Abstract
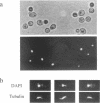
To investigate cell cycle regulation at the S or G2 phase in Saccharomyces cerevisiae, we have isolated mutants displaying supersensitivity to hydroxyurea (HU), a chemical that inhibits DNA replication. Such mutants, which we have named hydroxyurea sensitive (hys), defined four linkage groups and we characterized the hys2 mutation in this study. The hys2-1 mutant displays temperature sensitive growth and a constellation of phenotypes indicating defective DNA metabolism. At the restrictive temperature, hys2-1 cells arrest as large budded cells with a single nucleus at the neck of the bud and a short spindle. The hys2-1 mutant exhibits increased rates of chromosome loss and recombination. Additionally, hys2-1 appears to accumulate incompletely replicated DNA that can be detected by a pulse field electrophoresis assay. Finally, deletion of RAD9 in a hys2-1 strain decreases the percentage of arrested cells, suggesting that an intact RAD9-checkpoint is required for the cell cycle arrest in hys2-1 cells. HYS2 encodes a 55 kDa protein that is essential for viability at all temperatures. Taken together, these data suggest that Hys2 plays a role in DNA replication.
Full text
PDF



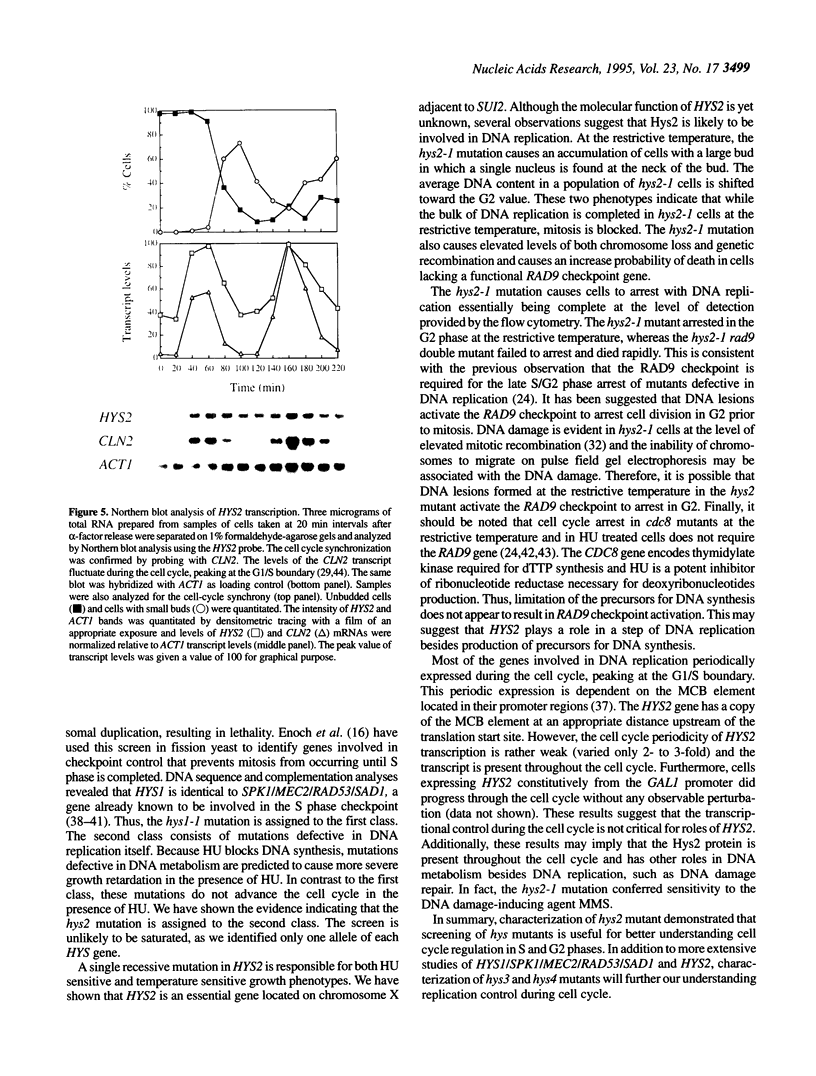

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amon A., Surana U., Muroff I., Nasmyth K. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature. 1992 Jan 23;355(6358):368–371. doi: 10.1038/355368a0. [DOI] [PubMed] [Google Scholar]
- Baker T. A., Kremenstova E., Luo L. Complete transposition requires four active monomers in the mu transposase tetramer. Genes Dev. 1994 Oct 15;8(20):2416–2428. doi: 10.1101/gad.8.20.2416. [DOI] [PubMed] [Google Scholar]
- Barker D. G., White J. H., Johnston L. H. The nucleotide sequence of the DNA ligase gene (CDC9) from Saccharomyces cerevisiae: a gene which is cell-cycle regulated and induced in response to DNA damage. Nucleic Acids Res. 1985 Dec 9;13(23):8323–8337. doi: 10.1093/nar/13.23.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank A., Kim B., Loeb L. A. DNA polymerase delta is required for base excision repair of DNA methylation damage in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1994 Sep 13;91(19):9047–9051. doi: 10.1073/pnas.91.19.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet A., Simon M., Faye G., Bauer G. A., Burgers P. M. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 1989 Jun;8(6):1849–1854. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M. E., Wittrup K. D., Bailey J. E., Campbell J. L. DNA polymerase I is required for premeiotic DNA replication and sporulation but not for X-ray repair in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):365–376. doi: 10.1128/mcb.9.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., Feng L., Donahue T. F. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. R., Tinkelenberg A. H. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991 May 31;65(5):875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- Doi K., Gartner A., Ammerer G., Errede B., Shinkawa H., Sugimoto K., Matsumoto K. MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 1994 Jan 1;13(1):61–70. doi: 10.1002/j.1460-2075.1994.tb06235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990 May;4(5):740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- Enoch T., Carr A. M., Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992 Nov;6(11):2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- Enoch T., Nurse P. Coupling M phase and S phase: controls maintaining the dependence of mitosis on chromosome replication. Cell. 1991 Jun 14;65(6):921–923. doi: 10.1016/0092-8674(91)90542-7. [DOI] [PubMed] [Google Scholar]
- Enoch T., Nurse P. Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication. Cell. 1990 Feb 23;60(4):665–673. doi: 10.1016/0092-8674(90)90669-6. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Hadwiger J. A., Wittenberg C., Richardson H. E., de Barros Lopes M., Reed S. I. A family of cyclin homologs that control the G1 phase in yeast. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I. M., Hyams J. S. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988 Mar;89(Pt 3):343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976 Jul 15;104(4):803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985 Jul;110(3):381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989 Nov 3;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hennessy K. M., Lee A., Chen E., Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991 Jun;5(6):958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- Hisamoto N., Sugimoto K., Matsumoto K. The Glc7 type 1 protein phosphatase of Saccharomyces cerevisiae is required for cell cycle progression in G2/M. Mol Cell Biol. 1994 May;14(5):3158–3165. doi: 10.1128/mcb.14.5.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell E. A., McAlear M. A., Rose D., Holm C. CDC44: a putative nucleotide-binding protein required for cell cycle progression that has homology to subunits of replication factor C. Mol Cell Biol. 1994 Jan;14(1):255–267. doi: 10.1128/mcb.14.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. H., Lowndes N. F. Cell cycle control of DNA synthesis in budding yeast. Nucleic Acids Res. 1992 May 25;20(10):2403–2410. doi: 10.1093/nar/20.10.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. H., Nasmyth K. A. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978 Aug 31;274(5674):891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- Lucchini G., Mazza C., Scacheri E., Plevani P. Genetic mapping of the Saccharomyces cerevisiae DNA polymerase I gene and characterization of a pol1 temperature-sensitive mutant altered in DNA primase-polymerase complex stability. Mol Gen Genet. 1988 Jun;212(3):459–465. doi: 10.1007/BF00330850. [DOI] [PubMed] [Google Scholar]
- Lucchini G., Muzi Falconi M., Pizzagalli A., Aguilera A., Klein H. L., Plevani P. Nucleotide sequence and characterization of temperature-sensitive pol1 mutants of Saccharomyces cerevisiae. Gene. 1990 May 31;90(1):99–104. doi: 10.1016/0378-1119(90)90444-v. [DOI] [PubMed] [Google Scholar]
- Lundgren K., Walworth N., Booher R., Dembski M., Kirschner M., Beach D. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell. 1991 Mar 22;64(6):1111–1122. doi: 10.1016/0092-8674(91)90266-2. [DOI] [PubMed] [Google Scholar]
- Murray A. W. Creative blocks: cell-cycle checkpoints and feedback controls. Nature. 1992 Oct 15;359(6396):599–604. doi: 10.1038/359599a0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Reynolds P., Prakash S., Prakash L. Cloning and sequence analysis of the Saccharomyces cerevisiae RAD9 gene and further evidence that its product is required for cell cycle arrest induced by DNA damage. Mol Cell Biol. 1989 May;9(5):1882–1896. doi: 10.1128/mcb.9.5.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick K. S., Carr A. M. Feedback controls and G2 checkpoints: fission yeast as a model system. Bioessays. 1993 Dec;15(12):775–782. doi: 10.1002/bies.950151202. [DOI] [PubMed] [Google Scholar]
- Sitney K. C., Budd M. E., Campbell J. L. DNA polymerase III, a second essential DNA polymerase, is encoded by the S. cerevisiae CDC2 gene. Cell. 1989 Feb 24;56(4):599–605. doi: 10.1016/0092-8674(89)90582-5. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Murray A. W. S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28. Nature. 1992 Jan 23;355(6358):365–368. doi: 10.1038/355365a0. [DOI] [PubMed] [Google Scholar]
- Stern D. F., Zheng P., Beidler D. R., Zerillo C. Spk1, a new kinase from Saccharomyces cerevisiae, phosphorylates proteins on serine, threonine, and tyrosine. Mol Cell Biol. 1991 Feb;11(2):987–1001. doi: 10.1128/mcb.11.2.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993 May;134(1):63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988 Jul 15;241(4863):317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Kiser G. L., Hartwell L. H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994 Mar 15;8(6):652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Wittenberg C., Sugimoto K., Reed S. I. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990 Jul 27;62(2):225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]