Abstract
Four new depsipeptides, mirabamides E-H (1–4), and the known depsipeptide mirabamide C (5) have been isolated from the sponge Stelletta clavosa, collected from the Torres Strait. The planar structures were determined on the basis of extensive 1D and 2D NMR and HRESIMS. The absolute configurations were established by the advanced Marfey’s method, NMR and GC-MS. The four new compounds all showed strong inhibition of HIV-1 in a neutralization assay with IC50 values of 121, 62, 68 and 41 nM, respectively.
Marine sponges have yielded a number of unique peptides and depsipeptides with a broad spectrum of biological activities.1–3 Among these sponge depsipeptides, callipeltin A,4 neamphamide,5 papuamides A-D,6 and mirabamides A-D7 are well known for their potent HIV-inhibitory activity and their structurally unique features incorporating several uncommon amino acid residues.
As part of a collaborative project with the Queensland Museum to investigate the intraspecific chemical variability of marine sponges sampled as part of the marine surveys of northeast Australia,8 we studied Stelletta clavosa collected by epibenthic sled from inter-lagoon seabed areas in the Torres Strait. Previous studies with S. clavosa led to the isolation of dimeric macrolide glycosides,9 polymethoxydienes,10 phosphorus containing clavosines,11 cyclic peptides,12 porphyrin analogs, swinholide polyketides, carotenoids, and diketopiperazines.13 Our investigation of S. clavosa resulted in the isolation of four new cyclic depsipeptides (1–4), along with the known compound mirabamide C.7 This is the first report of depsipeptides from Stelletta clavosa. Herein, we report the isolation and structure elucidation of these new compounds, as well as their HIV-1 inhibition activity.
Results and Discussion
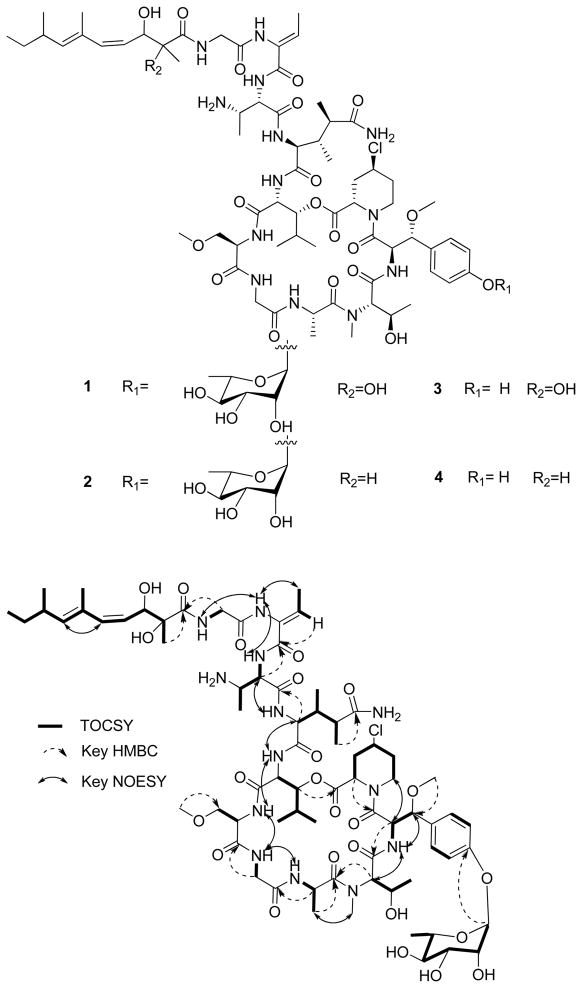
The molecular formula of 1 was determined as C72H112ClN13O24 on the basis of an observed pseudomolecular ion at m/z 1578.7705 Da by HRESIMS ([M+H]+, Δ−0.3 ppm). NMR experiments were performed in CD3CN-H2O (4:1) with and without 0.1% TFA-d. The presence of a large number of signals for exchangeable amide NH protons (δH 6.64–9.24 ppm) and carbonyl signals (δC 166.4 – 180.0 ppm) in the 1H and 13C NMR spectra of 1 reflected characteristic features of a peptide. Detailed analysis of 2D NMR data enabled us to identify 11 amino acid residues: β-methoxytyrosine (β-OMeTyr), N-methylthreonine (NMeThr), alanine, two glycine residues, 3-methoxyalanine (3-OMeAla), 3-hydroxyleucine (3-OHLeu), 4-chloro-homoproline (4-ClHpr), 3,4-dimethylglutamine (3,4-DiMeGln), 2,3-diaminobutanoic acid (Dab), and 2-amino-2-butenoic acid (Aba) (Table 1). In addition, a 2,3-dihydroxy-2,6,8-trimethyldeca-(4Z,6E)-dienoic acid (Dhtda) moiety and a rhamnose unit were apparent (Table 1).
Table 1.
NMR Spectroscopic Data for Mirabamide E (1) in (CD3CN-H2O 4:1, 0.1% TFA-d)a
| position | δC, mult | δH (J in Hz) | HMBC | NOESY |
|---|---|---|---|---|
| ClHpr | ||||
| 1 | 170.6, C | |||
| 2 | 49.5, CH | 5.01, br d (6.6) | 1, 3, 4, 6, 1β-OMeTyr | 3a, Me-33,4-DiMeGln |
| 3a | 32.7, CH2 | 2.49, br d (15.5) | 4, 5 | 2, 3b, 4 |
| 3b | 2.24, m | 1, 2 | 3a, 4, 5 | |
| 4 | 55.8, CH | 4.52, br s | 3a, 3b, 5 | |
| 5 | 32.2, CH2 | 1.90, m | 3b, 4, 6a, 6b | |
| 6a | 38.2, CH2 | 4.05, m | 5 | 5, 6b |
| 6b | 3.51, m | 5 | 5, 6a | |
| β-OMeTyr | ||||
| 1 | 171.6, C | |||
| 2 | 51.6, CH | 5.31, t (9.5) | 1, 3, 1′, 1NMeThr | 3, 2′, 6′, NH, 6ClHpr, |
| 3 | 84.0, CH | 4.29, d (9.5) | 1, 2, 1′, 2′, OMe-3 | 2, 2′, 6′ |
| 1′ | 131.3, C | |||
| 2′, 6′ | 130.1, CH | 7.32, d (8.4) | 3, 6′, 3′, 4′ | 2, 3, OMe-3, 3′, 4 NMeThr |
| 3′, 5′ | 116.7, CH | 7.01, d (8.4) | 1′, 6′, 4′ | 4 NMeThr, 1Rha |
| 4′ | 156.7, C | |||
| OMe-3 | 56.7, CH3 | 3.03, s | 3 | 3 |
| NH | 7.73, d (9.5) | 1NMeThr | 2, 2′, 6′, 4 NMeThr, 2 NMeThr | |
| NMeThr | ||||
| 1 | 168.4, C | |||
| 2 | 63.4, CH | 4.33, d (10.0) | 1, 1Ala, 3, 4, NMe | NH β-OMeTyr |
| 3 | 63.2, CH | 3.86, m | 2, 4 | 4, NMe |
| 4 | 19.2, CH3 | 0.38, d (5.9) | 2, 3 | 2, NHβ-OMeTyr |
| NMe | 30.5, CH3 | 3.04, s | 2, 1Ala | 3, 2Ala, 3Ala |
| Ala | ||||
| 1 | 174.0, C | |||
| 2 | 47.9, CH | 4.46, m | 1, 3 | 3, NH, NMeNMeThr |
| 3 | 15.0, CH3 | 1.41, d (6.9) | 1, 2 | NH, 2, NMeNMeThr |
| NH | 6.93, d (5.0) | 2, 3, 1Gly1 | 2, 3, NHGly1 | |
| Gly 1 | ||||
| 1 | 171.0, C | |||
| 2a | 42.5, CH2 | 3.87, dd (17.9, 5.5) | 1 | 2b |
| 2b | 3.55, dd (17.9, 5.5) | 1 | 2a | |
| NH | 8.29, br t (5.5) | 13-OMeAla | 2b, 23-OMeAla, NHAla, NH3-OMeAla | |
| 3-OMeAla | ||||
| 1 | 171.2, C | |||
| 2 | 56.3, CH | 4.21, m | 1, 3 | 3a |
| 3a | 70.7, CH2 | 3.81, m | 1, 2, OMe-3 | |
| 3b | 3.71, m | 1, 2, OMe-3 | ||
| OMe-3 | 58.8, CH3 | 3.31, s | 3 | |
| NH | 8.03, d (4.0) | 2, 3, 13-OHLeu | 2, 3, NH3-OHLeu, NHGly1, 23-OHLeu | |
| 3-OHLeu | ||||
| 1 | 172.2, C | |||
| 2 | 55.0, CH | 5.14, d (10.1) | 1, 3, 4, 13,4-DiMeGln | 4, NH, NH3-OMeAla |
| 3 | 80.3, CH | 5.19, m | 4, 1ClHpr | 5, 6, 4, 3 Ala |
| 4 | 28.4, CH | 1.92, m | 3, 5, 6 | 3, 5, 6, NH |
| 5 | 20.0, CH3 | 0.95, d (6.7) | 3, 4, 6 | 3, 4, 3ClHpr |
| 6 | 18.1, CH3 | 0.80, d (6.7) | 3, 4, 5 | NH, NH3-OMeAla, 3, 4 |
| NH | 8.67, d (10.9) | 13,4-DiMeGln, 2 | 2, 4, NH3-OMeAla, 23,4-DiMeGln | |
| 3,4-DiMeGln | ||||
| 1 | 174.7, C | |||
| 2 | 58.1, CH | 4.17, (dd, 10, 4.6) | 1, 3, 4, 1Dab | 3, Me-3 |
| 3 | 36.1, CH | 2.19, m | 5 | 2, 4, Me-3, NH |
| 4 | 42.0, CH | 2.60, m | 3, 5, Me-3, Me-4 | 3, Me-4, NH |
| 5 | 180.0, C | |||
| Me-3 | 13.0, CH3 | 1.01, d (6.9) | 2, 3, 4 | 2, 3, 4, 3 ClHpr, 43-OHLeu, 2ClHpr |
| Me-4 | 14.8, CH3 | 1.12, d (6.9) | 3, 4, 5 | 2, 3, 4 |
| NH | 9.24, d (4.6) | 2, 3, 1Dab | 2, 3, 4, NHDab, 2 Dab | |
| NH2-5 | 6.64, br s | 5 | 4 | |
| 7.11, br s | 5 | 4 | ||
| Dab | ||||
| 1 | 171.3, C | |||
| 2 | 54.8, CH | 4.38, t (7.7) | 1, 3, 4, 1Aba | |
| 3 | 48.6, CH | 3.86, m | 2 | 2 |
| 4 | 15.3, CH3 | 1.23, d (7.0) | 2, 3 | NH, 2, 3 |
| NH | 8.39, d (8.0) | 1, 2, 1Aba | 2, NH3,4-DiMeGln, NHAba | |
| NH2-3 | Na | |||
| Aba | ||||
| 1 | 166.4, C | |||
| 2 | 128.8, C | |||
| 3 | 133.5, CH | 6.59, q (6.7) | 1, 2, 4 | 4 |
| 4 | 12.9, CH3 | 1.68, d (6.7) | 2, 3, 1Gly2 | 3, NH |
| NH | 8.95, s | 1, 3, 1Gly2 | 4, NHDab, NH Gly2, 2 Gly2 | |
| Gly 2 | ||||
| 1 | 171.3, C | |||
| 2a | 43.2, CH2 | 4.13, m | 1, 1Dhtda | 2b |
| 2b | 3.90, m | 1, 1Dhtda | 2a | |
| NH | 8.26, t (6.2) | 2, 1Dhtda | 2a, 2b, NHAba | |
| Dhtda | ||||
| 1 | 177.3, C | |||
| 2 | 78.1, C | |||
| 3 | 72.2, CH | 4.77, t (10.6) | 1, Me-2, 4, 5 | 4, Me-2 |
| 4 | 124.5, CH | 5.41, m | 2, 5, 6, 7 | 3, 5 |
| 5 | 138.3, CH | 6.12, br d (12.0) | 3, 4, Me-6, 6, 7, | 4, 7 |
| 6 | 131.2, C | |||
| 7 | 139.0, CH | 5.22, m | Me-6, Me-8, 5, 8, 9 | 3, 5, 8, 9b, Me-8 |
| 8 | 34.3, CH | 2.33, m | 6, 7, 9, Me-8, 10 | 7, Me-6, Me-8 |
| 9a | 30.4, CH2 | 1.34, m | 10, Me-8, 8, 7 | 10 |
| 9b | 1.21, m | 10, Me-8, 8, 7 | 10 | |
| 10 | 11.8, CH3 | 0.81, t (7.0) | 9, Me-8, 8 | 8, Me-6 |
| Me-2 | 21.9, CH3 | 1.20, s | 1, 2, 3 | 2, 3 |
| Me-6 | 16.2, CH3 | 1.75, br s | 6, 7, Me-8 | 3, 5, 8, 9b |
| Me-8 | 20.3, CH3 | 0.91, d (6.5) | 7, 8, 9, 10 | 7, 8, 9a, 9b, Me-6 |
| Rha | ||||
| 1 | 98.2, CH | 5.40, br s | 3, 5, 4′β-OMeTyr | 3′, 5′ |
| 2 | 70.7, CH | 3.98, br s | 1, 3, 4 | |
| 3 | 71.1, CH | 3.79, m | 2, 4, 5 | |
| 4 | 72.4, CH | 3.40, t (8.5) | 2, 3, 5, 6 | |
| 5 | 69.7, CH | 3.57, m | 2, 3, 6 | 6 |
| 6 | 17.3, CH3 | 1.15, d (6.4) | 4, 5 | |
Spectra were recorded at 500 MHz for 1H and 125 MHz for 13C.
The amino acid sequence of 1 was assigned from a combination of inter-residue NOESY and HMBC correlations (Figure 1). The Dhtda group was linked to the N-terminus of the Gly2 residue by the HMBC correlations between the NHGly2 (δH 8.26), H-2Gly2 (δH 3.90, 4.13) and the carbonyl (δC 177.3) of Dhtda, and rhamnose was located at the C-4′ hydroxy of β-OMeTyr by an HMBC correlation between the anomeric proton (δH 5.40) and C-4′β-OMeTyr (δC 156.7).
Figure 1.
TOCSY and key HMBC and NOESY correlations for compound 1.
The structure of 1 was very similar to that of mirabamide A,7 the only difference being replacement of threonine with its dehydration product 2-amino-2-butenoic acid (Aba). This substitution was most apparent in the 13C NMR spectrum by signals at 12.9 ppm (C-4), 128.8 and 133.5 ppm (C-2 and C-3, respectively), and 166.4 ppm (C-1). NOESY cross-peaks between NHAba (δH 8.95) and NHDab (δH 8.39) and between NHAba (δH 8.95) and NHGly2 (δH 8.26) located Aba between Gly2 and Dab. Z geometry was indicated by a strong NOESY correlation between Me-4Aba (δH 1.68) and NHAba (δH 8.95).
As in the papuamides and mirabamides,6,7 the C-4/C-5 olefin of Dhtda was assigned a Z geometry on the basis of a JH-H of 12 Hz between H-4 and H-5. The C-6/C-7 double bond was assigned an E geometry on the basis of NOESY correlations between H-5 and H-7 and between Me-6 and Me-8.
By using the advanced Marfey’s method,14,15 it was possible to assign the absolute configuration of the α carbons for the following residues (Supporting Information): D-3- OHLeu, D-3-OMeAla, L-Ala, L-NMeThr, L-ClHpr, 3,4-dimethyl-L-glutamine, and L-Dab. The absolute configuration of C-3 in D-3-OHLeu was determined as 3R based on the large coupling constant between H-2 and H-3 (3J = 10 Hz) and by a NOESY correlation between H-4 and NH (Supporting Information). Similarly for Dab, a large coupling between H-2 and H-3 (3J = 7.7 Hz) and a NOESY correlation between H-4 and NH revealed the absolute configuration of C-3 to be S. Likewise, large coupling between H-2 and H-3 (3J = 10.0 Hz) in NMeThr and a NOESY correlation observed between 4-CH3 and NHβ-OMeTyr and the absence of NOESY correlation between NMe and 4-CH3 determined the absolute configuration of C-3 to be R. In 3,4-dimethyl-L-glutamine, a large coupling interaction between H-2 and H-3 (3J = 10.0 Hz) and the presence of NOESY correlations between Me-3 and H-2 and between H-4 and NH likewise suggested an absolute configuration of S for C-3. A small coupling interaction between H-3 and H-4 (3J = 4.6 Hz) indicated a gauche relationship between the two hydrogens. However, internal inconsistencies among the remaining NOESY correlations suggested that rotational averaging might be occurring around the C-3/C-4 bond. To unambiguously establish the configuration of C-4, derivatization of synthetic 2S,3S,4R- and 2S,3S,4S-dimethylglutamine with Marfey’s reagent was carried out and the retention times for the two diastereomers compared with the corresponding residue in 1. Naturally occurring 3,4-dimethylglutamine was identical in retention time to the derivative of 2S,3S,4R-dimethylglutamine, both of which eluted one minute earlier than the derivative of 2S,3S,4S-dimethylglutamine.
Similar to mirabamides A-D,7 an equatorial location for H-4ClHpr was suggested by uniformly small 3JH-H values to all of its coupling partners (δH 4.52 br s). On the basis of this similarity and coelution of the L-FDLA derivatives of ClHpr from the hydrolysates of 1 and mirabamide C, respectively (Supporting Information), the corresponding residue was assigned as (2S,4S)-4-chlorohomoproline. Comparison of the 1H NMR signals for H-2β-OMeTyr (δH 5.31, dd, J = 9.5, 9.5 Hz) and H-3β-OMeTyr (δH 4.29, d, J = 9.5 Hz) with the corresponding signals reported for (R)-β-OMe-D-Tyr in papuamide B17 (δH 5.18, dd, J = 9.4, 9.4 Hz; δH 4.37, d, J = 9.5 Hz, respectively) versus those of (R)-β-OMe-L-Tyr in neamphamide A5,17 (δH 4.67, s; δH 5.03, s, respectively), supported assignment of this residue in 1 as R-β-OMe-D-Tyr. Lastly, the carbohydrate unit was identified as L-rhamnose by hydrolyzing the glycosidic bond and comparing the retention time by GC/MS of the pertrimethylsilylated sugar to that of the corresponding derivative of authentic L-rhamnose. Occurrence of L-rhamnose in 1 as the α anomer was confirmed by a 1JC-H of 172 Hz for H-1/C-1.18
The HRESIMS spectrum of compound 2 suggested a molecular formula of C72H112ClN13O23 which differs from that of 1 by loss of an oxygen atom. The 1H and 13C NMR spectra of 2 (Tables 2 and 3) were very similar to those of 1 except for the absence of the quaternary carbon at 78.1 ppm (C-2Dhtda in 1), the presence of an additional methine at 47.1 ppm, the downfield shift of C-4Dhtda (from 124.5 ppm to 128.2 ppm), and the upfield shift of Me-2Dhtda (from 21.9 ppm to 13.2 ppm). This suggested that the amide-linked 2,3-dihydroxy-2,6,8-trimethyldeca-(4Z,6E)-dienoic acid in 1 was instead 3-hydroxy-2,6,8-trimethyldeca-(4Z,6E)-dienoic acid (Htda) in 2. This assignment was confirmed by a TOCSY correlation between Me-2Htda (δH 0.95) and H-5Htda (δH 6.00) and by HMBC correlations from Me-2Htda (δH 0.95) to C-1Htda (δC 177.3), C-2Htda (δC 47.1) and C-3Htda (δC 70.1). Thus, the planar structure of 2 was established.
Table 2.
1H NMR Data for Compounds 1–4 (500 MHz) in (CD3CN-H2O 4:1, 0.1% TFA-d)
| position | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| ClHpr | ||||
| 2 | 5.01, br d (6.6) | 4.97, br d (6.8) | 4.98, br d (6.8) | 4.98, br d (7.1) |
| 3a | 2.49, br d (15.5) | 2.45, br d (15.5) | 2.50, br d (15.2) | 2.49, br d (14.8) |
| 3b | 2.24, m | 2.21, m | 2.23, m | 2.23, m |
| 4 | 4.52, br s | 4.51, br s | 4.52, br s | 4.52, br s |
| 5 | 1.90, m | 1.89, m | 1.91, m | 1.91, m |
| 6a | 4.05, m | 4.03, m | 4.07, m | 4.07, m |
| 6b | 3.51, m | 3.48, m | 3.53, m | 3.53, m |
| β-OMeTyr | ||||
| 2 | 5.31, t (9.5) | 5.30, t (9.4) | 5.31, t (9.5) | 5.31, t (9.5) |
| 3 | 4.29, d (9.5) | 4.27, d (9.5) | 4.24, d (9.5) | 4.23, d (9.5) |
| 2′, 6′ | 7.32, d (8.4) | 7.30, d (8.2) | 7.22, d (7.4) | 7.21, d (8.2) |
| 3′, 5′ | 7.01, d (8.4) | 7.00, d (8.2) | 6.76, d (7.4) | 6.76, d (8.2) |
| OMe-3 | 3.03, s | 3.01, s | 3.03, s | 3.02, s |
| NH | 7.73, d (9.5) | 7.87, d (9.5) | 7.84, d (8.7) | 7.87, d (9.5) |
| NMeThr | ||||
| 2 | 4.33, d (10.0) | 4.33, d (10.0) | 4.35, d (9.8) | 4.35, d (10.0) |
| 3 | 3.86, m | 3.84, m | 3.87, m | 3.88, m |
| 4 | 0.38, d (5.9) | 0.34, d (5.9) | 0.47, d (5.2) | 0.47, d (5.9) |
| NMe | 3.04, s | 3.01, s | 3.07, s | 3.07, s |
| Ala | ||||
| 2 | 4.46, m | 4.47, m | 4.47, m | 4.47, m |
| 3 | 1.41, d (6.9) | 1.39, d (6.8) | 1.40, d (6.9) | 1.40, d (7.3) |
| NH | 6.93, d (5.0) | 6.98, d (5.0) | 7.03, d (3.5) | 7.04, d (5.0) |
| Gly 1 | ||||
| 2a | 3.87, dd (17.9, 5.5) | 3.87, dd (17.9, 5.5) | 3.86, m | 3.97, dd (17.9, 4.2) |
| 2b | 3.55, dd (17.9, 5.5) | 3.59, dd (17.9, 5.5) | 3.69, m | 3.70, dd (17.9, 4.2) |
| NH | 8.29, br t (5.5) | 8.39, br t (5.5) | 8.33, br s | 8.31, br t (4.7) |
| 3-OMeAla | ||||
| 2 | 4.21, m | 4.19, m | 4.21, m | 4.20, m |
| 3a | 3.81, m | 3.77, m | 3.98, m | 3.81, m |
| 3b | 3.71, m | 3.67, m | 3.65, m | 3.63, m |
| OMe-3 | 3.31, s | 3.29, s | 3.34, s | 3.33, s |
| NH | 8.03, d (4.0) | 8.00, br s | 7.92, br s | 7.90, br s |
| 3-OHLeu | ||||
| 2 | 5.14, d (10.1) | 5.08, d (10.4) | 5.03, d (8.8) | 5.01, m |
| 3 | 5.19, m | 5.16, m | 5.19, d (11.0) | 5.19, m |
| 4 | 1.92, m | 1.90, m | 1.91, m | 1.95, m |
| 5 | 0.95, d (6.7) | 0.91, d (6.7) | 0.92, d (6.6) | 0.92, d (6.6) |
| 6 | 0.80, d (6.7) | 0.77, d (6.7) | 0.83, d (6.6) | 0.83, d (6.6) |
| NH | 8.67, d (10.9) | 8.73, d (10.4) | 8.91, d (7.5) | 8.96, d (7.5) |
| 3,4-DiMeGln | ||||
| 2 | 4.17, m | 4.21, m | 4.25, m | 4.25, m |
| 3 | 2.19, m | 2.16, m | 2.20, m | 2.20, m |
| 4 | 2.60, m | 2.59, m | 2.65, m | 2.67, m |
| Me-3 | 1.01, d (6.9) | 0.97, d (7.0) | 0.99, d (7.1) | 0.99, d (7.0) |
| Me-4 | 1.12, d (6.9) | 1.09, d (6.8) | 1.11, d (7.1) | 1.11, d (6.8) |
| NH | 9.24, d (3.0) | 8.96, br s | 8.75, br s | 8.67, br s |
| NH2-5 | 6.64, br s | 6.67, br s | 6.65, br s | 6.69, br s |
| 7.11, br s | 7.16, br s | 7.17, br s | 7.17, br s | |
| Dab | ||||
| 2 | 4.38, t (7.7) | 4.45, m | 4.05, m | 4.54, m |
| 3 | 3.86, m | 3.96, m | 3.93, m | 4.00, m |
| 4 | 1.23, d (7.0) | 1.22, d (7.0) | 1.27, d (6.3) | 1.27, d (6.3) |
| NH | 8.39, d (8.0) | 8.45, d (6.1) | 8.62, br s | 8.61, d (6.1) |
| NH2-3 | Na | Na | Na | Na |
| Aba | ||||
| 3 | 6.59, q (6.7) | 6.61, q (7.9) | 6.64, q (7.2) | 6.63, q (7.3) |
| 4 | 1.68, d (6.7) | 1.68, d (7.9) | 1.71, d (7.2) | 1.71, d (7.3) |
| NH | 8.95, s | 9.11, s | 9.09, s | 9.20, s |
| Gly 2 | ||||
| 2a | 4.13, m | 4.07, m | 4.19, m | 4.18, m |
| 2b | 3.90, m | 3.86, m | 3.94, m | 3.88, m |
| NH | 8.26, t (6.2) | 8.09, t (6.2) | 8.32, br s | 8.08, br s |
| Dhtda/Htda | ||||
| 2 | 2.39, m | 2.44, m | ||
| 3 | 4.77, t (10.6) | 4.58, t (9.5) | 4.77, t (10.1) | 4.59, t (9.5) |
| 4 | 5.41, m | 5.18, m | 5.43, m | 5.21, m |
| 5 | 6.12, br d (12.0) | 6.00, dd (11.7, 3.8) | 6.12, dd (11.9, 1.7) | 6.02, dd (11.7, 3.8) |
| 7 | 5.22, m | 5.17, m | 5.22, m | 5.20, m |
| 8 | 2.33, m | 2.29, m | 2.32, m | 2.33, m |
| 9a | 1.34, m | 1.31, m | 1.34, m | 1.34, m |
| 9b | 1.21, m | 1.17, m | 1.21, m | 1.20, m |
| 10 | 0.81, t (7.0) | 0.78, t (7.0) | 0.82, t (7.0) | 0.82, t (7.0) |
| Me-2 | 1.20, s | 0.95, d (7.0) | 1.21, s | 0.97, d (6.5) |
| Me-6 | 1.75, br s | 1.71, br s | 1.75, br s | 1.75, br s |
| Me-8 | 0.91, d (6.5) | 0.88, d (7.1) | 0.92, d (7.1) | 0.91, d (7.1) |
| Rha | ||||
| 1 | 5.40, br s | 5.40, br s | ||
| 2 | 3.98, br s | 3.99, br s | ||
| 3 | 3.79, m | 3.81, m | ||
| 4 | 3.40, t (8.5) | 3.40, t (9.6) | ||
| 5 | 3.57, m | 3.57, m | ||
| 6 | 1.15, d (6.4) | 1.13, d (6.4) | ||
Table 3.
13C NMR Data for Compounds 1–4 (125 MHz) in (CD3CN-H2O 4:1)
| position | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| ClHpr | ||||
| 1 | 170.6 | 170.0 | 170.4 | 170.5 |
| 2 | 49.5 | 49.1 | 49.9 | 49.9 |
| 3 | 32.7 | 32.2 | 32.9 | 33.0 |
| 4 | 55.8 | 55.3 | 55.9 | 56.0 |
| 5 | 32.2 | 31.8 | 32.5 | 32.7 |
| 6 | 38.2 | 37.8 | 38.6 | 38.8 |
| β-OMeTyr | ||||
| 1 | 171.6 | 171.3 | 172.4 | 172.7 |
| 2 | 51.6 | 51.1 | 51.9 | 51.9 |
| 3 | 84.0 | 83.5 | 84.3 | 84.3 |
| 1′ | 131.3 | 130.8 | 128.7 | 128.8 |
| 2′, 6′ | 130.1 | 129.6 | 130.2 | 130.2 |
| 3′, 5′ | 116.7 | 116.3 | 115.7 | 115.7 |
| 4′ | 156.7 | 156.0 | 157.8 | 158.0 |
| OMe-3 | 56.7 | 56.4 | 56.7 | 56.8 |
| NMeThr | ||||
| 1 | 168.4 | 168.1 | 168.7 | 168.7 |
| 2 | 63.4 | 63.0 | 63.9 | 63.9 |
| 3 | 63.2 | 62.8 | 63.5 | 63.6 |
| 4 | 19.2 | 18.6 | 19.4 | 19.6 |
| NMe | 30.5 | 30.1 | 30.8 | 30.9 |
| Ala | ||||
| 1 | 174.0 | 173.5 | 174.2 | 174.4 |
| 2 | 47.9 | 47.4 | 47.9 | 48.0 |
| 3 | 15.0 | 14.5 | 15.2 | 15.3 |
| Gly 1 | ||||
| 1 | 171.0 | 170.6 | 171.1 | 171.1 |
| 2 | 42.5 | 41.0 | 41.2 | 42.8 |
| 3-OMeAla | ||||
| 1 | 171.2 | 171.2 | 171.4 | 172.0 |
| 2 | 56.3 | 55.8 | 56.0 | 56.2 |
| 3 | 70.7 | 70.0 | 70.8 | 71.1 |
| OMe-3 | 58.8 | 58.4 | 59.1 | 59.3 |
| 3-OHLeu | ||||
| 1 | 172.2 | 171.8 | 172.1 | 172.2 |
| 2 | 55.0 | 55.3 | 55.0 | 56.0 |
| 3 | 80.3 | 79.8 | 80.2 | 80.2 |
| 4 | 28.4 | 27.8 | 28.7 | 28.6 |
| 5 | 20.0 | 19.3 | 19.7 | 19.7 |
| 6 | 18.1 | 17.8 | 18.6 | 18.8 |
| 3,4-DiMeGln | ||||
| 1 | 174.7 | 174.0 | 174.3 | 174.2 |
| 2 | 58.1 | 57.6 | 58.4 | 58.5 |
| 3 | 36.1 | 36.1 | 36.8 | 37.1 |
| 4 | 42.0 | 41.1 | 41.0 | 41.0 |
| 5 | 180.0 | 179.3 | 179.4 | 179.3 |
| Me-3 | 13.0 | 13.2 | 13.2 | 13.0 |
| Me-4 | 14.8 | 14.1 | 15.2 | 15.2 |
| Dab | ||||
| 1 | 171.3 | 170.8 | 172.0 | 172.2 |
| 2 | 54.80 | 54.3 | 55.0 | 55.2 |
| 3 | 48.6 | 48.1 | 48.8 | 48.8 |
| 4 | 15.3 | 14.5 | 15.4 | 15.6 |
| Aba | ||||
| 1 | 166.4 | 166.2 | 166.7 | 166.7 |
| 2 | 128.8 | 128.2 | 129.1 | 129.2 |
| 3 | 133.5 | 133.6 | 133.9 | 133.9 |
| 4 | 12.9 | 12.6 | 13.1 | 13.2 |
| Gly 2 | ||||
| 1 | 171.3 | 171.3 | 172.0 | 172.3 |
| 2 | 43.2 | 43.0 | 43.5 | 43.9 |
| Dhtda/Htda | ||||
| 1 | 177.3 | 177.3 | 178.0 | 177.5 |
| 2 | 78.1 | 47.1 | 78.4 | 48.0 |
| 3 | 72.2 | 70.1 | 72.6 | 71.2 |
| 4 | 124.5 | 128.2 | 124.9 | 129.2 |
| 5 | 138.3 | 136.5 | 138.4 | 137.2 |
| 6 | 131.2 | 130.6 | 131.4 | 131.6 |
| 7 | 139.0 | 138.8 | 139.0 | 139.4 |
| 8 | 34.3 | 33.7 | 34.7 | 34.7 |
| 9 | 30.4 | 29.7 | 30.3 | 30.6 |
| 10 | 11.8 | 11.3 | 12.0 | 12.0 |
| Me-2 | 21.9 | 13.2 | 22.1 | 13.8 |
| Me-6 | 16.2 | 15.9 | 16.7 | 16.7 |
| Me-8 | 20.3 | 19.8 | 20.4 | 20.6 |
| Rha | ||||
| 1 | 98.2 | 97.7 | ||
| 2 | 70.7 | 70.1 | ||
| 3 | 71.1 | 70.5 | ||
| 4 | 72.4 | 71.9 | ||
| 5 | 69.7 | 69.1 | ||
| 6 | 17.3 | 16.7 | ||
The HRESIMS spectrum of compound 3 suggested the molecular formula C66H102ClN13O20 which differs from that of 1 by loss of C6H10O4. Comparing the 13C NMR spectrum of 3 with 1, the signals corresponding to the rhamnose unit were all absent. This suggested that 3 was the desrhamnosyl analogue of 1. Analysis of the 2D NMR data (HSQC, HMBC and NOESY) established the structure of 3.
The HRESIMS spectrum of compound 4 provided the molecular formula C66H102ClN13O19 indicating one less oxygen atom relative to 3, suggesting 4 should bear the same relationship to 2 that 3 does to 1. Investigation of the 13C NMR spectrum of 4 clearly indicated the absence of rhamnose and the existence of an amide-linked 3-hydroxy-2,6,8-trimethyldeca-(4Z,6E)-dienoic acid. Comparison of the 2D NMR data of 4 to those of 2 and 3 confirmed the structure of 4.
HRESIMS of the fifth compound provided the molecular formula C66H104ClN13O21 that was identical with mirabamide C. This was confirmed by comparison of the NMR data with published values.7
As with compound 1, LC-MS analysis of the D/L-FDLA derivatives of the acid hydrolysates of 2–4 and comparison of the retention times with those of 1 revealed that all the amino acid residues possessed identical configurations to 1. The configuration and glycosidic bond configuration for L-rhamnose in 2 were determined in the same manner as for 1.
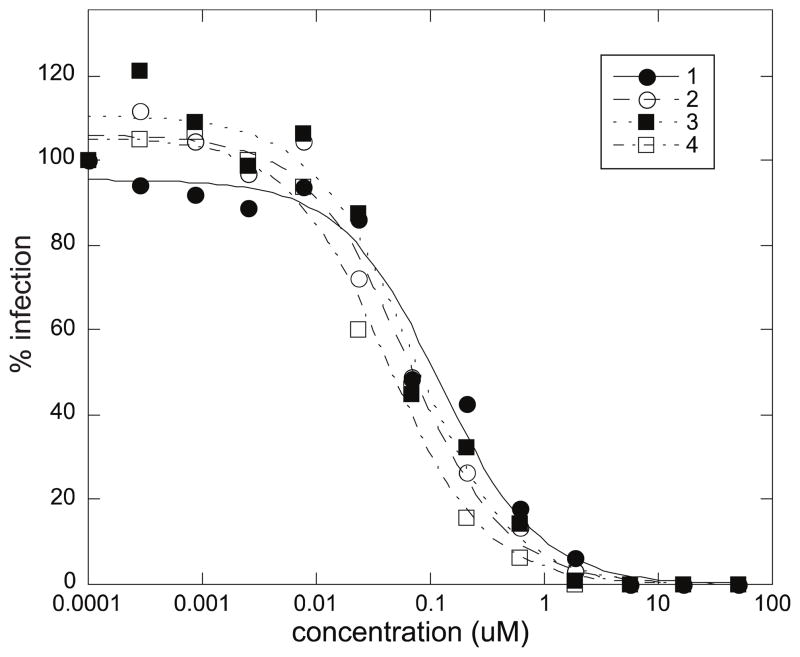
Compounds 1–4 are structurally similar to known HIV-1 entry inhibitors such as the papuamides6 and mirabamides A-D7. To test whether mirabamides E-H also possessed anti-HIV activity, 1–4 were tested in single round HIV-1 infectivity assays in parallel with the known HIV-1 inhibitor mirabamide C using methods described previously.7,19–20 All four depsipeptides were found to potently inhibit HIV-1 entry, with compound 4 being the most potent and displaying an IC50 value around 40 nM (Table 4). Compounds 2 and 3 (both at ~65 nM), followed by 1 and mirabamide C (both at ~120 nM) were slightly less potent.
Table 4.
IC50 Values (nM) for HIV-1 Neutralization Assays.
| Compound | IC50 (nM) |
|---|---|
| 1 | 121 ± 20 |
| 2 | 62 ± 9 |
| 3 | 68 ± 13 |
| 4 | 42 ± 5 |
| Mirabamide C | 123 ± 40 |
In comparing the HIV inhibitory activities of 1–4 with those previously determined for mirabamides A-D,7 some interesting trends emerge. The primary feature that distinguishes 1–4 from mirabamides A-D is the presence of 2-amino-2-butenoic acid in place of threonine. The assay results suggest that this change improves activity as evidenced by the two-fold increase in potency of 3 compared to mirabamide C whose structures are otherwise identical. Similarly, replacement of Dhtda with Htda also leads to a two-fold increase in potency as seen by comparing 1 to 2 and 3 to 4. By contrast, the conversion of 2,3-diaminobutanoic acid to 2-amino-2-butenoic acid led to a drastic decrease in potency for mirabamide B compared to mirabamide A. Overall these results suggest that increasing hydrophobicity in the side chain up to but not including Dab improves potency. A potential model that could account for this trend could involve insertion of the hydrophobic tail into the plasma membrane to a depth dictated by the presence of the polar head group (Dab). The role of the rhamnose residue in potency is less clear. For 1–4 the absence of rhamnose is correlated with improved activity (~ two-fold) in neutralization assays, whereas the absence of rhamnose was associated with an increase and a decrease in activity for mirabamide C vs. A and mirabamide D vs. papuamide A, respectively, in HIV-1 Envelope-mediated fusion assays.7
The isolation of mirabamides from phylogenetically distinct sponges (Siliquariaspongia mirabilis and Stelletta clavosa), and the fact that both sponges play host to diverse populations of bacteria, suggest a possible microbial origin for the mirabamides.
Experimental Section
General Experimental Procedures
Optical rotations were measured on a JASCO DIP-370 digital polarimeter. UV spectra were acquired in spectroscopy grade MeOH using a Hewlett-Packard 8452A diode array spectrophotometer. 1H and 13C NMR spectra were recorded on a Varian INOVA spectrometer operating at 500 MHz and 125 MHz, respectively. Chemical shifts are reported in ppm and were referenced to residual acetonitrile (δC 118.31; δH 1.94) in CD3CN-water (4:1). gCOSY, gHSQC, z-DIPSI-TOCSY, gHMBC and wgNOESY (τm = 200 ms) experiments were recorded using standard pulse sequence, all of which included water suppression. HMBC experiments were optimized for 2,3JC-H = 6 Hz. High-resolution ESIMS analyses were performed on a Bruker (Billerica, MA) APEXII FTICR mass spectrometer equipped with an actively shielded 9.4 T superconducting magnet (Magnex Scientific Ltd., UK), and an external Bruker APOLLO nanospray ESI source. Typically, a 5 μL sample was loaded into the nanoelectrospray tip (New Objective, Woburn, MA) with a potential of 1000 V applied between the nanoelectrospray tip and the capillary. Mass spectra were internally calibrated using HP tuning mix. LC-MS analyses were carried out using a Waters Micromass Q-tof micro integrated LC-MS system employing negative ion ESI mode with an ion source temperature of 130 °C, a desolvation temperature of 400 °C, and desolvation with nitrogen gas at a flow rate of 600 L/h. Analytical and semipreparative HPLC was accomplished utilizing a Beckman System Gold 126 solvent module equipped with a 168 PDA detector.
Sponge Material
Stelletta clavosa Ridley, 1884 was collected by epibenthic sled from inter-lagoon seabed areas in the Torres Strait, and frozen immediately after collection. The sample was identified by M. K. Harper. A voucher specimen is maintained at the Queensland Museum under accession number G329301.
Extraction and Isolation
The frozen sponge (365 g wet wt) was exhaustively extracted with MeOH to yield 7.3 g of extract. The extract was separated on HP20SS resin using a gradient of H2O to IPA in 25% steps, and a final wash of 100% MeOH, to yield five fractions. The third fraction (50/50 H2O/IPA) was further fractionated on Sephadex LH-20 with 1:1 CH3Cl: MeOH to give six fractions (Fr3.1–3.6). Fr3.2 was chromatographed by HPLC using a Phenomenex Luna C18 column (250 × 10 mm) employing 40% CH3CN/60% 0.2 M NaCl in H2O at 4 mL/min to yield compound 1 (7.0 mg, tR = 10.1 min), compound 2 (10.0 mg, tR = 12.8 min), compound 3 (10.0 mg, tR = 18.0 min), compound 4 (15.0 mg, tR = 23.9 min), and mirabamide C (3.5 mg, tR = 14.6 min) after desalting by C18 solid-phase extraction.
Mirabamide E (1): colorless, amorphous powder; [α]20 D –4 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 216 (4.30), 236 (4.41), 276 (3.51) nm; 1H and 13C NMR data Table 1; HRESIMS m/z 1578.7705 [M+H]+ (calcd for C72H113 35ClN13O24, 1578.7710).
Mirabamide F (2): colorless, amorphous powder; [α]20 D –6 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 218 (4.15), 236 (4.26), 276 (3.35) nm; 1H and 13C NMR data Tables 2 and 3; HRESIMS m/z 1562.7765 [M+H]+ (calcd for C72H113 35ClN13O23, 1562.7761).
Mirabamide G (3): colorless, amorphous powder; [α]20 D +13 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 218 (4.11), 236 (4.21), 276 (3.32) nm; 1H and 13C NMR data Tables 2 and 3; HRESIMS m/z 1432.7136 [M+H]+ (calcd for C66H103 35ClN13O20, 1432.7131).
Mirabamide H (4): colorless, amorphous powder; [α]20 D +12 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 218 (3.95), 236 (4.06), 276 (3.29) nm; 1H and 13C NMR data Tables 2 and 3; HRESIMS m/z 1416.7180 [M+H]+ (calcd for C66H103 35ClN13O19, 1416.7182).
Acid Hydrolysis of Peptides
Compounds 1–4, 300 μg each, were separately dissolved in degassed 6 N HCl (600 μL) and heated in sealed glass vials at 110 °C for 17 h. The solvent was removed in vacuo.
LC-MS Analysis of D/L-FDLA Derivatives14,15
The acid hydrolysates of 1–4 were dissolved in H2O (100 μL) separately. To a 50 μL aliquot of each were added 1 N NaHCO3 (20 μL) and 1% 1-fluoro-2,4-dinitrophenyl-5-L-leucinamide (L-FDLA or D/L-FDLA solution in acetone, 100 μL) and then heated to 40 °C for 50 min. The solutions were cooled to room temperature, neutralized with 1 N HCl (20 μL) and then dried in vacuo. The residues were dissolved in 1:1 CH3CN: H2O and then analyzed by LC-MS. The analysis of the L- and D/L-FDLA (mixture of D- and L-FDLA) derivatives was performed on a Supelcosil LC-18 column (150 × 4.6 mm, 5 μm) employing a linear gradient of 25% CH3CN/75% 0.01 M formic acid to 70% CH3CN/30% 0.01 M formic acid at 0.5 mL/min over 45 min. Based on the elution order of the D- and L-FDLA derivatives for each residue,14,15 it is possible to assign whether the configuration of the α-carbon is D or L: amino acids in which the L-FDLA analogue elutes first have an L configuration; those for which the D-FDLA analogue elutes first have a D configuration. The retention times of the D/L-FDLA mixtures (with the L-FDLA tR underlined) were: D- 3-OHLeu: 23.93, 29.94, m/z 440 [M-H]−; D-3-OMeAla: 23.10, 27.95, m/z 412 [M-H]−; L-NMeThr 20.63, 23.27, m/z 426 [M-H]−; L-Ala 24.25, 28.37, m/z 382 [M-H]−; L-ClHpr 30.30, 31.30, m/z 456 [M-H]−; 3,4-DiMe-L-Gln 23.41, 25.26 m/z 468 [M-H]−; L-Dab 37.02, 42.85 m/z 705 [M-H]−. Based on these determinations and analysis of the NMR data, the following absolute configurations were assigned: 2R,3R-OHLeu, 2R-3-OMeAla, 2S,3R-NMeThr, 2S-Ala, 2S,4S-ClHpr, 2S,3S,4R-DiMeGln, 2S,3S-Dab.
Absolute Configuration of Rhamnose
Compounds 1 and 2 (0.2 mg each) were separately dissolved in 1 N HCl (250 μL) and heated at 80 °C for 4h with stirring. After cooling, the solvent was removed in vacuo and the residue was dissolved in 1- (trimethylsilyl)imidazole (40 μL) and pyridine (160 μL). The solution was stirred at 60 °C for 20 min. The solvent was removed by blowing with nitrogen. The residue was partitioned with 1:1 CH2Cl2: H2O in 1 mL. The CH2Cl2 layer was analyzed by GC-MS. A 30 m × 0.25 mm ID Restek RT-bDEXm column was used for GC with the starting temperature at 75 °C and final temperature at 230 °C at a rate of 5 °C/min. A Waters GCT Premier time-of-flight mass spectrometer was employed for analysis. The retention time of the trimethylsilyl derivative of L-rhamnose standard was 14.69 min. The trimethylsilyl derivatives of L-rhamnose in the hydrolysates of 1 and 2 had retention times of 14.68 and 14.68 min, respectively.
HIV infectivity assays
Single round HIV-1 infectivity assays were performed as described previously19,20 using viruses pseudotyped with HIV-1 YU2-V3 Envelope, a CCR5-using strain. Briefly, virus stocks were added to wells in a 96-well plate format in the presence of buffer, vehicle, or increasing amounts of depsipeptide, followed by addition of TZM-bl target cells that constitutively express HIV receptors CD4 and CCR5. Plates were incubated at 37 °C, 5% CO2, for 18 h after which fresh media was added to each of the wells. Following incubation for 24 h, cells were lysed and luciferase activity was measured using a BrightGlo kit (Promega, Madison, WI) with a Synergy 2 luminescence plate reader (BioTek, Winooski, VT). Inhibition curves were fit to Equation 1 using Kaleidagraph software, and results normalized to positive and negative controls and plotted.
| (1) |
Supplementary Material
Figure 2.
Effects of mirabamides E-H in HIV-1 neutralization assays (standard deviations averaged 15%).
Acknowledgments
The authors wish to acknowledge Professor Y. Hamada, Chiba University, for providing standard 2S,3S,4R-dimethylglutamine and 2S,3S,4Sdimethylglutamine and Dr. P. Krishna and Dr. C. Nelson, University of Utah Mass Spectrometry and Proteomics Core Facility, for performing some of the HRESIMS experiments. The AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, is acknowledged for reagents and cell lines used in the HIV-1 neutralization assays. Funding for the Varian INOVA 500 MHz NMR spectrometer was provided through NIH grant RR06262. This work was supported by NIH grant CA36622 (C.M.I.) and the NIH Intramural AIDS Targeted Antiviral Program (C.A.B.).
Footnotes
Supporting Information Available: 1H NMR, 13C NMR, HMBC and wgNOESY spectra for compounds 1–4. Ion chromatograms of the D/L-FDLA derivatives and the LFDLA derivatives of the hydrolysis product of 1, papuamide A and mirabamide C in negative ion mode monitoring for specific amino acid residues. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Bewley CA, Faulkner DJ. Angew Chem Int Ed. 1998;37:2162–2178. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2162::AID-ANIE2162>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 2.Fusetani N, Matsunaga S. Chem Rev. 1993;93:1793–1806. [Google Scholar]
- 3.Shankar G, Andavan B, Lemmens-Gruber R. Mar Drugs. 2010;8:810–834. doi: 10.3390/md8030810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zampella A, D’Auria MV, Paloma LG, Casapullo A, Minale L, Debitus C, Henin Y. J Am Chem Soc. 1996;118:6202–6209. [Google Scholar]
- 5.Oku N, Gustafson KR, Cartner LK, Wilson JA, Shigematsu N, Hess S, Pannell LK, Boyd MR, McMahon JB. J Nat Prod. 2004;67:1407–1411. doi: 10.1021/np040003f. [DOI] [PubMed] [Google Scholar]
- 6.Ford PW, Gustafson KR, McKee TC, Shigematsu N, Maurizi LK, Pannell LK, Williams DE, de Silva ED, Lassota P, Allen TM, Van Soest R, Andersen RJ, Boyd MR. J Am Chem Soc. 1999;121:5899–5909. [Google Scholar]
- 7.Plaza A, Gustchina E, Baker HL, Kelly M, Bewley CA. J Nat Prod. 2007;70:1753–1760. doi: 10.1021/np070306k. [DOI] [PubMed] [Google Scholar]
- 8.Pitcher CR, et al. CSIRO/QM/QDPI CRC Torres Strait Task Final Report. CSIRO Marine and Atmospheric Research; Cleveland: 2007. Mapping and Characterisation of Key Biotic & Physical Attributes of the Torres Strait Ecosystem. CRC-TS Task Number T2.1; pp. 1–175. ( http://www.cmar.csiro.au/e-print/open/2007/pitchercr_a.pdf) [Google Scholar]
- 9.Erickson K, Gustafson KR, Pannell LK, Beutler JA, Boyd MR. J Nat Prod. 2002;65:1303–1306. doi: 10.1021/np020193z. [DOI] [PubMed] [Google Scholar]
- 10.Rao MR, Faulkner DJ. J Nat Prod. 2002;65:1201–1203. doi: 10.1021/np020040b. [DOI] [PubMed] [Google Scholar]
- 11.Fu X, Schmitz FJ, Kelly-Borges M, McCready TL, Holmes CFB. J Org Chem. 1998;63:7957–7963. [Google Scholar]
- 12.Erickson K, Gustafson KR, Milanowski DJ, Pannell LK, Klose JR, Boyd MR. Tetrahedron. 2003;59:10231–10238. [Google Scholar]
- 13.Wegerski CJ, France D, Cornell-Kennon S, Crews P. Bioorg Med Chem. 2004;12:5631–5637. doi: 10.1016/j.bmc.2004.07.061. [DOI] [PubMed] [Google Scholar]
- 14.Fujii K, Ikai Y, Mayumi T, Oka H, Suzuki M, Harada K. Anal Chem. 1997;69:3346–3352. [Google Scholar]
- 15.Fujii K, Ikai Y, Mayumi T, Oka H, Suzuki M, Harada K. Anal Chem. 1997;69:5146–5151. [Google Scholar]
- 16.Xie WQ, Ding D, Zi W, Li G, Ma D. Angew Chem Int Ed. 2008;47:2844–2848. doi: 10.1002/anie.200705557. [DOI] [PubMed] [Google Scholar]
- 17.Oku N, Krishnamoorthy R, Benson AG, Ferguson RL, Lipton MA, Phillips LR, Gustafson KR, McMahon JB. J Org Chem. 2005;70:6842–6847. doi: 10.1021/jo0508853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pederson AT, Andersen ØM, Aksnes DW, Nerdal W. Phytochem Anal. 1995;6:313–316. [Google Scholar]
- 19.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Svehla K, Mathy NL, Voss G, Mascola JR, Wyatt R. J Virol. 2006;80:1414–1426. doi: 10.1128/JVI.80.3.1414-1426.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.