Abstract
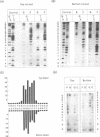
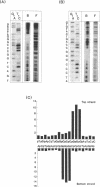
The XmaI endonuclease recognizes and cleaves the sequence C decreases CCGGG. Magnesium is required for catalysis, however, the enzyme forms stable, specific complexes with DNA in the absence of magnesium. An association constant of 1.2 x 10(9)/M was estimated for the affinity of the enzyme for a specific 195 bp fragment. Competition assays revealed that the site-specific association constant represented an approximately 10(4)-fold increase in affinity over that for non-cognate sites. Missing nucleoside analyses suggested an interaction of the enzyme with each of the cytosines and guanines within the recognition site. Recognition of each of the guanines was also indicated by dimethylsulfate interference footprinting assays. The phosphates 5' to the guanines within the recognition site appeared to be the major sites of interaction of XmaI with the sugar-phosphate backbone. No significant interaction of the protein was observed with phosphates flanking the recognition sequence. Comparison of the footprinting patterns of XmaI with those of the neoschizomer SmaI (CCC decreases GGG) revealed that the two enzymes utilize the same DNA determinants in their specific interaction with the CCCGGG recognition site.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiken C. R., Fisher E. W., Gumport R. I. The specific binding, bending, and unwinding of DNA by RsrI endonuclease, an isoschizomer of EcoRI endonuclease. J Biol Chem. 1991 Oct 5;266(28):19063–19069. [PubMed] [Google Scholar]
- Blackwell T. K., Huang J., Ma A., Kretzner L., Alt F. W., Eisenman R. N., Weintraub H. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993 Sep;13(9):5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Brunelle A., Schleif R. F. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Balendiran K., Schildkraut I., Anderson J. E. Structure of PvuII endonuclease with cognate DNA. EMBO J. 1994 Sep 1;13(17):3927–3935. doi: 10.1002/j.1460-2075.1994.tb06708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. L., Halay E. D., Lai E., Burley S. K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993 Jul 29;364(6436):412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Dang C. V., Dolde C., Gillison M. L., Kato G. J. Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker N., Cox M., Boelens R., Verrijzer C. P., van der Vliet P. C., Kaptein R. Solution structure of the POU-specific DNA-binding domain of Oct-1. Nature. 1993 Apr 29;362(6423):852–855. doi: 10.1038/362852a0. [DOI] [PubMed] [Google Scholar]
- Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992 Dec 24;71(7):1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Prendergast G. C., Ziff E. B., Burley S. K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993 May 6;363(6424):38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Heidmann S., Seifert W., Kessler C., Domdey H. Cloning, characterization and heterologous expression of the SmaI restriction-modification system. Nucleic Acids Res. 1989 Dec 11;17(23):9783–9796. doi: 10.1093/nar/17.23.9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Lesser D. R., Kurpiewski M. R., Jen-Jacobson L. The energetic basis of specificity in the Eco RI endonuclease--DNA interaction. Science. 1990 Nov 9;250(4982):776–786. doi: 10.1126/science.2237428. [DOI] [PubMed] [Google Scholar]
- Lubys A., Menkevicius S., Timinskas A., Butkus V., Janulaitis A. Cloning and analysis of translational control for genes encoding the Cfr9I restriction-modification system. Gene. 1994 Apr 8;141(1):85–89. doi: 10.1016/0378-1119(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Mayer A. N., Barany F. Interaction of TaqI endonuclease with the phosphate backbone. Effects of stereospecific phosphate modification. J Biol Chem. 1994 Nov 18;269(46):29067–29076. [PubMed] [Google Scholar]
- McLaughlin L. W., Benseler F., Graeser E., Piel N., Scholtissek S. Effects of functional group changes in the EcoRI recognition site on the cleavage reaction catalyzed by the endonuclease. Biochemistry. 1987 Nov 17;26(23):7238–7245. doi: 10.1021/bi00397a007. [DOI] [PubMed] [Google Scholar]
- Nikolov D. B., Hu S. H., Lin J., Gasch A., Hoffmann A., Horikoshi M., Chua N. H., Roeder R. G., Burley S. K. Crystal structure of TFIID TATA-box binding protein. Nature. 1992 Nov 5;360(6399):40–46. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Papp P. P., Chattoraj D. K. Missing-base and ethylation interference footprinting of P1 plasmid replication initiator. Nucleic Acids Res. 1994 Jan 25;22(2):152–157. doi: 10.1093/nar/22.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993 Sep 24;261(5129):1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Selent U., Rüter T., Köhler E., Liedtke M., Thielking V., Alves J., Oelgeschläger T., Wolfes H., Peters F., Pingoud A. A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry. 1992 May 26;31(20):4808–4815. doi: 10.1021/bi00135a010. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siksnys V., Pleckaityte M. Catalytic and binding properties of restriction endonuclease Cfr9I. Eur J Biochem. 1993 Oct 1;217(1):411–419. doi: 10.1111/j.1432-1033.1993.tb18260.x. [DOI] [PubMed] [Google Scholar]
- Somers W. S., Phillips S. E. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992 Oct 1;359(6394):387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- Suckow M., Schwamborn K., Kisters-Woike B., von Wilcken-Bergmann B., Müller-Hill B. Replacement of invariant bZip residues within the basic region of the yeast transcriptional activator GCN4 can change its DNA binding specificity. Nucleic Acids Res. 1994 Oct 25;22(21):4395–4404. doi: 10.1093/nar/22.21.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. Common features in DNA recognition helices of eukaryotic transcription factors. EMBO J. 1993 Aug;12(8):3221–3226. doi: 10.1002/j.1460-2075.1993.tb05991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. D., Badcoe I. G., Clarke A. R., Halford S. E. EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry. 1991 Sep 10;30(36):8743–8753. doi: 10.1021/bi00100a005. [DOI] [PubMed] [Google Scholar]
- Terry B. J., Jack W. E., Rubin R. A., Modrich P. Thermodynamic parameters governing interaction of EcoRI endonuclease with specific and nonspecific DNA sequences. J Biol Chem. 1983 Aug 25;258(16):9820–9825. [PubMed] [Google Scholar]
- Thielking V., Selent U., Köhler E., Wolfes H., Pieper U., Geiger R., Urbanke C., Winkler F. K., Pingoud A. Site-directed mutagenesis studies with EcoRV restriction endonuclease to identify regions involved in recognition and catalysis. Biochemistry. 1991 Jul 2;30(26):6416–6422. doi: 10.1021/bi00240a011. [DOI] [PubMed] [Google Scholar]
- Vermote C. L., Halford S. E. EcoRV restriction endonuclease: communication between catalytic metal ions and DNA recognition. Biochemistry. 1992 Jul 7;31(26):6082–6089. doi: 10.1021/bi00141a018. [DOI] [PubMed] [Google Scholar]
- Winkler F. K., Banner D. W., Oefner C., Tsernoglou D., Brown R. S., Heathman S. P., Bryan R. K., Martin P. D., Petratos K., Wilson K. S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993 May;12(5):1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers B. E., Ambroso L. A., Dunbar J. C. Structure and evolution of the XcyI restriction-modification system. Nucleic Acids Res. 1992 Dec 11;20(23):6267–6273. doi: 10.1093/nar/20.23.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers B. E., Dunbar J. C. Sequence-specific DNA recognition by the SmaI endonuclease. J Biol Chem. 1995 Mar 24;270(12):6496–6504. doi: 10.1074/jbc.270.12.6496. [DOI] [PubMed] [Google Scholar]
- Withers B. E., Dunbar J. C. The endonuclease isoschizomers, SmaI and XmaI, bend DNA in opposite orientations. Nucleic Acids Res. 1993 Jun 11;21(11):2571–2577. doi: 10.1093/nar/21.11.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. Y., Schildkraut I. Isolation of BamHI variants with reduced cleavage activities. J Biol Chem. 1991 Mar 5;266(7):4425–4429. [PubMed] [Google Scholar]
- Zebala J. A., Choi J., Trainor G. L., Barany F. DNA recognition of base analogue and chemically modified substrates by the TaqI restriction endonuclease. J Biol Chem. 1992 Apr 25;267(12):8106–8116. [PubMed] [Google Scholar]