Mutations in the human cytomegalovirus UL27 gene confer resistance to an inhibitor of the viral protein kinase UL97. Terhune and colleagues demonstrate that UL27 destabilizes the Tip60 histone acetyltransferase, setting forth intriguing new mechanisms for antiviral drug resistance and for viral regulation of the cell cycle.
Well before the term chemical biology became popular, much was learned about the biology and biochemistry of viruses from studying small-molecule antiviral drugs and mutants resistant to them. These discoveries included the finding that the influenza virus M2 protein forms an ion channel from studies of amantadine-resistant mutant viruses, and the finding that the human cytomegalovirus (HCMV) UL97 (or pUL97) protein kinase can phosphorylate nucleoside analogs from studies of ganciclovir-resistant mutant viruses. But, UL97 is important for more than phosphorylating antiviral drugs. A major physiologic role of UL97 is to act as a viral mimic of cyclin dependent kinase complexes (CDKs). HCMV ordinarily arrests the cell cycle of productively infected cells at the G1/S boundary. Upon infection of quiescent cells, UL97 phosphorylates retinoblastoma tumor suppressor (Rb) family pocket proteins, which in turn results in expression of S-phase genes that likely are crucial for HCMV DNA synthesis (Hume et al., 2009; Kamil et al., 2009).
An inhibitor of UL97 kinase activity, maribavir, has antiviral activity in cell culture, and showed promise as an antiviral drug in humans in phase I and phase II clinical trials. As with other antiviral agents, drug resistant viruses emerge during serial passage of HCMV in the presence of maribavir. As expected, many of these viruses contain mutations in the UL97 open reading frame, which confer high level resistance to maribavir. However, others of these viruses contain mutations in the UL27 open reading frame, which confer modest resistance to maribavir (Komazin et al., 2003).This observation is highly unusual, as nearly all virus drug resistance mutations directly alter drug targets. The UL27 open reading frame encodes a nuclear protein, UL27 (or pUL27). There have been precious few clues, if any, to the function of UL27. The murine CMV (MCMV) UL27 homolog, M27, is responsible for degradation of STAT2, which provides a mechanism for viral interference with alpha/beta IFN receptor signaling and evasion of innate immunity. However, degradation of STAT2 occurs in a UL27-independent manner in HCMV infected cells (Le et al., 2008).
To investigate the function of UL27, Reitsma et al. (this issue) used mass spectrometry to detect proteins from infected cells that co-purified with epitope-tagged UL27 expressed from an engineered HCMV. This was not an easy experiment, as UL27 is not abundant. Among the putative UL27 interaction partners they detected were five members of the Tip60 histone acetyltransferase (HAT) core complex, yet not Tip60 itself. Nonetheless, the authors examined Tip60 expression during HCMV infection and observed that Tip60 undergoes rapid, proteasome-dependent degradation during the early phase of HCMV infection, and that UL27 is necessary and sufficient for this effect. Viruses lacking UL27 expression were unable to cause Tip60 degradation and in the presence of maribavir, consistent with previous studies, were able to replicate to higher yield and synthesize higher levels of viral DNA than wild-type HCMV.
Tip60, a MYST family HAT, was first identified by virtue of its interaction with HIV-1 Tat (Kamine et al., 1996). Later, Tip60 was shown to be part of a multimeric complex with DNA helicase, ATPase and DNA binding properties, and to play roles in a wide array of cellular processes, including transcription, the DNA damage response, cell cycle control, and apoptosis (reviewed in Sapountziet al., 2006). Strikingly, Terhune and colleagues find that expression of Tat, which can also destabilize Tip60 expression, can restore sensitivity to maribavir of HCMV mutants lacking UL27 expression. This strongly argues that it is the Tip60 degradation function of UL27 that is crucial for its role in maribavir action. The authors also find that expression of UL27 induces heightened mRNA and protein expression levels of the CDK-inhibitor protein p21WAF/CIP, which was attributed to loss of Tip60 expression. Thus, in the context of HCMV infection, Tip60 apparently plays a role in repressing p21WAF/CIP.The authors also used shRNA to disrupt expression of another putative UL27 interacting protein, the ubiquitin-independent proteasome activator protein, PSME3. This treatment, too, caused elevated expression of p21WAF/CIP, which likewise restored maribavir sensitivity to UL27-null HCMV. This suggests that p21WAF/CIP is relevant to UL27's role in maribavir sensitivity.
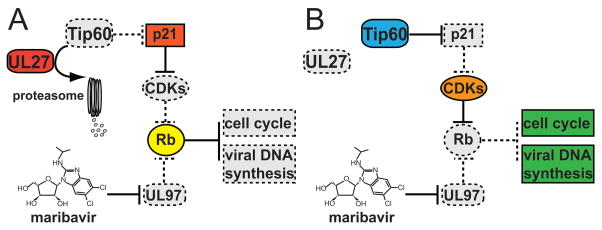
The authors hypothesize (Fig. 1) that UL27 ordinarily acts to block cell cycle progression towards the G1/S phase boundary. Specifically, they suggest that UL27-mediated destruction of Tip60 leads to enhanced expression of p21WAF/CIP, which can potently inhibit cellular CDK activity, thus blocking cell cycle progression. In contrast, UL97 kinase activity, which is relatively insensitive to p21WAF/CIP, ordinarily promotes progression towards S phase, at least in quiescent cells, by phosphorylation and inactivation of Rb family members (Hume et al., 2008, Kamil et al., 2009). Therefore, when UL97 is inhibited by maribavir, increased levels of p21, such as those induced by UL27-mediated destruction of Tip60, would likely block cellular CDK activity, and thus, Rb would remain active and S-phase genes necessary for viral DNA synthesis would remain silent. Thus, the loss of UL27 would compensate for the loss of UL97 function imposed by maribavir, explaining the mechanism of resistance.
Figure 1. Model for the Roles of UL27 and the Tip60 Histone Acetyltransferase in Human Cytomegalovirus Antiviral Drug Resistance.
A) UL27 promotes proteasome-dependent degradation of Tip60, leading to increased expression of p21WAF/CIP(p21), an inhibitor of cellular cyclin-dependent kinase complexes (CDKs). Thus, in the presence of maribavir, an inhibitor of the UL97 protein kinase, wild-type human cytomegalovirus (HCMV) lacks any efficient mechanism, cellular or viral, by which to inactivate the retinoblastoma tumor suppressor protein (Rb). Because active Rb represses cellular genes necessary for cell cycle progression and for viral DNA synthesis, wild-type HCMV replication is impaired in the presence of maribavir. B)UL27-null viruses are unable to promote degradation of Tip60. In the absence of Tip60 destruction, p21 expression remains low enough for cellular CDKs to inactivate Rb, thus compensating for maribavir inhibition of UL97 activity, and providing for resistance of UL27 mutant viruses to maribavir. In both A) and B), active functions are shown as solid-bordered, colored shapes, while inactive functions are shown as dashed-bordered, gray shapes.
Terhune and colleagues have made considerable progress towards solving the riddle of UL27 mutations in maribavir resistance and in doing so, have shed new light on the biological function of a mysterious viral gene. Nonetheless, several matters merit further inquiry. First, Tip60 degradation during wild-type HCMV infection appears to be a transitory phenomenon that does not correlate neatly with UL27 expression (Reitsma et al., this issue). UL27 is detectable starting at 4–5 hours post infection (hpi), after which its expression is relatively constant through at least 72 hpi. Tip60, however, is destabilized as early as 2 hpi, is undetectable at 6 hpi, and then its levels recover dramatically by 24 hpi, and appear to increase further by 72 hpi. Why UL27 is capable of degrading Tip60 only at early, but not late times during infection is unclear. One explanation would be that UL27’s function or substrate specificity is altered at later time points. It will be interesting to determine whether UL27 becomes post-translationally modified as infection proceeds. Regardless, UL27 affects the impact of maribavir on viral DNA synthesis and yield of infectious particles at up to 96 and 144 hpi, respectively, so how UL27's very early, apparently transitory effects on Tip60 set the stage for later events remains to be elucidated.
As mentioned above, MCMV M27 is responsible for degradation of STAT2. Thus, MCMV M27 and HCMV UL27 exhibit a conserved general function in protein degradation, but have diverged in their substrate specificity to target distinct cellular factors for destruction. Though their specificities do not seem to overlap, one wonders whether either of these two viral proteins target any additional proteins for degradation. The function of the HCMV tegument protein pp71 in protein degradation was originally identified as targeting Rb family proteins, but later was also found to be crucial for destabilization of Daxx (reviewed in Kalejta, R.F., 2008).
Since Reitsma et al. lean heavily on the use of relatively high concentrations of maribavir to inhibit UL97, it will be important to confirm whether Tat and/or p21 exacerbate the replication defective phenotypes of UL97-null or UL97 kinase-deficient HCMV mutants. Also, it remains to be seen whether direct ectopic overexpression of p21WAF/CIP can restore maribavir sensitivity to HCMV lacking UL27 expression. Notably, in HCMV-infected cells, p21WAF/CIP expression appears to be regulated mainly by cathepsins rather than by the proteasome, supporting the notion that HCMV actively modifies the expression of this molecule (Chen et al., 2001). The mechanisms by which HCMV orchestrates its regulation of p21WAF/CIP warrant further inquiry.
UL97 mimicry of CDKs is important not only for viral DNA synthesis, but also plays a role at the stage of nuclear egress, where UL97 is required for disassembly of the nuclear lamina, evidently by phosphorylating lamin A/C (Hamirally et al., 2009). The effects of UL27 mutations on maribavir inhibition of viral DNA synthesis that Reitsma et al. observed do not fully account for the corresponding effects on yield of infectious virus. Therefore, it would be worthwhile to determine at which phase(s) UL27 arrests cell cycle progression and whether viral nuclear egress and lamin A/C phosphorylation are enhanced during replication of UL27-null viruses in the presence of maribavir.
The literature implicates Tip60 in processes as varied as chromatin regulation, DNA repair and apoptosis. Moreover, Tip60 is posited to play roles not only as a HAT, but also in acetylation and regulation of other biologically complex transcriptional regulators, including p53 (reviewed in Kruse and Gu, 2009). Thus, as is inevitable when a new function has been identified, the work of Reitsma et al. has opened the door for a plethora of studies that are sure to tell us much more about how HCMV regulates the cell cycle during infection. The study represents an excellent example of how using drugs in the laboratory can be beneficial, and it is not out of the question that its results might have implications for the use of such drugs in the clinic.
Acknowledgments
The authors apologize that due to space constraints it was not possible to cite all relevant literature. J.P.K. is supported by an NIH NCRR COBRE award (P20-RR018724). D.M.C. gratefully acknowledges NIH support (RO1 AI026077).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jeremy P. Kamil, Email: jkamil@lsuhsc.edu, Department of Microbiology and Immunology, Louisiana State University Health Sciences Center, Shreveport, LA. 1501 Kings Hwy, Shreveport, LA 71130, USA.
Donald M. Coen, Email: don_coen@hms.harvard.edu, Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 250 Longwood Ave, Boston, MA 02115, USA.
References
- Chen Z, Knutson E, Kurosky A, Albrecht T. J Virol. 2001;75:3613–3625. doi: 10.1128/JVI.75.8.3613-3625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume AJ, Finkel JS, Kamil JP, Coen DM, Culbertson MR, Kalejta RF. Science. 2008;320:797–799. doi: 10.1126/science.1152095. [DOI] [PubMed] [Google Scholar]
- Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, Jahng WJ, Baek MC, Noton S, Silva LA, Simpson-Holley M, Knipe DM, Golan DE, Marto JA, Coen DM. PLoS Pathog. 2009;5:e1000275. doi: 10.1371/journal.ppat.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalejta RF. Microbiol Mol Biol Rev. 2008;72:249–65. doi: 10.1128/MMBR.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamil JP, Hume AJ, Jurak I, Münger K, Kalejta RF, Coen DM. Proc Natl Acad Sci USA. 2009;106:16823–16828. doi: 10.1073/pnas.0901521106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- Komazin G, Ptak RG, Emmer BT, Townsend LB, Drach JC. J Virol. 2003;77:11499–11506. doi: 10.1128/JVI.77.21.11499-11506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le VT, Trilling M, Wilborn M, Hengel H, Zimmermann A. J Gen Virol. 2008;89:2416–2426. doi: 10.1099/vir.0.2008/001669-0. [DOI] [PubMed] [Google Scholar]
- Reitsma, et al. This issue. [Google Scholar]
- Sapountzi V, Logan IR, Robson CN. Int J Biochem Cell Biol. 2006;38:1496–509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]