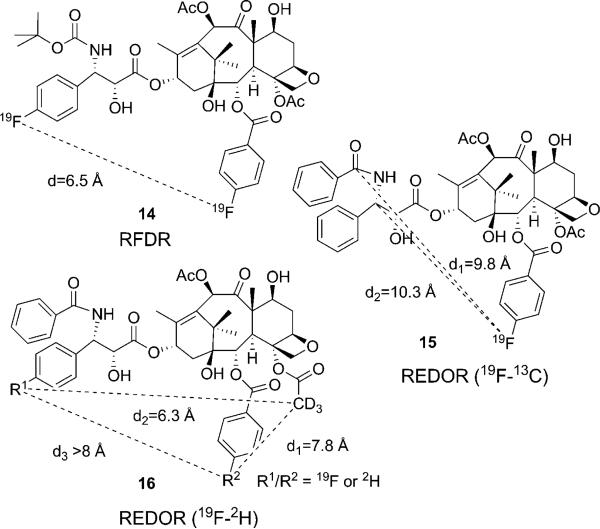
Application of 19F NMR advanced techniques is a very powerful tool to study dynamic conformational equilibria of fluorine-containing bioactive molecules. In fact, the wide dispersion of 19F chemical shifts simplifies the observation of molecular structures at low temperature. To study the binding conformation of paclitaxel and other taxoids, the fluorine-containing taxanes were successfully used as probes for NMR analysis in conjunction with molecular modeling.[49] The solid-state magic angle spinning (SSMAS) 19F NMR technique and the radio-frequency driven dipolar recoupling (RFDR) technique were applied to measure the F–F distance for the microtubule-bound conformation of F2-10-Ac-docetaxel (14, Figure 4).[50] In the mean time, another breakthrough in the investigation into the structure of the tubulin-bound paclitaxel was achieved by the application of the rotational echo double resonance (REDOR) NMR spectroscopy of the 19F/13C/15N-labeled paclitaxel–microtubule complex in the solid state in 2000.[51] The REDOR NMR data provided two 13C–19F intramolecular distances of the microtubule-bound 2-(4-fluorobenzoyl)paclitaxel (F-taxol) (15, Figure 4). Since real microtubules, i.e., not the Zn2+-stabilized tubulin dimer model, were used in this experiment, the results were critically important to probe the relevance of the cryo-EM structure (1TUB, 1JFF).
Figure 4.
Solid state NMR studies on microtubule-bound fluorine probes.
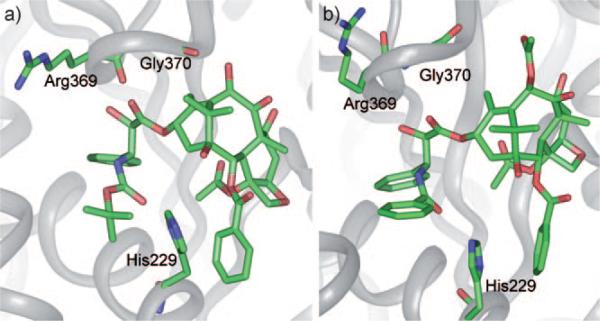
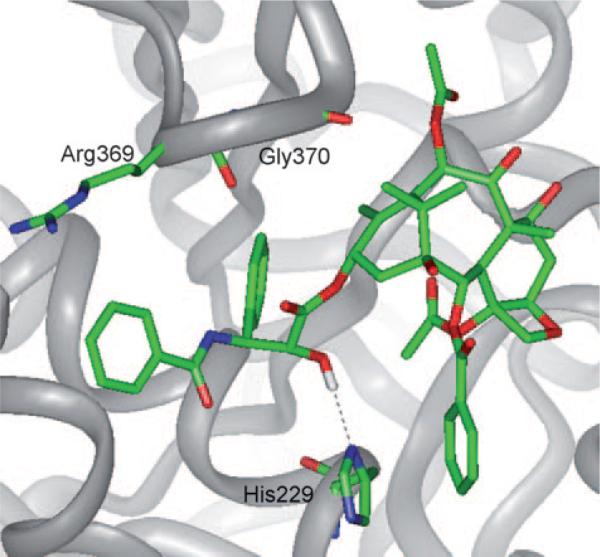
On the basis of the REDOR distances, MD analysis of paclitaxel conformers, the photoaffinity labeling and molecular modeling using the 1TUB coordinates,[28] Ojima et al. proposed the “REDOR-taxol” structure as the most plausible microtubule-bound paclitaxel structure in 2005.[8] Unlike the T-taxol structure, the C2′-OH group interacts with His 229 as the H-bond donor in the REDOR-taxol structure (Figure 5).[8]
Figure 5.
REDOR-taxol in 1TUB.
In 2007, three additional intramolecular distances between key atoms in the microtubule-bound 19F/2H-labeled paclitaxel were determined using solid state REDOR NMR spectroscopy (16, Figure 4).[52] All five known intramolecular distances were used to examine the five proposed conformations (polar, nonpolar, 1JFF-taxol, T-taxol and REDOR-taxol), and only the REDOR-taxol and T-taxol were found to be fully consistent with the experimental data.[52]
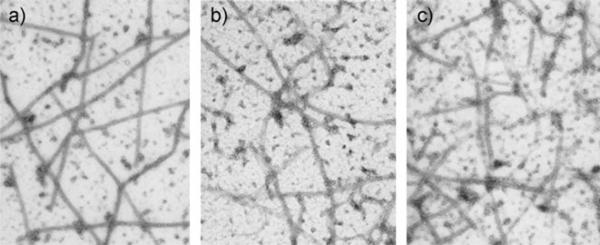
The REDOR-taxol structure was successfully used for the design and synthesis of a series of highly active C14–C3′NBz-linked macrocyclic taxoids (17).[8,53] SB-T-2053 inhibits the growth of human breast cancer cells (wild type and drug resistant) in the same order of magnitude as paclitaxel, and induces in vitro tubulin polymerization at least as well as paclitaxel.[8] The isomer, SB-T-2054, possesses virtually the same potency as that of paclitaxel in both cytotoxicity and tubulin polymerization assays.[53] Also, the microtubules formed with SB-T-2054 and paclitaxel are very similar, while those formed with GTP are longer and more uniform (Figure 6). 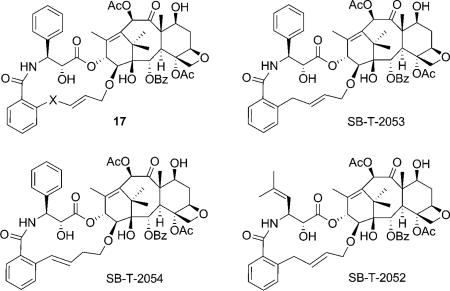
Figure 6.
Electromicrographs of microtubule: a) GTP; b) paclitaxel; c) SB-T-2054.
The C3′-isobutenyl analogue, SB-T-2052, however, showed much lower activity. Since the replacement of the 3′-phenyl moiety of paclitaxel and docetaxel with a 2-methylpropen-2-yl group has been shown to increase the potency,[54,55] the results suggest that the second generation taxoids bearing a 3′-(2-methylpropen-2-yl) moiety have a slightly different binding site from that of paclitaxel, or that these taxoids have a different bioactive conformation than that of paclitaxel.
Comparison of REDOR-taxol and T-taxol
The positions of the C-3′ phenyl rings in the REDOR-taxol and T-taxol structures are close to each other. Essentially all SAR studies,[56–58] photoaffinity labeling[26–28] and REDOR NMR results[51,52] support both structures. The critical difference between these two structures is the orientation of the C2′-OH group. In the REDOR-taxol structure, the C2′-OH group interacts with His 229 as the hydrogen donor (Figure 5),[8] while the H bond is formed between C2′-O and the backbone NH of Gly 370 in the T-taxol structure.[33]
As mentioned above, the paclitaxel-bound Zn2+-stabilized α,β-tubulin dimer sheet structure was refined by Lowe et al. in 2001 to provide the 1JFF structure. Since the 1JFF co-crystal structure has higher resolution than the 1TUB, and the T-taxol structure was updated by adopting the 1JFF structure,[37,59] the REDOR-taxol–1JFF complex was recently constructed to see possible differences between this complex and the REDOR-taxol–1TUB and also to perform detailed comparison with T-taxol structure.[53]
The REDOR-taxol structure generated in the 1TUB coordinates was manually docked into the β-tubulin unit of the 1JFF protein wherein the 1JFF-taxol substrate had been removed from the coordinates prior to docking. The resulting drug–protein complex structure (REDOR-taxol–1JFF) was minimized by using Insight II 2000 (CVFF force field). The H bond between the C2′-OH group and His 229, a key feature of the REDOR-taxol model, was very stable during the process, with a final H–N distance of 2.25 Å (Figure 7).[53]
Figure 7.
REDOR-taxol in 1JFF.
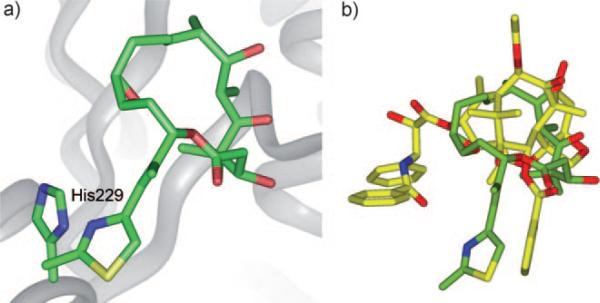
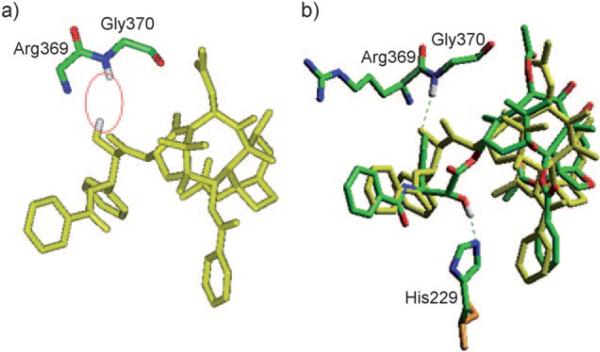
For fair and accurate comparison of the REDOR-taxol structure with the T-taxol structure,[37,59] the coordinates of the T-taxol–1JFF complex structure were obtained directly from the Emory University group,[37] and subjected to computational analyses. Unexpectedly, there was no H bond between the C2′-OH and the C=O of Arg 369 in the obtained T-taxol–1JFF coordinates although this particular H bond was reported in the original T-taxol paper.[33] Additionally, no H bond between the C2′-OH and the nitrogen of the Gly 370 NH group was observed; in this structure the C2′-OH points towards the hydrogen of the Gly 370 NH rather than the lone pair (Figure 8 a).
Figure 8.
a) T-taxol–1JFF structure as received; b) Overlay of the minimized REDOR-taxol–1JFF (green; H bond with His 229) and T-taxol–1JFF (yellow; H bond with Gly 370) structures. For clarity, only heavy atoms and C2′OH of Taxol and a few nearby residues are shown.
After energy minimization (Insight II 2000, CVFF), a H bond (3.4 Å) was formed between the C2′-O (H-bond acceptor) and H-N of Gly 370.[37] This H-bond formation caused a slight change in the T-taxol conformation in the 1JFF protein. It should be noted that the mode of this H bond (i.e., reverse H bond) is not consistent with the well-established SAR studies, which indicate that the free hydroxy group at the C2′ position is critically important, i.e., the C2′-OH serves as an H-bond donor.[10,60] The overlay of REDOR-taxol and T-taxol in 1JFF protein structure is shown in Figure 8 b wherein the key H bonds are highlighted.
The five key intramolecular atom–atom distances in the energy minimized T-taxol were measured in the same manner as that for the REDOR-taxol and results are listed in Table 1. The corresponding distances reported for the original T-taxol structure are also shown. On the basis of the comparison of the five key atom–atom distances in the REDOR-taxol, T-taxol and the experimental data, it can be safely concluded that both REDOR-taxol and T-taxol structures are consistent with the REDOR NMR data.
Table 1.
Intramolecular atomic distances of paclitaxel measured using 19F/13C/15N/2H-labeled paclitaxels
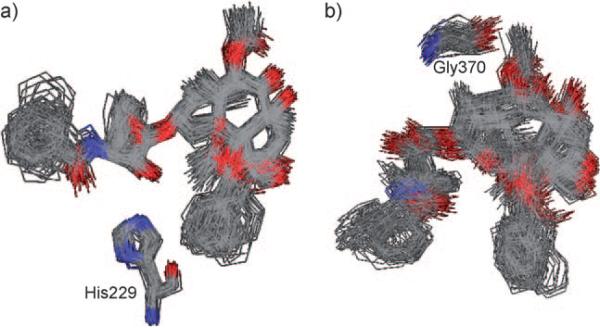
To further examine the validity of the minimized REDOR-taxol–1JFF and T-taxol–1JFF structures, a 20 ps MD simulation was performed using the Macromodel program (MMFF94 force field[61]); all atoms farther than 10 Å from the binding site were held fixed following our previously reported protocol.[8] The stability of the C2′-OH N (His 229) H bond and C2′-O-HN (Gly 370) H bond was monitored during the whole simulation. The overlay of 100 snapshots (sampled every 0.2 ps) of the taxol conformations in each case is shown in Figure 9 a.
Figure 9.
MD simulation of a) REDOR-taxol in 1JFF and b) T-taxol in 1JFF. For clarity, only heavy atoms are shown.
As the overlay clearly shows, the MD simulations of both structures are very stable and do not cause any substantial structural change. The C2′-OH–N(His 229) H bond is very stable throughout the MD simulation process, maintaining an average distance of 2.0±0.2 Å. The simulation confirms that the REDOR-taxol conformation is a stable local minimum in the tubulin-binding site. However, the H bond in the 1JFF-T-taxol structure was unstable throughout the simulation, with an average C2′-O–HN(Gly 370) distance of 5.0±0.8 Å.
Therefore, the REDOR-taxol structure was confirmed to be a valid model for the microtubule-bound bioactive conformation of paclitaxel, although the T-taxol structure has been claimed to be the only valid bioactive paclitaxel structure.[37,41,52,59] The paclitaxel SAR data clearly indicates the critical importance of a free hydroxy group at the C2′ position. However, it was found that the originally proposed critical C2′-OH–N(Arg 369) H bond in the T-taxol model (1TUB) does not exist in the 1JFF-T-taxol model. Although the C2′-O–HN(Gly 370) H bond is possible, this reverse H bond wherein the oxygen of the C2′-OH serves as a hydrogen acceptor, is inconsistent with the well-established SAR of the C2′-OH moiety.[10,60] Accordingly, the comparison of the REDOR-taxol with the T-taxol by MD simulations for stability has revealed that the C2′-OH–N(His 229) bond in REDOR-taxol is very stable, while the C2′-O–HN(Gly 370) H bond in T-taxol is not stable during the simulations.
Active macrocyclic taxoids and their tubulin-binding conformations
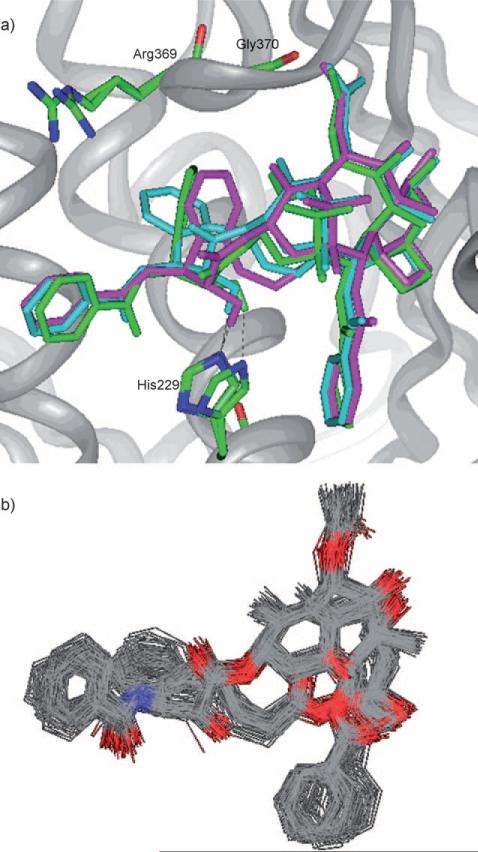
The C4–C3′ linked macrocyclic paclitaxel analogues reported by Kingston et al. were designed based on the T-taxol structure.[40,41] In order to claim that the REDOR-taxol structure is a valid model for the bioactive paclitaxel conformation, it is necessary to examine whether those analogues can be predicted by the REDOR-taxol conformation in the 1JFF protein. Two macrocyclic analogues K1 and K2 were selected, the paclitaxel molecule in the REDOR-taxol–1JFF complex was modified directly to include the K1 and K2 linkers and these structures energy minimized (Insight II 2000, CVFF).
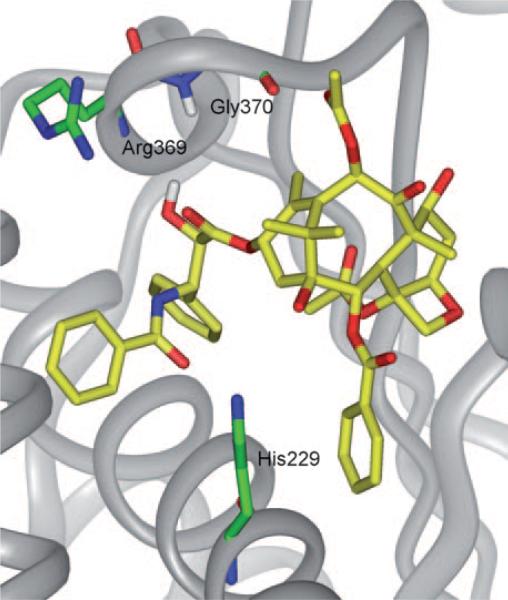
As Figure 10 a shows, K1 and K2 adopt the REDOR-taxol structure very well, keeping the critical H bond between their C2′-OH and His 229. Next, a 20 ps MD simulation of the “REDOR-K2–1JFF” structure was performed to examine the stability of this complex. This MD simulation revealed that the “REDOR-K2” structure was very stable and the C2′-O-H–N-(His 229) H-bond distance remained at 2.0±0.3 Å during the whole MD simulation (Figure 10 b).[62]
Figure 10.
a) Overlay of the REDOR-taxol (green) with “REDOR-K1” (blue) and “REDOR-K2” (magenta) structures; b) MD simulation of the “REDOR-K2” in 1JFF. For clarity, only heavy atoms and C2′OH of taxoids and a few residues are shown.
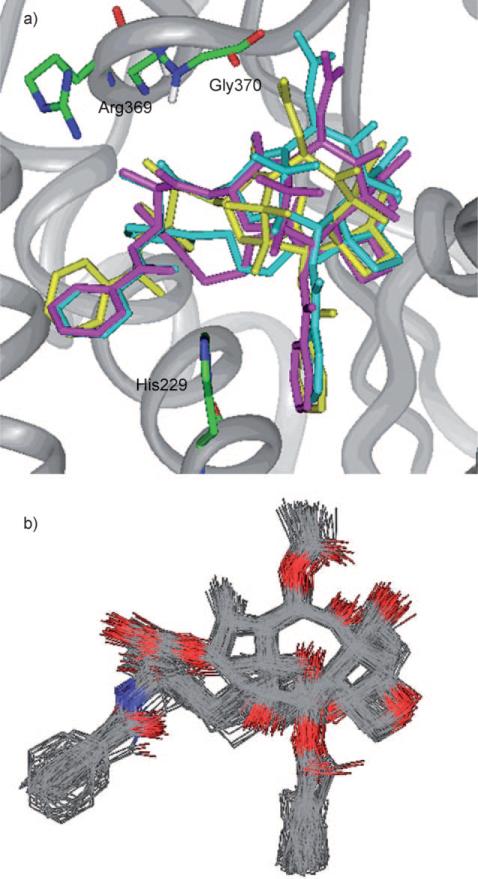
In the same manner, the “T-K2” structure was created by directly introducing the linker to the paclitaxel molecule in the T-taxol–1JFF structure, followed by energy minimization (Insight II 2000, CVFF; Figure 11 a). The MD simulation of the “T-K2–1JFF” was performed in the same manner as that for the “REDOR-K2–1JFF”. The simulation showed a stable structure, but the C2′-O–HN(Gly 370) H bond was not stable with an average distance of 5.1±0.5 Å.
Figure 11.
a) Overlay of the T-taxol structure (yellow) with “T-K1” (blue) and “T-K2” (magenta) structures; b) MD simulation of the “T-K2” in 1JFF. For clarity, only heavy atoms and C2′OH of taxoids and a few residues are shown.
Several C2–C3′ linked macrocyclic taxoids were reported to mimic the T-taxol structure, including a taxoid QT, which showed the same level of activity as paclitaxel in the tubulin-polymerization assay, although the cytotoxicity of QT was much weaker than that of paclitaxel.[44]
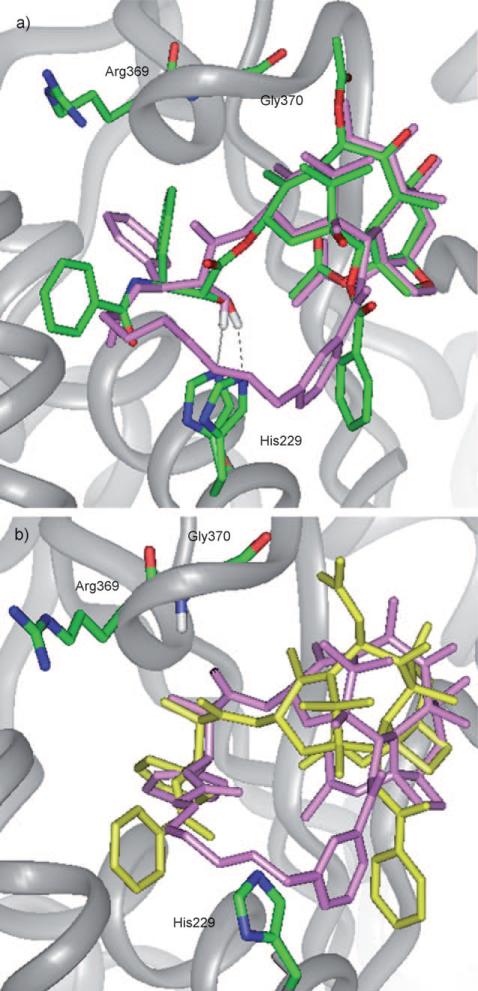
To predict the tubulin-bound structure of QT, an eight-atom linker was introduced between the C3′N and C2-meta position of paclitaxel molecule in the REDOR-taxol–1JFF and T-taxol–1JFF complexes and these structures energy minimized. This linker is long and flexible enough to avoid the collision with His 229, which exists in some inactive C2–C3′ linked taxoids with short linkers.[8,33] There were two possible orientations for the QT linker.[44] Thus, the orientation in which the C3′ phenyl group was in a similar position as that of the REDOR- or T-taxol structure was selected for overlay. The overlay of QT with the REDOR-taxol (“REDOR-QT”) and that of QT with the T-taxol (“T-QT”) after energy minimization (Insight II 2000) are shown in Figure 12 a and b, respectively.
Figure 12.
a) Overlay of the REDOR-taxol structure (green) with “REDOR-QT” (light magenta); b) overlay of the T-taxol structure (yellow) with “T-QT” (light magenta).
In the tubulin-bound QT structure, the distance between C2′-OH and N(His 229) is 1.8 Å (Figure 12 a), while the distance between C2′-O and HN(Gly 370) is 3.7 Å (Figure 12 b). In both overlays, the 2-benzoate moiety of QT substantially deviates from that of the REDOR-taxol or the T-taxol structure.
Because the C14–C3′NBz-linked macrocyclic taxoids, designed based on the REDOR-taxol structure, showed a similar activity as paclitaxel,[53] both T-taxol and REDOR-taxol conformations were examined. The overlay of SB-T-2053 and SB-T-2054 with the REDOR-taxol is shown in Figure 13 a. The H-bond distance between the C2′-OH of SB-T-2054 and His 229 is 2.7 Å, while that between C2′-O and HN(Gly 370) is 3.7 Å in the T-taxol (Figure 13 b).
Figure 13.
a) Overlay of the REDOR-taxol (green) with “REDOR-2053” (blue) and “REDOR-2054” (magenta) structures; b) overlay of the T-taxol structure (yellow) with “T-2053” (blue) and “T-2054” (magenta) structures.
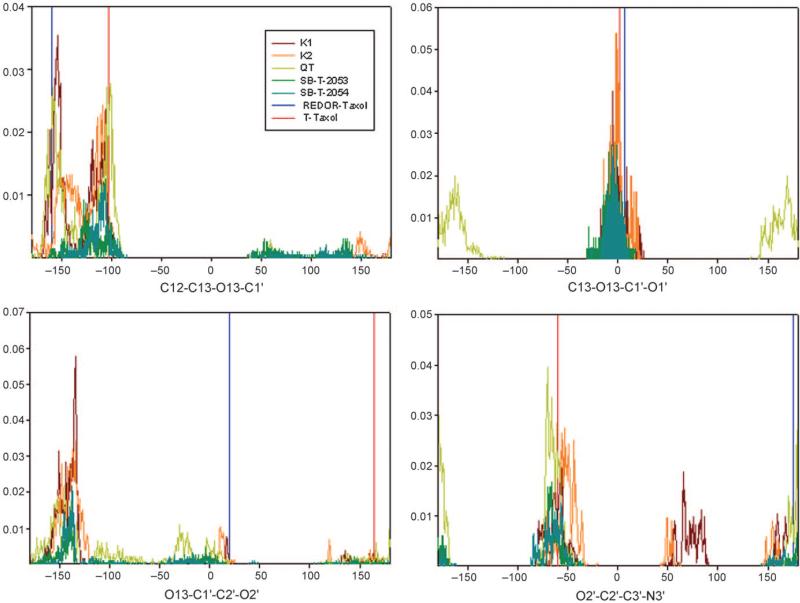
In order to examine how the conformationally restricted C13 side chains in the macrocyclic taxoids fit to the REDOR-taxol or the T-taxol structure, we conducted a Monte Carlo conformational search in aqueous medium on SB-T-2053, SB-T-2054, K1, K2 and QT. We focused on the dihedral angles involved in the C13 side chain since those should be the angles most affected by the introduction of the constraint. Figure 14 shows the dihedral angle distributions for the three macrocyclic taxoids and comparisons of those to the reference values from the REDOR-taxol and the T-taxol structures.
Figure 14.
Conformational diversity in macrocyclic taxoids: K1, brown; K2, orange; QT, yellow; SB-T-2053, green; SB-T-2054, dark green. The reference value for REDOR-taxol (blue) and T-taxol (red) are indicated by vertical lines.
K2 has a very similar dihedral angle distribution to that of K1, while paclitaxel has a wider distribution than the macrocyclic taxoids as expected. All the taxoids shared the same range between –100° and –150° for the C13–O13 torsion angle. SB-T-2053 has a similar range to K1 and K2 in the second torsion angle (O13–C1′), but the torsion angle distribution of QT is very different. SB-T-2054 and SB-T-2053 show a very similar dihedral angle distribution since they have the same length linkers; however, SB-T-2054 is more rigid. For the C1′–C2′ dihedral angle, however, neither REDOR-taxol nor T-taxol is in the major dihedral angle distributions, while the two structures are consistent with the distributions of the last torsion angle (C2′–C3′). The results indicate that both the REDOR-taxol and the T-taxol structures can be used to predict the tubulin-bound structures of macrocyclic taxoids equally well, while leaving the critical difference in the C2′-OH H-bonding mode and stability, as described above.
While the molecular mechanics (MM), MD simulation and Monte Carlo structural search studies of the macrocyclic taxoids indicate that both REDOR-taxol and T-taxol structures are equally predictive of these active taxoids, the dihedral angles of C13–C1′–C2′–O2′ of all the taxoids did not match those of REDOR-taxol or T-taxol. These findings suggest that both structures need further refinement.
Recent advances in the MD simulations of T-taxol in tubulin conformations
Long time MD simulations were widely used in computational chemistry and structural biology with the rapid development of computer and computational software. Recently, three MD simulations of paclitaxel and other taxoids in tubulin were reported based on the T-taxol structure.
In 2006, Botta et al. reported the simulation of paclitaxel, IDN-5390 and epothilone A in human class I and class III β-tubulins to study the resistance against paclitaxel caused by the tubulin isoform.[63] The T-taxol–1JFF complex[33,37] was used as the template for the complexes and 2 ns simulations were performed by using Macromodel (v 7.2)[64] and AMBER united atom force field,[65] with a force constant of 23.9 kcal mol Å–1 applied to the protein backbone. Solvent effects were taken into account by means of the implicit GB/SA water model.[66] Similar to the proposed T-taxol–1JFF complex, a H bond formed between the C2′-O and H-N of Gly 370. The direct interaction of paclitaxel and the M loop could explain the higher binding affinity between paclitaxel and class I β-tubulin than class III β-tubulin. The epothilone A–1JFF (1TVK) structure was used as the template to create epothilone–class I/III complexes, and the smaller and more flexible epothilone molecule moves from its original position during the simulation. 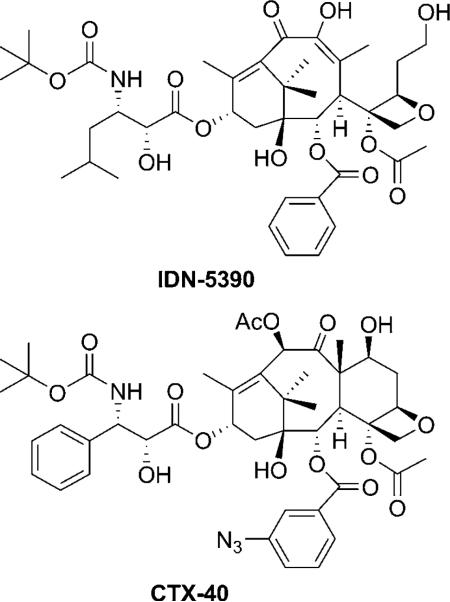
In 2007, Snyder et al. reported > 5 ns simulations of T-taxol, K1 and K2 in α,β-tubulin dimer with the GROMACS (v 3.2.1)[67] and the GROMOS96 united atom force field[68] to explain the higher binding affinity of the bridged taxoids with stabilized microtubule compared with paclitaxel.[43] Each α,β-tubulin–ligand complex was solvated in a box of 35 000–39 000 SPC water molecules with two magnesium cations associated with the phosphates of the nucleotides and 36 sodium cations added to each system to neutralize the overall charge. The K1–1JFF and K2–1JFF complexes were created by inserting the bridges into the T-taxol molecule followed by energy minimization and MD simulation. The taxoids could keep the T-taxol conformation during the process. Although the detailed interaction of individual groups with β-tubulin was not shown, it was found that the bridged taxoids caused greater conformational change in the β-tubulin M loop than paclitaxel with a conformation that strengthens the contact between adjacent microtubule protofilaments.
In 2008, a 10 ns MD simulation (AMBER force field[69])was performed for paclitaxel, docetaxel and CTX-40 (IC50/IC50,paclitaxel = 5.5 (A2780), 0.01 (A2780AD)) in β-tubulin (1JFF) to study the binding mode of the taxoids.[70] Conditions used for the simulation (pH 6.5) led to the double protonation of His 229 and therefore a H bond was formed between His 229 and the C3′N carbonyl, rather than π–π interactions with the C2 benzoate.[37] The relative orientation of these groups was claimed to be in agreement with structural data obtained for microtubule-bound docetaxel by means of the transfer nuclear Overhauser enhancement (TR-NOESY) experiment.[70]
Progress in the development of paclitaxel mimics
On the basis of the binding conformation of paclitaxel in tubulin, it may be possible to design paclitaxel mimics with simpler structure and similar or even higher activity.[7,8] The common pharmacophore analysis for the microtubule-stabilizing agents[18] suggested that the role of the baccatin core was to serve as a rigid scaffold that secures the proper orientation of the C2, C3′, and C3′N side chains and the baccatin core could be replaced by a much simpler scaffold that retains most of its three-dimensional features, but without its structural complexity.[71]
In 2004, Ojima et al. reported four paclitaxel mimics with bicyclic scaffold based on the common pharmacophore model.[71] Two hydroxy groups were included in the design to mimic the C2 and C13 hydroxy groups on baccatin with similar 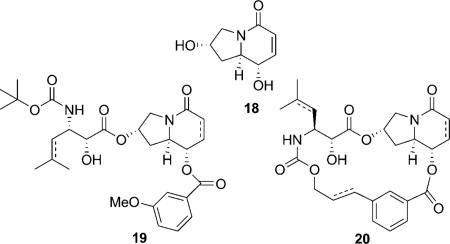 dihedral angles. The mimics showed micromolar activity against human breast cancer cell lines, and the open-chain taxoid mimic 19 is a little more potent than the macrocyclic taxoid mimic 20. However, none of the mimics showed appreciable ability to promote tubulin polymerization.
dihedral angles. The mimics showed micromolar activity against human breast cancer cell lines, and the open-chain taxoid mimic 19 is a little more potent than the macrocyclic taxoid mimic 20. However, none of the mimics showed appreciable ability to promote tubulin polymerization.
Roussi et al. also synthesized four steroidal compounds bearing the phenylisoerine and benzoate side chains to mimic the T-form of docetaxel. Two hydroxy groups were used to mimic the C13 and C2 hydroxy groups in baccatin. The compounds showed at least 10 000 times less activity than docetaxel, and no activity in a microtubule disassembly inhibitory assay. However, unexpectedly, two compounds (21 and 22) showed inhibitory activity against microtubule assembly.[72]
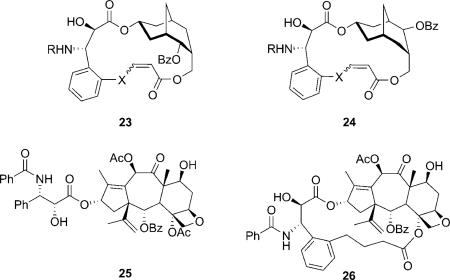
In 2006, Kingston et al. reported the macrocyclic paclitaxel mimics based on the active C4–C3′ linked macrocyclic taxoids, which were designed from the T-taxol structure.[40,73] The same bridge could converted the inactive open-chain nor-paclitaxel derivative (25) to a highly active macrocyclic one (26).[74] The three hydroxy groups on the bicyclic ring were designed to mimic the C2, C4 and C13 hydroxy groups. Achiral 2-adamantanone was used as the starting material in the synthesis giving rise to two diastereomers (23 and 24). The compounds showed micromolar activity, slightly promoting tubulin polymerization. Interestingly, 24 showed somewhat higher activity than 23, indicating the relative regiochemistry of the benzoate group does not affect the activity significantly, although 23 is a better mimic than 24. The macrocyclic paclitaxel mimics can adopt the essential elements of the T-taxol conformation according to a molecular modeling study.[73]
Outlook
Due to the lack of resolution in the electron crystallographic structure, molecular modeling, synthetic chemistry and solid-state 19F NMR became the most important methods in the study of the protein-bound conformation of paclitaxel in β-tubulin. The structures of paclitaxel itself obtained in crystal or solution (polar conformation and nonpolar conformation), which were proposed to be possible tubulin-bound conformations earlier, were proven incorrect based on these studies.
The T-taxol structure, obtained from the molecular modeling study of the electron crystallographic structure, was claimed to be the only valid bioactive paclitaxel structure. Many structural constrained taxoids were designed based on the T-taxol conformation; few of these constrained taxoids possess higher or equal activity compared with paclitaxel in cytotoxicity or tubulin polymerization assays. The T-taxol structure has been serving as a common template in many molecular modeling studies. On the other hand, the REDOR-taxol structure, proposed based on the molecular modeling study of the REDOR NMR data, is also proven to be a valid model for the microtubule-bound bioactive conformation of paclitaxel. The REDOR-taxol structure was used to design a series of highly active macrocyclic taxoids. The C2′-OH–N(His 229) H bond in the REDOR-taxol–1JFF structure was found to be very stable in MD simulation, while the C2′-O–HN(Gly 370) H bond in the T-taxol–1JFF structure is unstable in MD simulation. Also, the fact that the oxygen of the C2′-OH serves as hydrogen acceptor in T-taxol is inconsistent with the known SAR data. The Monte Carlo structural search of the macrocyclic taxoids suggests that both proposed bioactive conformations of paclitaxel need further refinement.
With the increasing availability of ultrafast computers, and new and more advanced computational methods, MD simulation is widely used in the study of the bioactive conformation of paclitaxel in tubulin, although different results appear to be obtained by using different methods and parameters. Novel paclitaxel mimics with simpler structures have been designed and examined but their activities are still much weaker than paclitaxel. Nevertheless, it is still possible that new paclitaxel mimics with high activity emerge based on innovative design and syntheses by fully exploiting the tubulin-bound paclitaxel structure in combination with those of other microtubule-stabilizing agents.
Acknowledgements
Some of the work done in the laboratories of authors described was supported by grants from the National Institutes of Health (CA103314 and GM42798 to I.O.) and the National Science Foundation (PACI-MCA02N028 to C.S.). The authors are grateful to Dr. James P. Snyder, Emory University for providing the coordinates of the T-taxol–1JFF complex structure.
References
- 1.Jemal A, Ward E, Hao Y, Thun M. JAMA J. Am. Med. Assoc. 2005;294:1255–1259. doi: 10.1001/jama.294.10.1255. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, M J. Thun. CA-Cancer J. Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 3.Rowinsky EK. Annu. Rev. Med. 1997;48:353–374. doi: 10.1146/annurev.med.48.1.353. [DOI] [PubMed] [Google Scholar]
- 4.Mekhail TM, Markman M. Expert Opin. Pharmacother. 2002;3:755–766. doi: 10.1517/14656566.3.6.755. [DOI] [PubMed] [Google Scholar]
- 5.Schiff PB, Fant J, Horwitz SB. Nature. 1979;277:665–667. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- 6.Jordan MA, Wilson L. Nat. Rev. Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 7.Kingston DG, Bane S, Snyder JP. Cell Cycle. 2005;4:279–289. [PubMed] [Google Scholar]
- 8.Geney R, Sun L, Pera P, Bernacki Ralph J, Xia S, Horwitz Susan B, Simmerling Carlos L, Ojima I. Chem. Biol. 2005;12:339–348. doi: 10.1016/j.chembiol.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Dubois J, Guenard D, Gueritte-Voegelein F, Guedira N, Potier P, Gillet B, Beloeil JC. Tetrahedron. 1993;49:6533–6544. [Google Scholar]
- 10.Williams HJ, Scott AI, Dieden RA, Swindell CS, Chirlian LE, Francl MM, Heerding JM, Krauss NE. Tetrahedron. 1993;49:6545–6560. [Google Scholar]
- 11.Vander Velde DG, Georg GI, Grunewald GL, Gunn CW, Mitscher LA. J. Am. Chem. Soc. 1993;115:11650–11651. [Google Scholar]
- 12.Mastropaolo D, Camerman A, Luo Y, Brayer GD, Camerman N. Proc. Natl. Acad. Sci. USA. 1995;92:6920–6924. doi: 10.1073/pnas.92.15.6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S, Ojima I. Expert Opin. Ther. Pat. 2000;10:869–889. [Google Scholar]
- 14.Bollag DM, McQueney PA, Zhu J, Hensens O, Koupal L, Liesch J, Goetz M, Lazarides E, Woods CM. Cancer Res. 1995;55:2325–2333. [PubMed] [Google Scholar]
- 15.Giannakakou P, Kowalski RJ, Hamel E. J. Biol. Chem. 1997;272:2534–2541. doi: 10.1074/jbc.272.4.2534. [DOI] [PubMed] [Google Scholar]
- 16.Lindel T, Jensen PR, Fenical W, Long BH, Casazza AM, Carboni J, Fairchild CR. J. Am. Chem. Soc. 1997;119:8744–8745. [Google Scholar]
- 17.ter Haar E, Kowalski RJ, Hamel E, Lin CM, Longley RE, Gunasekera SP, Rosenkranz HS, Day BW. Biochemistry. 1996;35:243–250. doi: 10.1021/bi9515127. [DOI] [PubMed] [Google Scholar]
- 18.Ojima I, Chakravarty S, Inoue T, Lin S, He L, Horwitz SB, Kuduk SD, Danishefsky SJ. Proc. Natl. Acad. Sci. USA. 1999;96:4256–4261. doi: 10.1073/pnas.96.8.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boge TC, Wu Z-J, Himes RH, Vander Velde DG, Georg GI. Bioorg. Med. Chem. Lett. 1999;9:3047–3052. doi: 10.1016/s0960-894x(99)00522-3. [DOI] [PubMed] [Google Scholar]
- 20.Ojima I, Lin S, Inoue T, Miller ML, Borella CP, Geng X, Walsh JJ. J. Am. Chem. Soc. 2000;122:5343–5353. [Google Scholar]
- 21.Ojima I, Geng X, Lin S, Pera P, Bernacki RJ. Bioorg. Med. Chem. Lett. 2002;12:349–352. doi: 10.1016/s0960-894x(01)00747-8. [DOI] [PubMed] [Google Scholar]
- 22.Geng X, Miller ML, Lin S, Ojima I. Org. Lett. 2003;5:3733–3736. doi: 10.1021/ol0354627. [DOI] [PubMed] [Google Scholar]
- 23.Querolle OD, Thoret J, Dupont S, Gueritte C, Guenard F. Eur. J. Org. Chem. 2003:542–550. [Google Scholar]
- 24.Querolle O, Dubois J, Thoret S, Roussi F, Montiel-Smith S, Gueritte F, Guenard D. J. Med. Chem. 2003;46:3623–3630. doi: 10.1021/jm030770w. [DOI] [PubMed] [Google Scholar]
- 25.Nogales E, Wolf SG, Downing KH. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 26.Rao S, Krauss NE, Heerding JM, Swindell CS, Ringel I, Orr GA, Horwitz SB. J. Biol. Chem. 1994;269:3132–3134. [PubMed] [Google Scholar]
- 27.Rao S, Orr GA, Chaudhary AG, Kingston DG, Horwitz SB. J. Biol. Chem. 1995;270:20 235–20 238. doi: 10.1074/jbc.270.35.20235. [DOI] [PubMed] [Google Scholar]
- 28.Rao S, He L, Chakravarty S, Ojima I, Orr GA, Horwitz SB. J. Biol. Chem. 1999;274:37990–37994. doi: 10.1074/jbc.274.53.37990. [DOI] [PubMed] [Google Scholar]
- 29.Lowe J, Li H, Downing KH, Nogales E. J. Mol. Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 30.Nettles JH, Li H, Cornett B, Krahn JM, Snyder JP, Downing KH. Science. 2004;305:866–869. doi: 10.1126/science.1099190. [DOI] [PubMed] [Google Scholar]
- 31.Buey RM, Barasoain I, Jackson E, Meyer A, Giannakakou P, Paterson I, Mooberry S, Andreu JM, Diaz JF. Chem. Biol. 2005;12:1269–1279. doi: 10.1016/j.chembiol.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Canales A, Matesanz R, Gardner NM, Andreu JM, Paterson I, Diaz JF, Jimenez-Barbero J. Chem. Eur. J. 2008;14:7557–7569. doi: 10.1002/chem.200800039. [DOI] [PubMed] [Google Scholar]
- 33.Snyder JP, Nettles JH, Cornett B, Downing KH, Nogales E. Proc. Natl. Acad. Sci. USA. 2001;98:5312–5316. doi: 10.1073/pnas.051309398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gueritte-Voegelein F, Guenard D, Lavelle F, Le Goff MT, Mangatal L, Potier P. J. Med. Chem. 1991;34:992–998. doi: 10.1021/jm00107a017. [DOI] [PubMed] [Google Scholar]
- 35.Swindell CS, Krauss NE, Horwitz SB, Ringel I. J. Med. Chem. 1991;34:1176–1184. doi: 10.1021/jm00107a042. [DOI] [PubMed] [Google Scholar]
- 36.Williams HJ, Moyna G, Scott AI, Swindell CS, Chirlian LE, Heerding JM, Williams DK. J. Med. Chem. 1996;39:1555–1559. doi: 10.1021/jm950796q. [DOI] [PubMed] [Google Scholar]
- 37.Alcaraz AA, Mehta AK, Johnson SA, Snyder JP. J. Med. Chem. 2006;49:2478–2488. doi: 10.1021/jm051119r. [DOI] [PubMed] [Google Scholar]
- 38.Metaferia BB, Hoch J, Glass TE, Bane SL, Chatterjee SK, Snyder JP, Lakdawala A, Cornett B, Kingston DGI. Org. Lett. 2001;3:2461–2464. doi: 10.1021/ol016124d. [DOI] [PubMed] [Google Scholar]
- 39.Liu C, Schilling JK, Ravindra R, Bane S, Kingston DGI. Bioorg. Med. Chem. 2004;12:6147–6161. doi: 10.1016/j.bmc.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 40.Ganesh T, Guza RC, Bane S, Ravindra R, Shanker N, Lakdawala AS, Snyder JP, Kingston DGI. Proc. Natl. Acad. Sci. USA. 2004;101:10006–10011. doi: 10.1073/pnas.0403459101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ganesh T, Yang C, Norris A, Glass T, Bane S, Ravindra R, Banerjee A, Metaferia B, Thomas SL, Giannakakou P, Alcaraz AA, Lakdawala AS, Snyder JP, Kingston DGI. J. Med. Chem. 2007;50:713–725. doi: 10.1021/jm061071x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snyder JP, Nevins N, Cicero DO, Jansen J. J. Am. Chem. Soc. 2000;122:724–725. [Google Scholar]
- 43.Shanker N, Kingston DGI, Ganesh T, Yang C, Alcaraz AA, Geballe MT, Banerjee A, McGee D, Snyder JP, Bane S. Biochemistry. 2007;46:11514–11527. doi: 10.1021/bi700753y. [DOI] [PubMed] [Google Scholar]
- 44.Querolle O, Dubois J, Thoret S, Roussi F, Gueritte F, Gu nard D. J. Med. Chem. 2004;47:5937–5944. doi: 10.1021/jm0497996. [DOI] [PubMed] [Google Scholar]
- 45.Larroque A-L, Dubois J, Thoret S, Aubert G, Guenard D, Gueritte F. Bioorg. Med. Chem. Lett. 2005;15:4722–4726. doi: 10.1016/j.bmcl.2005.07.069. [DOI] [PubMed] [Google Scholar]
- 46.Larroque A-L, Dubois J, Thoret S, Aubert G, Chiaroni A, Gueritte F, Guenard D. Bioorg. Med. Chem. 2007;15:563–574. doi: 10.1016/j.bmc.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 47.Barboni L, Lambertucci C, Appendino G, Vander Velde DG, Himes RH, Bombardelli E, Wang M, Snyder JP. J. Med. Chem. 2001;44:1576–1587. doi: 10.1021/jm001103v. [DOI] [PubMed] [Google Scholar]
- 48.Liu C, Tamm M, Notzel MW, Rauch K, de Meijere A, Schilling JK, Lakdawala A, Snyder JP, Bane SL, Shanker N, Ravindra R, Kingston DGI. Eur. J. Org. Chem. 2005:3962–3972. [Google Scholar]
- 49.Ojima I, Kuduk SD, Chakravarty S, Ourevitch M, Begue J-P. J. Am. Chem. Soc. 1997;119:5519–5527. [Google Scholar]
- 50.Ojima I, Kuduk SD, Chakravarty S. Adv. Med. Chem. 1999;4:69–124. [Google Scholar]
- 51.Li Y, Poliks B, Cegelski L, Poliks M, Cryczynski A, Piszcek G, Jagtap PG, Studelska DR, Kingston DGI, Schaefer J, Bane S. Biochemistry. 2000;39:281–291. doi: 10.1021/bi991936r. [DOI] [PubMed] [Google Scholar]
- 52.Paik Y, Yang C, Metaferia B, Tang S, Bane S, Ravindra R, Shanker N, Alcaraz AA, Johnson SA, Schaefer J, O'Connor RD, Cegelski L, Snyder JP, Kingston DGI. J. Am. Chem. Soc. 2007;129:361–370. doi: 10.1021/ja0656604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun L, Geng X, Geney R, Li Y, Simmerling C, Li Z, Lauher JW, Xia S, Horwitz SB, Veith JM, Pera P, Bernacki RJ, Ojima I. J. Org. Chem. 2008;73:9584–9593. doi: 10.1021/jo801713q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ojima I, Slater JC, Michaud E, Kuduk SD, Bounaud P-Y, Vrignaud P, Bissery M-C, Veith J, Pera P, Bernacki RJ. J. Med. Chem. 1996;39:3889–3896. doi: 10.1021/jm9604080. [DOI] [PubMed] [Google Scholar]
- 55.Ojima I, Chen J, Sun L, Borella CP, Wang T, Miller ML, Lin S, Geng X, Kuznetsova L, Qu C, Gallager D, Zhao X, Zanardi I, Xia S, Horwitz SB, Mallen-St. Clair J, Guerriero JL, Bar-Sagi D, Veith JM, Pera P, Bernacki RJ. J. Med. Chem. 2008;51:3203–3221. doi: 10.1021/jm800086e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kingston DGI. J. Nat. Prod. 2000;63:726–734. doi: 10.1021/np000064n. [DOI] [PubMed] [Google Scholar]
- 57.Barboni L, Giarlo G, Ricciutelli M, Ballini R, Georg GI, Vander-Velde DG, Himes RH, Wang M, Lakdawala A, Snyder JP. Org. Lett. 2004;6:461–464. doi: 10.1021/ol036204c. [DOI] [PubMed] [Google Scholar]
- 58.Ferjancic Z, Matovic R, Cekovic Z, Jiang Y, Snyder JP, Trajkovic V, Saicic RN. Tetrahedron. 2006;62:8503–8514. [Google Scholar]
- 59.Johnson SA, Alcaraz AA, Snyder JP. Org. Lett. 2005;7:5549–5552. doi: 10.1021/ol051780p. [DOI] [PubMed] [Google Scholar]
- 60.Kant J, Huang S, Wong H, Fairchild C, Vyas D, Farina V. Bioorg. Med. Chem. Lett. 1993;3:2471–2474. [Google Scholar]
- 61.Halgren TA. J. Comput. Chem. 1996;17:490–519. [Google Scholar]
- 62. It was claimed that the “REDOR-K2” structure was unstable and drastic conformational reorganization of the structure took place “to avoid steric clash (with Phe 272)” in the MD simulation done by the Emory group using the same force field (MMFF94) (Ref. [59]). However, such reorganization did not happen at all in the study performed by the Stony Brook group. The origin of this marked difference in the computational analysis is most likely due to the “reconstructed” REDOR-taxol structure in the Emory Group study in the absence of the exact coordinates from the Stony Brook group. [Google Scholar]
- 63.Magnani M, Ortuso F, Soro S, Alcaro S, Tramontano A, Botta M. FEBS J. 2006;273:3301–3310. doi: 10.1111/j.1742-4658.2006.05340.x. [DOI] [PubMed] [Google Scholar]
- 64.Mohamadi F, Richards NGJ, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC. J. Comput. Chem. 1990;11:440–467. [Google Scholar]
- 65.Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Jr., Weiner P. J. Am. Chem. Soc. 1984;106:765–784. [Google Scholar]
- 66.Still WC, Tempczyk A, Hawley RC, Hendrickson T. J. Am. Chem. Soc. 1990;112:6127–6129. [Google Scholar]
- 67.Lindahl E, Hess B, van der Spoel D. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- 68.Scott WRP, Huenenberger PH, Tironi IG, Mark AE, Billeter SR, Fennen J, Torda AE, Huber T, Krueger P, van Gunsteren WF. J. Phys. Chem. A. 1999;103:3596–3607. [Google Scholar]
- 69.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Jr., Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 70.Matesanz R, Barasoain I, Yang C-G, Wang L, Li X, de Ines C, Coderch C, Gago F, Barbero JJ, Andreu JM, Fang W-S, Diaz JF. Chem. Biol. 2008;15:573–585. doi: 10.1016/j.chembiol.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 71.Geng X, Geney R, Pera P, Bernacki RJ, Ojima I. Bioorg. Med. Chem. Lett. 2004;14:3491–3494. doi: 10.1016/j.bmcl.2004.04.060. [DOI] [PubMed] [Google Scholar]
- 72.Roussi F, Ngo QA, Thoret S, Gueritte F, Guenard D. Eur. J. Org. Chem. 2005:3952–3961. [Google Scholar]
- 73.Ganesh T, Norris A, Sharma S, Bane S, Alcaraz AA, Snyder JP, Kingston DGI. Bioorg. Med. Chem. 2006;14:3447–3454. doi: 10.1016/j.bmc.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 74.Tang S, Yang C, Brodie P, Bane S, Ravindra R, Sharma S, Jiang Y, Snyder JP, Kingston DGI. Org. Lett. 2006;8:3983–3986. doi: 10.1021/ol061438s. [DOI] [PMC free article] [PubMed] [Google Scholar]