Abstract
The ribosome converts genetic information into protein by selecting aminoacyl-tRNAs whose anticodons base pair to an mRNA codon. Mutations in the tRNA body can perturb this process and affect fidelity. The Hirsh suppressor is a well-studied tRNATrp harboring a G24A mutation that allows readthrough of UGA stop codons. Here we present crystal structures of the 70S ribosome complexed with EF-Tu and aminoacyl tRNA (native tRNATrp, G24A tRNATrp, or the miscoding A9C tRNATrp) bound to cognate UGG or near-cognate UGA codons, determined at 3.2 Å resolution. The A9C and G24A mutations lead to miscoding by facilitating the distortion of tRNA required for decoding. A9C accomplishes this by increasing tRNA flexibility, while G24A allows the formation of an additional hydrogen bond that stabilizes the distortion. Our results also suggest that each native tRNA will adopt a unique conformation when delivered to the ribosome that allows accurate decoding.
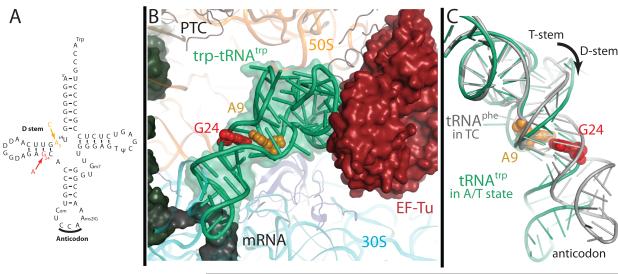
The elucidation of the genetic code in the 1960s revealed that each three-nucleotide mRNA sequence coded for a particular amino acid, and that this code determines the primary sequence of proteins. It was discovered that these codons are recognized by a complementary tRNA anticodon, which is covalently linked to the appropriate amino acid. Though tRNAs were initially assumed to be inert adaptor molecules, the discovery that a mutation (G24A) in the D-stem of tRNATrp allows suppression of UGA stop codons changed this view. G24 is far from the anticodon (Fig. 1A), so this mutation demonstrated that properties of the tRNA body are also critical for decoding1. G24 pairs with U11, so it is even more puzzling why its mutation to A24, which could form a canonical base pair with U11, would affect fidelity. Also intriguingly, the strength of the 24-11 pair does not correlate with miscoding efficiency 2.
Figure 1.
Overview of miscoding mutations and Trp-tRNATrp bound in the A/T state. A) The traditional cloverleaf diagram of Trp-tRNATrp showing the locations of the miscoding mutations A9C (orange) and G24A (red). B) When bound to the ribosome along with EF-Tu, the aminoacyl-tRNA (green) adopts the distorted A/T conformation. Structures were also determined for Trp-tRNATrp containing mutations at position 24 (red) and 9 (orange). C) The A/T conformation requires a ~30° bend in the tRNA body (green) compared to a canonical tRNA (grey)19. This bend is achieved through two isolated regions of distortion, first in the anticodon stem and the second in the D-stem, where both the A9C and G24A mutations are located.
Since the identification of this so-called Hirsh suppressor tRNA (from here on referred to as G24A tRNATrp), several other mutations in tRNATrp outside the anticodon have been shown to lead to miscoding, including A9C 3-5. Four decades of biochemical and kinetic experiments have attempted to explain in detail how such mutations in the tRNA body cause miscoding 2,3,5-7. These studies have led to many conflicting theories, including that the G24A mutation alters the tRNA flexibility 67, changes internal base pairing in the tRNA2, or allows additional interactions between the tRNA and the ribosome5,7. It is likely that these mutations affect the ability of tRNA to adopt the distorted A/T conformation during decoding, which allows the tRNA to continue to interact with EF-Tu while base pairing with the codon at the decoding center. However detailed structures of these mutant tRNAs trapped during decoding on the ribosome are needed to provide a molecular understanding of their loss of fidelity. This information will also provide insight into the native mechanisms employed by the translation machinery to ensure accurate decoding.
Native tRNATrp recognizes only its cognate UGG codon efficiently. However both A9C and G24A mutations in tRNATrp allows the aberrant recognition of the near-cognate UGA stop codon. Discrimination between UGA and UGG requires differentiation between an A and G at the wobble position, where Watson-Crick base pairing is not stringently monitored8. tRNATrp is the only elongator tRNA that discriminates between purines in the wobble position without the aid of a modification at residue 34 9 and UGA has a read-through rate as high as 10−2, compared to the more typical ribosomal error rate of 10−3−10−4 10,11. Furthermore, G24A and A9C tRNATrp are ~8 and 18-fold better at recognizing UGA than wild type tRNATrp 2. Additionally, although GTP hydrolysis by EF-Tu should only be efficiently triggered by binding to a cognate codon, the rate of GTPase activation is 5-fold faster with G24A tRNATrp on a non-cognate codon when compared to wild type tRNA7. The rate of GTPase activation by G24A tRNA on a stop codon is only 2-fold less than that of native tRNATrp on its cognate codon. Therefore, the energetic difference between binding of the G24A and native tRNATrp to a near-cognate codon is on the order of 4 kJ/mol, approximately the energy of a single additional hydrogen bond under solvated conditions 12.
During decoding, the combined energetic penalty of inducing the conformational changes required in the 30S subunit, the tRNA (A/T state), and EF-Tu is precisely balanced against the energy derived from cognate tRNA binding. This includes contributions from the interaction of 16S rRNA nucleotides A1492, A1493, and G530 with the minor groove of the codon-anticodon helix. Mutations in the tRNA body could cause miscoding through two possible mechanisms, i) by increasing the flexibility of the tRNA body or ii) by forming stabilizing interactions in the A/T state. Both strategies lower the energetic penalty for adoption of the A/T conformation that is required for GTPase activation. With the goal of understanding the error-prone phenotypes of both the G24A and A9C tRNATrp we have solved four crystal structures of aminoacyl-tRNAs bound with EF-Tu, to the 70S ribosome from Thermus thermophilus, stabilized by the antibiotic kirromycin. These four structures contain: wild type Trp-tRNATrp bound to the cognate UGG codon, G24A Trp-tRNATrp bound to UGG, G24A Trp-tRNATrp bound to the near-cognate UGA stop codon, and A9C Trp-tRNATrp bound to UGA (Fig. 1, Table 1). The structures reveal the detailed mechanisms by which the G24A and A9C mutations lead to miscoding. Furthermore, a comparison with the structure of 70S - EF-Tu - Thr-tRNAThr demonstrates how different tRNAs use distinct conformations to productively bind to the ribosome and thereby ensure accurate decoding.
Table 1.
Summary of crystallographic data and refinement
| Data collection | 70S-EF-Tu trp-tRNATrp (merged from 5 crystals) |
70S-EF-Tu G24A trp- tRNATrpUGG (from 2 crystals) |
70S-EF-Tu G24A trp- tRNATrpUGA (from 2 crystals) |
70S-EF-Tu A9C trp- tRNATrpUGA (from 2 crystals) |
|---|---|---|---|---|
| Space Group | P21 | P21 | P21 | P21 |
| Cell dimensions | ||||
| a, b, c (Å) |
a=290.2 b=269.2 c=404.0 |
a=289.8 b=269.1 c=404.0 |
a=289.9 b=269.4 c=404.5 |
a=289.9 b=268.5 c=403.6 |
| α, β, γ (°) | α=90.0 β=91.5 γ=90.0 |
α=90.0 β=91.2 γ=90.0 |
α=90.0 β=91.5 γ=90.0 |
α=90.0 β=91.6 γ=90.0 |
| Resolution (Å) | 50-3.1 (3.2-3.1) * | 50-3.1 (3.2-3.1) † | 50-3.1 (3.2-3.1) § | 50-3.1 (3.2-3.1) ‡ |
| Rsym | 19.9 (115.2) | 20.4 (100.9) | 26.8 (130.1) | 21.5 (103.3) |
| I/σI | 7.12 (1.24) * | 7.27 (1.35) † | 6.19 (1.23) § | 10.48 (1.69) ‡ |
| Completeness (%) |
99.8 (99.9) | 97.4 (92.7) | 98.7 (98.5) | 97.9 (93.2) |
| Redundancy | 6.3 (5.2) | 4.6 (4.1) | 6.4 (6.5) | 5.0 (4.4) |
| Refinement | ||||
| Resolution (Å) | 50.0-3.1 | 50.0-3.1 | 50.0-3.1 | 50.0-3.1 |
| No. unique reflections |
1,116,606 | 1,087,126 | 1,105,502 | 1,090,696 |
| Rwork/Rfree | 23.7/26.4 | 24.7/28.5 | 23.8/27.5 | 24.3/26.7 |
| No. atoms | ||||
| RNA | 205,448 | 205,202 | 205,200 | 205,442 |
| Protein | 100,888 | 100,888 | 100,888 | 100,888 |
| B-factors | ||||
| RNA | 99 | 82 | 90 | 76 |
| Protein | 103 | 86 | 92 | 81 |
| R.m.s deviations |
||||
| Bond lengths (Å) |
0.008 | 0.008 | 0.008 | 0.007 |
| Bond angles (°) | 1.2 | 1.3 | 1.3 | 1.2 |
I/σI =1.97 at 3.2 Å resolution
I/σI =2.08 at 3.2 Å resolution
I/σI =1.92 at 3.2 Å resolution
I/σI =2.58 at 3.2 Å resolution
Results
The overall conformation of ribosomal decoding complexes
Codon selection by the ribosome triggers GTP hydrolysis, therefore a ribosomal complex stalled at the point of GTP hydrolysis would provide the best structural insight into decoding. Comparison of the structures of the ternary complex on the ribosome directly before (stalled by a GTP analog) 13 and after (stalled by kirromycin, this study) GTP hydrolysis reveals an identical tRNA conformation (Supplemental Fig. S1). Therefore although the kirromycin-stalled structures presented here represent post-hydrolysis complexes, the observed conformations of the tRNA body are indeed relevant for understanding how the G24A and A9C mutations affect tRNA selection. All complexes reported here have Watson-Crick base pairs in the first two positions of the codon-anticodon helix, and thus the 16S rRNA residues A1492-3 and G530, which monitor these positions, are flipped out, and the 30S subunit is in the “closed” conformation. The conformational changes predicted to communicate codon matching from the ribosome to EF-Tu are observed, though some movements are less pronounced 14. Interestingly, the G24A and A9C mutations do not appear to alter the interactions between tRNATrp and the ribosome, in contrast to a recent hypothesis 5,7. Despite these similarities, changes at the anticodon are observed, as are functionally important differences in the tRNA body, which suggest a solution to these decades-old miscoding puzzles.
The decoding center
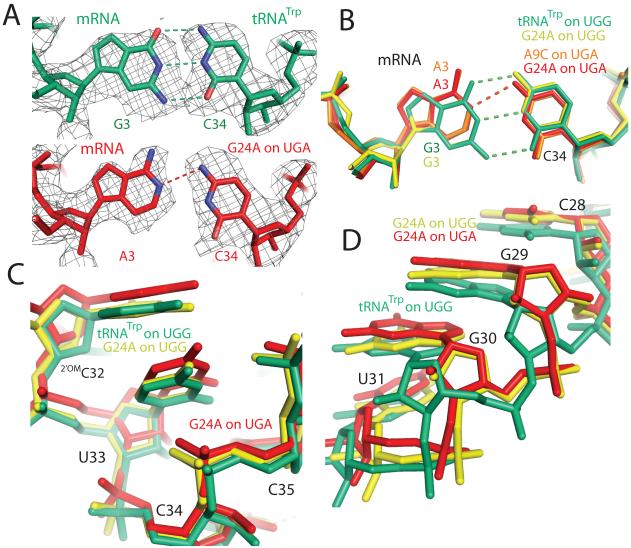
For both the G24A and A9C tRNAs bound to the UGA stop codon, a clear C34•A3 mismatch is observed at the wobble position (Fig. 2a, b). Formation of this hydrogen bonding interaction results in shifts in both the tRNA anticodon and mRNA codon. This distortion is propagated along the tRNA body through residue 31 (Fig. 2c, d).
Figure 2.
Comparison of cognate and near-cognate structures in the decoding center. A) Unbiased Fo - Fc electron density (displayed at 1.3σ within 2 Å of EF-Tu) is shown for residues in the wobble position of the codon-anticodon helix. B) Binding of a near-cognate tRNA (A9C on UGA: orange, G24A on UGA: red), compared to a cognate tRNA (G24A on UGG: yellow, tRNATrp on UGG: green), results in a shift in both the tRNA anticodon and mRNA codon. C) The distortion in the anticodon caused by this mismatch is propagated three residues, resulting in a shift in the tRNA backbone for the G24A on UGA when compared to the G24A or tRNATrp on UGG. D) However this distortion does not continue past residue 31; by residue 28 the backbone of the G24A on UGA has converged with that of the G24A on UGG.
Wild type and mutant tRNATrp in the A/T conformation
Outside the decoding center, the structure of Trp-tRNATrp is distorted to maintain interactions with EF-Tu (Fig. 1b, c) 14. The bodies of the wild type and mutant tRNAs Trp-tRNATrp are in a very similar conformation compared to one another, and to Thr-tRNAThr 14. However in all Trp-tRNATrp studied here, the phosphodiester backbone of residues 44-45 is positioned tightly into the tRNA body, near residues 25-26 (Fig. 3).
Figure 3.
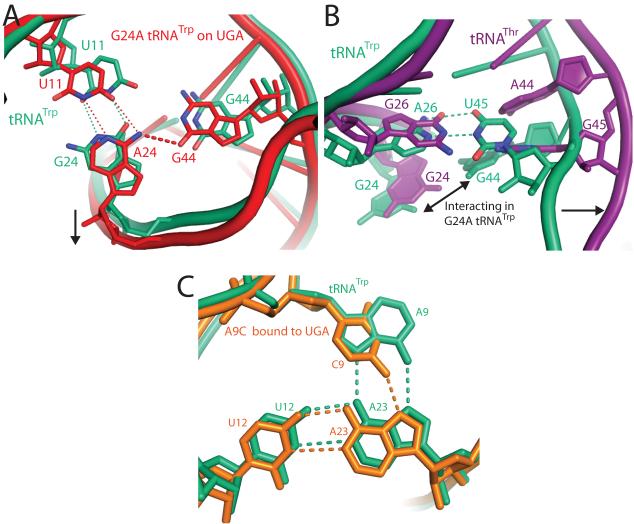
Miscoding by the G24A and A9C Trp-tRNATrp. A) Mutation of G24A (red) facilitates formation of a hydrogen bonding interaction that is not possible in native tRNATrp (green) between the exocylic amine of A24 and O6 of G44. This interaction is facilitated by a small shift in the RNA backbone between residues 20 to 30 and a commensurate movement between residues 9-16, B) The interaction between residues 24 and 44 in the G24A tRNATrp is only possible because of a base pair between residues A26-U45 in tRNATrp that dictates the backbone conformation in this region. In contrast, tRNAThr14(purple) contains two purines, G26 and G45, at these positions, which push the tRNA backbone apart and separate residues G24 and A44. That the A26-U45 base pair is not conserved even throughout bacterial tRNATrp further illustrates that evolution has fine-tuned each tRNA to find a unique solution to tRNA bending during decoding. (C) In native tRNATrp (shown in green) a base triple forms between residues 9:12:23, which is at the junction of three separate strands of the tRNA. Mutation of residue 9 to a cytosine (shown in orange) weakens both the packing and hydrogen-bonding of this base-triple, which could result in higher flexibility of the tRNA body and explain its ability to miscode.
The structures of G24A tRNATrp bound to both the UGG and UGA codons indicate that the mutation at residue 24 allows formation of an additional hydrogen bond in the A/T conformation. In both structures, the N6 of nucleotide A24, which is not present in the wild type G24, is within hydrogen bonding distance of the exocyclic O6 of G44 (Fig. 3a).
Conversely, the structure of A9C tRNATrp bound to a UGA codon show that this mutation disrupts hydrogen bonding in the A/T conformation. In wild type tRNATrp a base triple is formed between residues A9-A23-U12. When residue A9 is mutated to cytosine this base triple is weaker, as a cytidine is unable to form the two hydrogen bonds provided by adenosine (Fig. 3c).
Discussion
Conformational changes in the decoding center
One striking difference between the four complexes is at the decoding center where a C34•A3 mismatch is observed in the wobble position for both the G24A and A9C tRNAs bound to the UGA stop codon (Fig. 2a, b). In both cases, C34 in the tRNA anticodon and A3 in the mRNA codon each shift to facilitate this interaction. This is in contrast to previous studies where the mRNA remained stationary while interacting with several cognate anticodons15. The distortion at the anticodon required for binding near-cognate UGA is propagated three residues, causing a shift in the G24A and A9C tRNAs between residues 34-31 (Fig. 2c). Interestingly, mutations in this region are known to cause miscoding16, perhaps by altering the anticodon stem to allow binding to a mismatched codon. However, this movement is not communicated to the rest of the tRNA, as after residue 31 the structures of the G24A tRNA in the near-cognate and cognate complexes converge. Therefore the error-prone phenotype of the G24A and A9C tRNAs is not caused by a distortion propagated from the anticodon, but rather is a direct effect of these mutations on the tRNA body. This is consistent with kinetic data that shows a similar increase in the rate of GTPase activation for G24A tRNATrp compared to wild-type tRNATrp independent of the presence of mRNA or identity of the near-cognate codon7.
The mechanism for miscoding by G24A tRNATrp
Inspection of the body of G24A tRNATrp reveals that the G24A mutation promotes miscoding by preferentially stabilizing the distorted conformation through formation of an additional internal interaction. The mutation introduces an exocyclic amine at ring position 6 of nucleotide 24, which, in this distorted A/T state, is within hydrogen bonding distance of the exocyclic O6 of G44 (Fig. 3a). This hydrogen bond, which would not be possible in wild-type tRNATrp, or in the unbent G24A tRNATrp, could stabilize the bent form of the G24A tRNATrp. This mutant tRNA is therefore more easily able to adopt the A/T conformation than its wild-type counterpart, and thus more easily misreads the code. This 24-44 interaction could only form if residue 24 contains an exocyclic amine at position 6, consistent with the fact that mutations at residues 24 and 11 only cause miscoding when G24 is mutated to an A or C2.
It is therefore tempting to predict that in all bacterial species native tRNATrp would either not contain an A or C at position 24, or be unable to form a hydrogen bond between residues 24 and 44. Indeed of ~560 bacterial tRNATrp sequences, 97.5% would not be able to make this 24-44 hydrogen bond 17. However the remaining few bacterial tRNATrp that do contain an A or C at position 24 and a G at position 44 presumably do not miscode. These tRNAs have likely arrived at an energetic balance that includes this hydrogen bond, or have positioned residue 44 away from 24 in the A/T conformation.
The structural cause of miscoding by G24A tRNA was not predicted by the many previous biochemical and structural studies, perhaps because nucleotides 44 and 24 are far apart in unbent tRNAs (7.2 Å in yeast tRNAPhe18, Supplemental Fig. S2), when bound to EF-Tu (7.8 Å19) and even in the complex of distorted tRNAThr bound to the ribosome (8.2 Å14, Fig. 3b). The interaction is possible only in the bent form of G24A tRNATrp because of a base pair between residues A26 and U45, which brings the strands containing G44 and A24 together when bound to the ribosome (Fig. 3b). Direct observation of this interaction also explains why the strength of the 24:11 base pair does not correlate with miscoding efficiency2, an observation that discounted the simple explanation that the G24A mutation increases tRNA flexibility, consistent with recent biochemical work 5.
tRNA identity affects the details of tRNA conformation in the A/T state
In our previously reported structure of 70S - EF-Tu - Thr-tRNAThr, the tRNAThr contains guanosines at both positions 26 and 45. Comparing this structure with the structure containing Trp-tRNATrp, one of the most obvious differences is at nucleotides 44-45, which in Thr-tRNAThr are pushed away from the tRNA body, while in Trp-tRNATrp are drawn in by more than 5 Å due to the contact between 45 and 26 (Fig. 3b). The overall bends in the two tRNAs are extremely similar and the respective positions of the tRNA-EF-Tu interface are within approximately 1 Å of each other. To accommodate a larger amino acid, the backbone of EF-Tu around the amino acid binding pocket, and the 3′ end of the Trp-tRNATrp have shifted very slightly. These small differences, seen both in the structures reported here and the ribosome - EF-Tu - GDPCP- Trp-tRNATrp structure 13, are compensated for by slight rotations in EF-Tu of domain 1 with respect to domains 2 and 3 (Supplemental Fig S3), allowing EF-Tu to interact with the ribosome in the same way independent of tRNA identity. Similar small differences in the conformation of tRNA and EF-Tu during decoding have been reported from low-resolution studies 20. These subtle differences in tRNA conformation on the ribosome are understandable given that tRNAs have differences in sequence and structure that are required for fidelity of other processes such as aminoacylation.
Miscoding by A9C tRNATrp
From the structure of A9C tRNA, it seems evident that its error-prone phenotype is due to enhanced flexibility of the tRNA, which facilitates formation of the A/T conformation. Adenosine 9 in tRNATrp forms a trans Hoogsteen-Hoogsteen base pair21 with A23 of the A23-U12 pair in the D-stem. This base triple is at the junction between three separate strands of the tRNA, and is within the upper region of distortion of the A/T conformation. Mutation of A9 to cytosine weakens this triple, which leads to the destabilization of the tRNA (Fig. 3c). Indeed, the electron density for regions of this tRNA body is significantly weaker when compared to that for other structures, or even the anticodon and 3′ end of the A9C tRNA. This enhanced flexibility would allow A9C tRNA to more easily access the A/T state and position EF-Tu in conformation for GTP hydrolysis even in presence of the UGA stop codon. This increase in flexibility is likely sufficient to explain the ~18-fold increase in misreading that has been reported2.
Conclusions
Proper decoding by the ribosome requires a delicate balance between the energy derived from binding of a cognate tRNA, and the combined energy required for distortions in the tRNA, EF-Tu, and the 30S subunit that enable GTP hydrolysis. The two mutant tRNAs studied here cause miscoding via different mechanisms, A9C by increasing flexibility, and G24A by facilitating an additional internal interaction. Using these strategies the G24A and A9C mutations decrease the energetic penalty for tRNA distortion into the A/T state, which allows GTPase activation to occur on the near-cognate UGA stop codon. Comparison of Trp-tRNATrp and Thr-tRNAThr suggest that the exact details of the decoding conformations will be different for every tRNA, as the energetics of the tRNA body have been uniquely optimized to ensure accurate decoding while maintaining requirements for other processes such as aminoacylation and a uniform affinity for EF-Tu22. Any perturbation to the precise energetic balance in the tRNA, EF-Tu, and the ribosome can therefore lead to hyperaccuracy or miscoding.
Methods
Thermus thermophilus 70S ribosomes harboring a C-terminal truncation of protein L923 were purified as previously described24 from cells grown at the Bioexpression and Fermentation Facility at the University of Georgia. Model mRNAs were purchased from Dharmcon (Thermo Scientific) with sequences:
Cognate Trp mRNA: 5′GGCAAGGAGGUAAAAAUGUUCUGGAAA
UGA Stop mRNA: 5′GGCAAGGAGGUAAAAAUGUUCUGAAAA
tRNA expression and purification
Wildtype tRNATrp was expressed as described for tRNAPhe 24. Plasmid pRT33C and strain MY87 for expression of Hirsh tRNA7,25 was a kind gift from Rachel Green. Quikchange mutagenesis was performed to obtain a tRNATrp with an A9C mutation. Hirsh tRNA and A9C tRNA were expressed as described7.
The mutant and wildtype tRNATrp were phenol extracted from lysed cells as described for tRNAPhe24. Extracted tRNA was bound to a DEAE - Sepharose column equilibrated in 50 mM Tris-HCl pH 7.5, and eluted with a linear gradient to 50 mM Tris HCl pH7.5 and 1 M NaCl over 5 column volumes. Fractions containing tRNA are pooled and solid (NH4)2SO4 is added to a final concentration of 1.7 M, for loading onto a TSK 5PW hydrophobic column equilibrated in 10 mM ammonium acetate pH6.3, 1.7 M (NH4)2SO4. tRNA is eluted using a linear gradient 1.7- 0.85 M (NH4)2SO4 over 5 column volumes. tRNA from the resulting peak fractions are assayed for aminoacylation efficiency using 14C – tryptophan as described7.
The tRNA from the appropriate fractions is further purified using a C4 reverse phase column equilibrated in 20 mM NH4OAc pH5.5, 400 mM NaCl, 10 mM Mg(OAc)2, eluted with a 0-40% linear gradient over 8 column volumes using the same buffer containing 60% methanol, then aminoacylated with non-radioactive tryptophan. Separation of acylated from deacylated tRNA was accomplished using the TSK 5PW hydrophobic column using the same conditions described above. The sample was dialyzed into a solution of 10 mM NH4OAc, pH5 50 mM KCl, and stored at −80° C.
Complex formation
Complexes of Trp-tRNATrp-GTP-70S ribosome were prepared and purified by Ni-NTA affinity purification as described14, with the exception that the tRNA was pre-acylated before complex formation and the antibiotic paromomycin was not included in the reaction mixture. Paromomycin increases miscoding, and as such was not appropriate for the present study with non-cognate tRNA. However, we observe no significant differences in the cognate tRNATrp and tRNAThr complexes attributable to paromomycin.
Affinity purification for complexes containing Trp-tRNATrp and Hirsh tRNA bound to a cognate UGG codon both resulted in 45-55% yield of input ribosomes. Complexes with Hirsh tRNA and an mRNA containing a near-cognate UGA stop codon were somewhat lower, at ~20% yield. Control experiments with Trp-tRNATrp and UGA were very low, at ~3% yield.
Crystallization
Crystals were grown under conditions described14, with the modification of the reservoir solution to include, 5.3% PEG 5.2, and 60-100 mM KCl. Data was collected at beamline ID 14-4 of the European Synchrotron Light Source26, and processed as described14 using XDS27. Iterative rounds of model building and refinement were carried out in coot28 and CNS29 as previously described14. Sequence analysis of residues 24 and 44 in bacterial tRNATrp was performed on sequences obtained from the Genomic tRNA Database 17. All figures were made in Pymol30.
Supplementary Material
Acknowledgements
We thank R. Green and R. Ortiz-Meoz, Johns Hopkins University, for plasmids and bacterial strains for production of mutant tRNATrp, A. McCarthy and S. Brockhauser at ESRF ID14.4 for facilitating data collection, O. Uhlenbeck for helpful discussion, and F. Murphy for scripting. This work was supported by the Medical Research Council UK, the Wellcome Trust, the Agouron Institute and the Louis-Jeantet Foundation. RV received support from the Gates-Cambridge scholarship. TMS received support from HFSP and Emmanuel College.
Footnotes
Supplementary Information is linked to the online version of this paper
Accession codes: Protein Data Bank: 2Y0U, 2Y0V, 2Y0W, 2Y0X for A9C, 2Y0Y, 2Y0Z, 2Y12, 2Y13 for HS, 2Y14, 2Y15, 2Y16, 2Y17 for HT, and 2Y18, 2Y19, 2Y10, 2Y11 for TT
References
- 1.Hirsh D. Tryptophan tRNA of Escherichia coli. Nature. 1970;228:57. doi: 10.1038/228057a0. [DOI] [PubMed] [Google Scholar]
- 2.Smith D, Yarus M. Transfer RNA structure and coding specificity. II. A D-arm tertiary interaction that restricts coding range. J Mol Biol. 1989;206:503–511. doi: 10.1016/0022-2836(89)90497-x. [DOI] [PubMed] [Google Scholar]
- 3.Smith DW, Yarus M. Transfer RNA structure and coding specificity. I. Evidence that a D-arm mutation reduces tRNA dissociation from the ribosome. J Mol Biol. 1989;206:489–501. doi: 10.1016/0022-2836(89)90496-8. [DOI] [PubMed] [Google Scholar]
- 4.Schultz DW, Yarus M. tRNA structure and ribosomal function. I. tRNA nucleotide 27-43 mutations enhance first position wobble. J Mol Biol. 1994;235:1381–1394. doi: 10.1006/jmbi.1994.1095. [DOI] [PubMed] [Google Scholar]
- 5.Ortiz-Meoz RF, Green R. Functional elucidation of a key contact between tRNA and the large ribosomal subunit rRNA during decoding. RNA. 2010;16:2002–2013. doi: 10.1261/rna.2232710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favre A, Buchingham R, Thomas G. tRNA tertiary structure in solution as probed by the photochemically induced 8-13 cross-link. Nucleic Acids Res. 1975;2:1421–1431. doi: 10.1093/nar/2.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 9.Agris PF, Vendeix FA, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 10.Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. Embo J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fersht AR. The hydrogen bond in molecular recognition. Trends Biochem Sci. 1987;12:301–304. [Google Scholar]
- 13.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy F. V. t., Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat Struct Mol Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- 16.Olejniczak M, Uhlenbeck OC. tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie. 2006;88:943–950. doi: 10.1016/j.biochi.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA. 2000;6:1091–105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nissen P, et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 20.Li W, et al. Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM. Embo J. 2008;27:3322–3331. doi: 10.1038/emboj.2008.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fahlman RP, Dale T, Uhlenbeck OC. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 23.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 25.Eisenberg SP, Söll L, Yarus M. The purification and sequence of a temperature-sensitive tryptophan tRNA. J Biol Chem. 1979;254:5562–5566. [PubMed] [Google Scholar]
- 26.McCarthy AA, et al. A decade of user operation on the macromolecular crystallography MAD beamline ID14-4 at the ESRF. J Synchrotron Radiat. 2009;16:803–812. doi: 10.1107/S0909049509035377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–200. [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.DeLano WL. The PyMOL Molecular Graphics System. 2006. http://www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.