Abstract
Background
Copy number variations (CNVs) can contribute to variable degrees of fitness and/or disease predisposition. Recent studies show that at least 1% of any given genome is copy number variable when compared to the human reference sequence assembly. Homozygous deletions (or CNV nulls) that are found in the normal population are of particular interest because they may serve to define non-essential genes in human biology.
Results
In a genomic screen investigating CNV in Autism Spectrum Disorders (ASDs) we detected a heterozygous deletion on chromosome 10p12.1, spanning the Patched-domain containing 3 (PTCHD3) gene, at a frequency of ~1.4% (6/427). This finding seemed interesting, given recent discoveries on the role of another Patched-domain containing gene (PTCHD1) in ASD. Screening of another 177 ASD probands yielded two additional heterozygous deletions bringing the frequency to 1.3% (8/604). The deletion was found at a frequency of ~0.73% (27/3,695) in combined control population from North America and Northern Europe predominately of European ancestry. Screening of the human genome diversity panel (HGDP-CEPH) covering worldwide populations yielded deletions in 7/1,043 unrelated individuals and those detected were confined to individuals of European/Mediterranean/Middle Eastern ancestry. Breakpoint mapping yielded an identical 102,624 bp deletion in all cases and controls tested, suggesting a common ancestral event. Interestingly, this CNV occurs at a break of synteny between humans and mouse. Considering all data, however, no significant association of these rare PTCHD3 deletions with ASD was observed. Notwithstanding, our RNA expression studies detected PTCHD3 in several tissues, and a novel shorter isoform for PTCHD3 was characterized. Expression in transfected COS-7 cells showed PTCHD3 isoforms colocalize with calnexin in the endoplasmic reticulum. The presence of a patched (Ptc) domain suggested a role for PTCHD3 in various biological processes mediated through the Hedgehog (Hh) signaling pathway. However, further investigation yielded one individual harboring a homozygous deletion (PTCHD3 null) without ASD or any other overt abnormal phenotype. Exon sequencing of PTCHD3 in other individuals with deletions revealed compound point mutations also resulting in a null state.
Conclusion
Our data suggests that PTCHD3 may be a non-essential gene in some humans and characterization of this novel CNV at 10p12.1 will facilitate population and disease studies.
Background
Unbalanced submicroscopic genetic variation, known as copy number variation (or CNV), is now well studied [1-4]. CNV can potentially contribute to variable degrees of fitness and/or disease predisposition [5-7]. In a recent high-resolution microarray study of CNV in humans, it was determined that at least 0.78% of the genome is CNV between any two individuals [4]. Analyses of the sequence assembly of a diploid genome at higher resolution determined that approximately 1.28% was CNV [8,9]. While CNVs, and in particular CNV deletions, are more often observed in gene poor regions [3,4], in the highest resolution population study to date, 3,811 of the validated biallelic deletions detected across 41 healthy individuals were found to overlap 1,432 genes [4].
Homozygous CNV deletions, herein called CNV nulls, affecting genes have previously been reported in the human genome [10]. Though loss of gene functions mostly reduce fitness, under certain conditions complete loss of functions can be beneficial [11,12]. For example, it is reported that homozygous mutations affecting DARC or CCR5 result in resistance to certain categories of malaria [13] or AIDS [14,15], respectively, without posing any overt deleterious effect on the individuals carrying these mutations.
Here, we discover and characterize a CNV null region in the human genome encompassing the PTCHD3 gene that does not appear to be associated with an overt phenotype. Breakpoint mapping indicated a recurrent 102,624 bp deletion, suggesting a single ancestral event that is now present in ~0.6-1.6% of individuals of European ancestry. Expression analyses showed PTCHD3 in several tissues with the highest levels detected in lymph node, testes and tongue. A previously uncharacterized shorter isoform was also found and PTCHD3 isoforms were determined to co-localize with calnexin in the endoplasmic reticulum in transfected cells.
Methods
Sample Sets
The Ethics Board of Hospital for Sick Children approved this study and all individuals providing samples signed informed consents covering aspects of the experiments conducted. Population control samples originated from other studies and they were consented and collected precisely for the aforesaid purpose (manuscripts citing the source of the population control samples are mentioned below).
Initial screening of PTCHD3 deletions used an index sample set of 427 probands diagnosed with ASD, all of which met the criteria by the Autism Diagnostic Interview-Revised (ADI-R) and Autism Diagnostic Observation Schedule (ADOS) [16]. We also screened another 177 cases from the same ASD population for the deletion using qPCR. Additional control cohort microarray data include 1,152 individuals from Ontario of predominately (95%) European ancestry genotyped on Affymetrix 500 K arrays [17], 1,123 Northern Europeans from the German PopGen project [18] genotyped on the Affymetrix 6.0 SNP array, and 1,234 individuals of European decent from the Ottawa River Valley [19] genotyped on the Affymetrix 6.0 SNP array. DNA panels used for quantitative PCR screening include 1,043 individuals from the HGDP-CEPH Human Genome Diversity Panel [20], and a panel of 186 North American Caucasians (Coriell Institute, USA). Ancestry of deletion carriers was either self reported or in the cases where microarray results were available, through inference using the SNP genotypes clustered with the HapMap samples as described previously [16].
Affymetrix GeneChip Human Mapping 500 K SNP and 6.0 Microarrays and CNV analysis
Affymetrix 500 K experiments and CNV analysis for the ASD cases and Ontario controls were carried out as described previously [16]. CNV analysis of Affymetrix 6.0 arrays is described elsewhere [21].
PTCHD3 Deletion Validation and Population Screening
For all cases where DNA was available, putative PTCHD3 deletions were validated with SYBR-Green I-based real time quantitative PCR (qPCR) using the CFTR locus as a reference as described previously [16]. The same assay was used to screen the 177 additional ASD probands, the 186 from the North American Caucasian Panel, and the 1,043 individuals from the HGDP-CEPH for PTCHD3 deletions.
Deletion Breakpoint Analysis and Exon Sequencing
Putative PTCHD3 deletions in ASD and HGDP-CEPH individuals were amplified with Stratagene Taq2000 polymerase and sequenced for breakpoint analysis. PCRs product sizes of either 3 kb (primers PCR01F and PCR01R) or 2 kb (primers PCR01F and SEQ03R) were used for sequencing using one or more of the primers SEQ01F, SEQ01R, SEQ02R, SEQ03R, and SEQ04R (see Additional file 1 for primer sequences). Sanger dideoxy-DNA sequencing and Exon-sequencing of the PTCHD3 (NM_001034842) in deletion carrier families was carried out using 12 different amplicons. Primer sequences and PCR conditions used for amplification are available on request.
Antibodies
Mouse monoclonal anti-Myc (sc-40); rabbit polyclonal anti-calnexin (sc-11397) and goat anti-mouse HRP-conjugated secondary antibody (sc-2005) were purchased from Santa Cruz biotechnology (USA). Alexa Fluor 488 goat antimouse IgG1 (A-21121) and Alexafluor 555 goat anti-rabbit IgG (A-21429) secondary antibodies were purchased from Invitrogen.
PTCHD3 Cloning and Transfection
Using the primer set C (Additional file 2) the full-length isoform of PTCHD3 was amplified from a pCR-Bunt II-TOPO plasmid containing the full length human PTCHD3. Primer set C was designed so that the amplicons would have BamH1 and Xba1 restriction sites at their 5' and 3' ends, respectively. For the shorter isoform of PTCHD3, cDNA from human lymph node was initially amplified using the primer set D designed based on sequences at/or around 5'UTR and 3'UTR regions of PTCHD3, and the ~2,000 bp product of this amplification was then used as a PCR template for primer set C. The ~1,600 bp amplified product of the latter then was used for cloning and further characterisation. The amplified long and short isoforms were then cloned into TOPO A vectors (Invitrogen) for further amplification in Top 10 competent cells (Invitrogen) following the instruction provided by the manufacturer. Finally, the long and short isoforms were both cloned into the BamH I-Xba I site of a pcDNA3 vector (Invitrogen) which had already been cloned with a c-Myc expressing motif at its Kpn I-BamH I sites. COS-7 cells were grown in DMEM medium containing 10% FBS. For western blot assay, the cells were transfected with the plasmids expressing PTCHD3 using Genejuice transfection reagent (Novagen) according to the instructions provided by the manufacturer. 48 hours later the cells were lysed in western blot lysis buffer and transferred to -20°C until used for further analysis. For immunocytochemistry, COS-7 cells were grown in chamber slides (Lab-Tek, Miles Laboratories) and transfected with the plasmids using Genejuice as explained above. 48 hrs later cells were used for immunolabelling as described below.
Western Blotting
Cell lysates were loaded on a 12% Polyacryamide gel and the resolved proteins were transferred to a nitrocellulose membrane following the general protocol for western blotting. The membranes were blocked in 5% milk for 1 hour and then were incubated with the 1:1,000 dilution of anti-Myc primary antibody in blocking buffer (5% milk) for another hour. After three washes of 10 minutes each, the membrane was incubated with 1:4,000 dilution of anti-mouse HRP-conjugated secondary antibody for 45 minutes. Membranes were washed three times, 10 minutes each and were developed using western blotting chemiluminescence reagents (PerkinElmer).
Immunocytochemistry
Cells were briefly washed with PBS and fixed in 4% formaldehyde at room temperature for 20 minutes. The fixed cells were permeablized in 0.1% triton (Sigma) in PBS at room temperature for 10 minutes, washed twice with PBS and were blocked in 10% Bovine Serum Albumin (BSA) (Sigma) in PBS at room temperature for 1 hour. The cells were then incubated in 1:100 dilution of anti-Myc and anti-calnexin primary antibodies in 3% BSA at room temperature for 1 hour. After 3 washes of 3 minutes each in PBS, the cells were incubated with 1 μg/ml of each of Alexa Fluor 488 goat antimouse IgG1 and Alexafluor 555 goat anti-rabbit IgG in 3% BSA for 45 minutes. After three washes in PBS, cells were studied using confocal microscopy.
Multiple Tissue Northern (MTN) Blot
The probe was amplified using primer set shown in Additional file 2, and the cDNA prepared from human lymph nodes. Probe labelling was conducted using 32P-labelled dCTP (PerkinElmer) following the protocol explained below. 1.2 μl of 0.1 U/μl random hexamers mix (GE Healthcare) was added to 7.2 μl of DNA probe (11 ng/μl), boiled in a water bath for 2 minutes and placed immediately on ice. 10 μl of 2.5× random priming buffer (0.5 M HEPES pH 6.6, 12.5 mM MgCl2, 28.8 mM β-mercaptoethanol, 125 mMTris pH 8.0 and 0.05 mM dATP/dGTP/dTTP mix) and 1 μl of 10× BSA (New England Biolabs Inc.) were added to the probe reaction and incubated at 22°C for 10 minutes followed by addition of 4 U of DNA Poly I Klenow fragment (USB corp.) and 5 μl (1.85 MBq) of [α32P]dCTP. The reaction was incubated at 22°C for 4 hours. 25 μl of TE with 0.1% SDS was added to the labelled probe and the unincorporated nucleotides were removed by centrifugation at 700 g for 2 minutes on a ProbeQuant G-50 Sephadex micro column (GE Healthcare). The flow through (labelled probe) was collected and the specific activity was measured using a QC-2000 reader (BioScan). The labelled probe was then boiled for 10 minutes and added to 5 ml of warm (68°C) buffer (clontech). The probe mixture was then added to a Multiple Tissue Northen blot membrane (Clonetch) that had been prehybridized in ExpressHyb buffer for 1 hour at 68°C. The hybridization proceeded at 68°C for 16 hours. The membrane was washed and exposed overnight against Biomax XAR autoradiograph film (Kodak) at -80°C and developed.
Results and Discussion
Discovery and Population Distribution of PTCHD3 Deletion
Using Affymetrix 500K arrays to investigate structural variation in individuals with Autism Spectrum Disorder (ASD), we detected a recurrent heterozygous deletion at chromosome 10p12.1. From our initial assessment the deletion was found at a frequency of ~1.4% (6/427) and was observed to intersect a single annotated gene, Patched-domain containing 3 (PTCHD3). In all ASD cases the deletion was found to be inherited, but nonetheless was determined to be an interesting candidate gene since it had not previously been described in control populations. Additionally, we and others recently described deletions in the X-linked PTCHD1, a PTCHD3 homologue, to be associated with ASD and intellectual disability [16,21-23]. We then tested an additional 177 ASD probands and found two more heterozygous deletions, bringing the total to 1.3% (8/604) (Table 1). All probands were of European ancestry. To determine the frequency in population controls, we initially used a qPCR assay to screen a panel of North American Caucasians and found the heterozygous deletion at a similar frequency of ~1.6% (3/186). Subsequent analysis of Affymetrix array data in three populations of predominately European ancestry yielded heterozygous PTCHD3 deletion frequencies of ~0.67% (8/1,152) [17], 0.36% (4/1,123) [18], and 0.97% (12/1,234) [19] (Table 1). Though the deletions were at slightly elevated frequency in ASD, we did not see a statistically significant association over controls when considering all data (p = 0.13; Fisher's 2-tailed exact test) (Table 1).
Table 1.
PTCHD3 deletions frequencies in Autism and control populations
| Population | Origin | Method | Total Number unrelated |
PTCHD3 del count | Frequency (%) | Reference |
|---|---|---|---|---|---|---|
| Autism | Canada | 500 K array | 427 | 6 | 1.40 | Marshall [16] |
| Canada | qPCR | 177 | 2 | 1.13 | unpublished | |
| Total | 604 | 8 | 1.32 | |||
| Control | Canada | 500 K array | 1,152 | 8 | 0.69 | Zogopolous [17] |
| Other | qPCR | 1,043 | 7 | 0.67 | HGDP | |
| N.America | qPCR | 186 | 3 | 1.61 | Coriell, USA | |
| Germany | Affy6.0 | 1,123 | 4 | 0.36 | Krawczak [18] | |
| Canada | Affy6.0 | 1,234 | 12 | 0.97 | Stewart [19] | |
| Total | 4,552 | 31 | 0.68 | |||
p-value for cases versus controls is not significant (Fisher's extact two tail; P = 0.13). 8/604 and 31/3695 + 648 = 4343 excluding the African, East Asian, and Oceania populations from the HDGP samples.
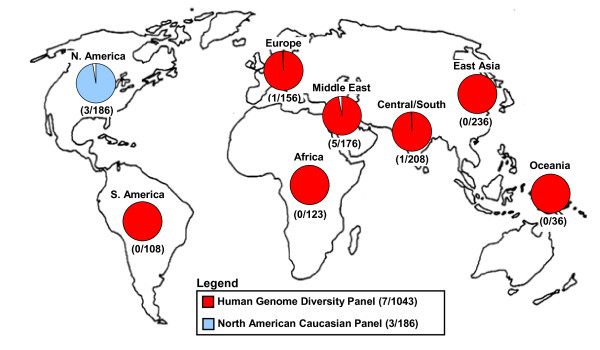
Although no obvious role for involvement in ASD was found, we sought to determine the population frequency and possible ancestral origin of the deletion by characterizing the human genome diversity panel (HGDP-CEPH), which is comprised of DNA samples from worldwide populations [20]. We found heterozygous PTCHD3 deletions in ~0.67% (7/1,043) individuals in the HGDP-CEPH (Figure 1) including three of Palestinian descent (from Israel Central), one of Druze descent (from Israel Carmel), one of Balochi descent (from Pakistan) and one of Northern Italian descent (from Italy Bergamo). Thus the deletion appeared to be present in those of Mediterranean/Middle East descent, suggesting this population as the likely origin of the ancestral CNV event. When we had sufficient family histories on the autism families to assess geneology, they were also found to have links to the Mediterranean/Middle East regions.
Figure 1.
Geographical population distribution of PTCHD3 deletions.
Breakpoint Characterization
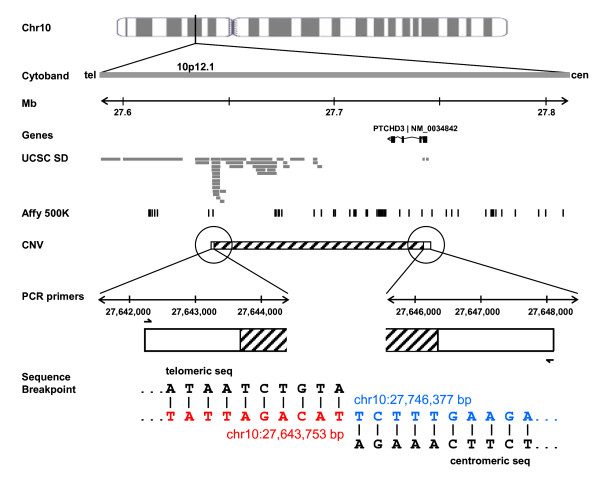
Using several PCR assays (Figure 2, Additional file 1) we defined and sequenced the breakpoints of the deletion, eventually determining it was 102,624 base pairs in size (spanning Chr.10: 27,643,753-Chr.10:27,746,377; NCBI Build 35). The breakpoint was found to be identical in the eight ASD probands and seven HGDP-CEPH individuals tested, suggesting a single ancestral event. The distal 10p12.1 breakpoint resides in a segmental duplication while the proximal end does not. Interestingly this PTCHD3 CNV region corresponds to a human-murine break of synteny with portions of murine chromosome 2 (inverted), chromosome 11 (inverted) and chromosome 18 mapping to human 10p12.1 [24] (Additional file 3).
Figure 2.
Genomic characterization of the PTCHD3 deletion variant. The cytoband and genomic coordinates (NCBI Build 35) are expanded from the chromosome 10 ideogram. The PTCHD3 gene (black), UCSC segmental duplications (grey bars), and Affymetrix 500 K SNP probes (black lines) are also shown. The deletion is denoted by a rectangle with hatched bars showing the sequenced breakpoints. Expansion of the region shows the location of primers (arrows) used for sequencing with the telomeric (red) and centromeric (blue) breakpoint sequences.
No Obvious Clinical Phenotype in PTCHD3 Nulls
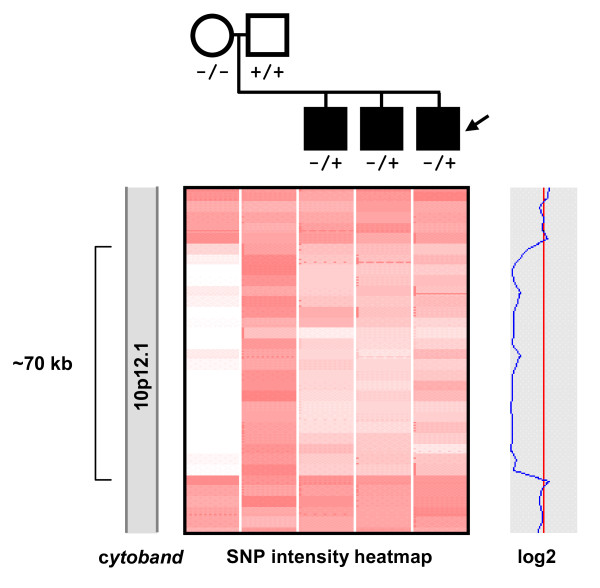
Of the eight ASD probands with deletions, we were able to determine maternal inheritance in four families (SK0191, SK0136, SK0257, MM0303) and paternal inheritance in two families (SK0145, MM0145). For two of the families, we did not have parental DNA. In one family, SK0191, all three offspring were found to carry the deletion and the mother was found to be homozygous for the deletion. We confirmed transmission by running the entire family (mother, father, proband, two affected siblings) on the Affymetrix 500 K array and subsequent PCR validation and breakpoint determination (Figure 3). The mother with the homozygous deletion has no apparent abnormal phenotype.
Figure 3.
Segregation of a homozygous PTCHD3 deletion. The '-' denotes the deleted allele and '+' the non-deleted wild type allele. Females are denoted by circles and males with squares. Individuals with a diagnosis of ASD are black with the proband denoted by an arrow. The heatmap profiles below show individual probe intensities covering the deleted region (top, telomeric end) with white, pink, and red signifying copy numbers of 0, 1, and 2, respectively. The blue line shows the smoothed log2 ratio for the mother.
To further characterize PTCHD3, we sequenced exons in the families harboring deletions (Additional file 4). We found two novel sequence variants that, in combination with the deletions, result in a null genotype. The father in family MM0145 has a deletion (CNV) on one allele compounded with a single base pair deletion at A1767. This single nucleotide deletion results in a frameshift introducing a stop codon at ~46 bp downstream of the deleted base suggesting no functional PTCHD3 copy in this individual. However, this nucleotide variant only affects the larger PTCHD3 isoform as the sequence harboring it is excluded from the novel shorter isoform we identified (see below). Family members from both SK0145 (father) and SK0136 (male proband) (Additional file 4) have CNV deletions and single base pair insertions at 923G. Again, this single nucleotide insertion, which in this case affects both the long and short PTCHD3 isoforms, results in a frameshift introducing a stop codon at ~75 bp downstream of the inserted base. Both fathers have no obvious clinical phenotype and have fathered children despite the suggested role of PTCHD3 in sperm motility [25], indicating PTCHD3 is not essential for fertility.
PTCHD3 Expression Profile and Functional Analysis
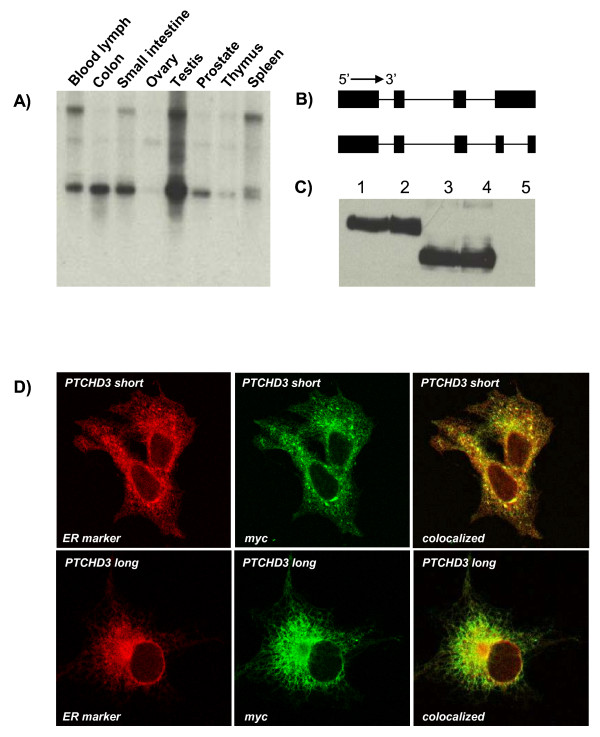
Due to the lack of an obvious phenotype in PTCHD3 nulls, we sought to further characterize the gene and rule out the possibility of PTCHD3 being a pseudogene. Using primer set A (Additional file 2) PTCHD3 expression was evaluated by PCR in cDNA panels of adult and fetal tissues. Although mouse Ptchd3 expression is testis-specific [25], the RT-PCR performed on a diverse panel of human cDNAs showed widespread expression in adult tissues (pancreas, placenta, salivary gland, skin, spleen, thymus, thyroid, trachea, bone marrow, brain, colon, heart, kidney, lung, lymph node, tongue, testis, ovary, spinal cord) and foetal sources (brain, bladder, kidney, lung, spleen and stomach) (data not shown). We did not detect expression of PTCHD3 in adult mammary gland, skeletal muscle, stomach, adrenal gland, cerebellum, fibroblasts, liver, uterus and foetal liver, skeletal muscle, thymus and aorta (data not shown). Expression appeared to be highest in adult lymph node, testes and tongue. Using a probe detecting both PTCHD3 isoforms, the PTCHD3 expression was further confirmed by northern blot on RNAs from 8 human tissues (Figure 4A). Despite apparent null genotypes in some individuals, PTCHD3 is expressed in human tissues.
Figure 4.
Expression and localization of PTCHD3 isoforms. A: A multiple tissue northern blot demonstrating the expression profile of PTCHD3 isoforms in different tissues. B: A schematic demonstration of two predominant isoforms of PTCHD3 showing truncation of the last exon of previously documented PTCHD3 isoform (coding sequence: 2,304 bps) in the newly identified isoform (coding sequence: 1,602 bps). C: Cloning and expression of PTCHD3 isoforms tagged with a Myc epitope in COS-7 cells resulted in detection (by western blot) of two proteins of ~115 KDa (lanes 1 and 2) and ~80 KDa (lanes 3 and 4); lane 5: run with the lysates prepared from untransfected cells. D: Both PTCHD3 isoforms are localised to the endoplasmic reticulum (ER). Immunostaining of COS-7 cells transfected with constructs expressing Myc-tagged short or longer isoforms of PTCHD3 using antibodies detecting calnexin (red) as an ER marker and Myc epitope (green) demonstrated co-localization of PTCHD3 isoforms with calnexin to the ER (Pearson coefficient 0.85).
Using primer set B (Additional file 2), expression in lymph nodes showed amplification of a long and a short isoform of PTCHD3 running on an agarose gel at ~2.3 kb and ~1.6 kb, respectively (Additional file 5). Both isoforms were cloned into and expressed from a pcDNA3 myc vector for further functional characterization. Plasmids containing either short (1,602 bp from, and including, start to stop codons) or long (2,304 bp from, and including, start to stop codons) PTCHD3 isoforms were transfected into COS-7 cell line resulting in the expression of a ~80 or ~115 kDa proteins, respectively, as determined by Western blotting (Figure 4C). Immunocytochemistry analysis using anti-Myc antibodies demonstrated co-localization of both of the PTCHD3 isoforms with calnexin to the endoplasmic reticulum (Pearson correlation coefficient 0.85) (Figure 4D). The sequence of the newly characterized PTCHD3 isoform has been submitted to the GenBank (Genbank accession number: JF332167).
Two Ptchd3 isoforms have been detected in mouse, Ptchd3a (AK017136) (coding for 410 amino acids) and Ptchd3b (AB235902) (coding for 906 amino acids), of which the first encoded 409 amino acids are identical [25]. In mice, Ptchd3 expression is developmentally regulated and detected exclusively in testes [25]. Human PTCHD3 shares ~63% amino acids identity with its mouse orthologue and it has been shown that in both species PTCHD3 is expressed and localized to the midpiece of sperm, suggesting possible involvement of PTCHD3 in sperm motility and hence fertility [25]. However, we report healthy offspring fathered from males in which both germline PTCHD3 alleles are apparently inactive. It is possible that PTCHD3 can improve fecundity, but its absence may not always be accompanied by infertility.
The hedgehog (Hh) signaling pathway (including Hh protein and its receptor Ptc) has important roles in embryonal patterning and development in both vertebrates and invertebrates [26]. PTCHD3 has Ptc and Sterol Sensing (SSD) domains suggesting a possible role for PTCHD3 in hedgehog signaling. Although, we cannot exclude the possibility of PTCHD3 having an important function in humans, our observation indicate that the absence of PTCHD3 has no overt effect on normal development. Nonetheless, whether there are some other molecules compensating for PTCHD3 loss, or whether this molecule might have beneficial effects under certain conditions remains to be determined. It is noteworthy that different individuals with the same heterozygous deletions, e.g. 16p11.2 microdeletions [27-30], can present a spectrum of phenotypes. In another example, a sib pair was recently reported to both have large homozygous chromosomal deletion (812-902 kb) at chromosome 12q21.1, but while one sib demonstrated dysmorphic features and developmental delay, the other child was unaffected [31]. The variable penetrance may arise due to unshared genes between the sibs, highlighting the need to interpret CNV data in a genome-wide context [7,32].
Conclusion
We identified a novel 102.6 kb CNV ancestral deletion on chromosome 10p12.1 that is predominantly present in European/Middle Eastern populations and encompasses the PTCHD3 gene. A novel shorter ~1.6 kb isoform of PTCHD3 (Genbank accession number: JF332167) was also characterized. Expression studies revealed that both the long and short PTCHD3 isoforms co-localize with calnexin to the endoplasmic reticulum. We demonstrate that a PTCHD3 null state can exist in humans through homozygous deletions, or combinations of deletions and single nucleotide mutations, with no overt abnormal phenotype being associated. Our data helps further define the essential human gene set.
List of Abbreviations
CNV: Copy Number Variation; ASD: Autism Spectrum Disorders; MTN: Multiple Tissue Northern Blot; Hh: Hedghog; HGDP: Human Genome Diversity Panel; BSA: Bovine Serum Albumin.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MMGHS, BK, KKML, RM, ACL, and CRM performed laboratory and computational analyses. MMGHS, RM, CRM and SWS designed the study, interpreted the data and wrote the manuscript. All authors read and approved the manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
A Table listing the primers used for breakpoint mapping.
A table listing primers used for RT-PCR, cloning and northern blot.
A figure depicting murine break of synteny at ptchd3 region versus human.
Pedigrees demonstrating compounded PTCHD3 mutations complexion in families with individuals affected with ASD.
A figure depicting PTCHD3 expression in human lymph node (L.N.) detected by RT-PCR.
Contributor Information
Mohammad M Ghahramani Seno, Email: mgseno@yahoo.com.
Benjamin YM Kwan, Email: BKWAN066@uottawa.ca.
Ka Ki M Lee-Ng, Email: mich.leeng@gmail.com.
Rainald Moessner, Email: rainald.moessner@ukb.uni-bonn.de.
Anath C Lionel, Email: anath.lionel@utoronto.ca.
Christian R Marshall, Email: crm@sickkids.ca.
Stephen W Scherer, Email: stephen.scherer@sickkids.ca.
Acknowledgements
This work is supported by The Centre for Applied Genomics, Genome Canada/Ontario Genomics Institute, the Canadian Institutes of Health Research (CIHR), the McLaughlin Centre, the Canadian Institute of Advanced Research, the Hospital for Sick Children (SickKids) Foundation, and the Autism Research Training Program. MMGHS is supported by a fellowship from Autism Speaks, USA. S.W.S holds the GlaxoSmithKline-CIHR Pathfinder Chair in Genetics and Genomics at the University of Toronto and the Hospital for Sick Children (Canada).
References
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y. et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P. et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD. et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y. et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Beckmann JS, Estivill X, Antonarakis SE. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet. 2007;8:639–646. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
- Lee C, Scherer SW. The clinical context of copy number variation in the human genome. Expert Rev Mol Med. 2010;12:e8. doi: 10.1017/S1462399410001390. [DOI] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP. et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang AW, Macdonald JR, Pinto D, Wei J, Rafiq MA, Conrad DF. et al. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 2010;11:R52. doi: 10.1186/gb-2010-11-5-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC. et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- Olson MV. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Grus WE, Zhang J. Gene losses during human origins. PLoS Biol. 2006;4:e52. doi: 10.1371/journal.pbio.0040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournamille C, Colin Y, Cartron JP, Le Van KC. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM. et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- Zimmerman PA, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C. et al. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J. et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogopoulos G, Ha KC, Naqib F, Moore S, Kim H, Montpetit A. et al. Germ-line DNA copy number variation frequencies in a large North American population. Hum Genet. 2007;122:345–353. doi: 10.1007/s00439-007-0404-5. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Nikolaus S, von Eberstein H, Croucher PJ, El Mokhtari NE, Schreiber S. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- Stewart AF, Dandona S, Chen L, Assogba O, Belanger M, Ewart G. et al. Kinesin family member 6 variant Trp719Arg does not associate with angiographically defined coronary artery disease in the Ottawa Heart Genomics Study. J Am Coll Cardiol. 2009;53:1471–1472. doi: 10.1016/j.jacc.2008.12.051. [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L. et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Noor A, Whibley A, Marshall CR, Gianakopoulos PJ, Piton A, Carson AR. et al. Disruption at the PTCHD1 Locus on Xp22.11 in Autism Spectrum Disorder and Intellectual Disability. Sci Transl Med. 2010;2:49ra68. doi: 10.1126/scitranslmed.3001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whibley AC, Plagnol V, Tarpey PS, Abidi F, Fullston T, Choma MK. et al. Fine-scale survey of X chromosome copy number variants and indels underlying intellectual disability. Am J Hum Genet. 2010;87:173–188. doi: 10.1016/j.ajhg.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengol L, Pujana MA, Cheung J, Scherer SW, Estivill X. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum Mol Genet. 2003;12:2201–2208. doi: 10.1093/hmg/ddg223. [DOI] [PubMed] [Google Scholar]
- Fan J, Akabane H, Zheng X, Zhou X, Zhang L, Liu Q. et al. Male germ cell-specific expression of a novel Patched-domain containing gene Ptchd3. Biochem Biophys Res Commun. 2007;363:757–761. doi: 10.1016/j.bbrc.2007.09.047. [DOI] [PubMed] [Google Scholar]
- Alcedo J, Noll M. Hedgehog and its patched-smoothened receptor complex: a novel signalling mechanism at the cell surface. Biol Chem. 1997;378:583–590. doi: 10.1515/bchm.1997.378.7.583. [DOI] [PubMed] [Google Scholar]
- Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K. et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez BA, Roberts W, Chung B, Weksberg R, Meyn S, Szatmari P. et al. Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J Med Genet. 2010;47:195–203. doi: 10.1136/jmg.2009.069369. [DOI] [PubMed] [Google Scholar]
- Sampson MG, Coughlin CR, Kaplan P, Conlin LK, Meyers KE, Zackai EH, Evidence for a recurrent microdeletion at chromosome 16p11.2 associated with congenital anomalies of the kidney and urinary tract (CAKUT) and Hirschsprung disease. Am J Med Genet A. 2010. [DOI] [PubMed]
- Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, Andersson J. et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry CJ, Mao R, Aston E, Mongia SK, Treisman T, Procter M. et al. Homozygous deletions of a copy number change detected by array CGH: a new cause for mental retardation? Am J Med Genet A. 2008;146A:1903–1910. doi: 10.1002/ajmg.a.32450. [DOI] [PubMed] [Google Scholar]
- Ali-Khan SE, Daar AS, Shuman C, Ray PN, Scherer SW. Whole genome scanning: resolving clinical diagnosis and management amidst complex data. Pediatr Res. 2009;66:357–363. doi: 10.1203/PDR.0b013e3181b0cbd8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A Table listing the primers used for breakpoint mapping.
A table listing primers used for RT-PCR, cloning and northern blot.
A figure depicting murine break of synteny at ptchd3 region versus human.
Pedigrees demonstrating compounded PTCHD3 mutations complexion in families with individuals affected with ASD.
A figure depicting PTCHD3 expression in human lymph node (L.N.) detected by RT-PCR.